Microbial Communities in and Around the Siboglinid Tubeworms from the South Yungan East Ridge Cold Seep Offshore Southwestern Taiwan at the Northern South China Sea
Abstract
:1. Introduction
2. Materials and Methods
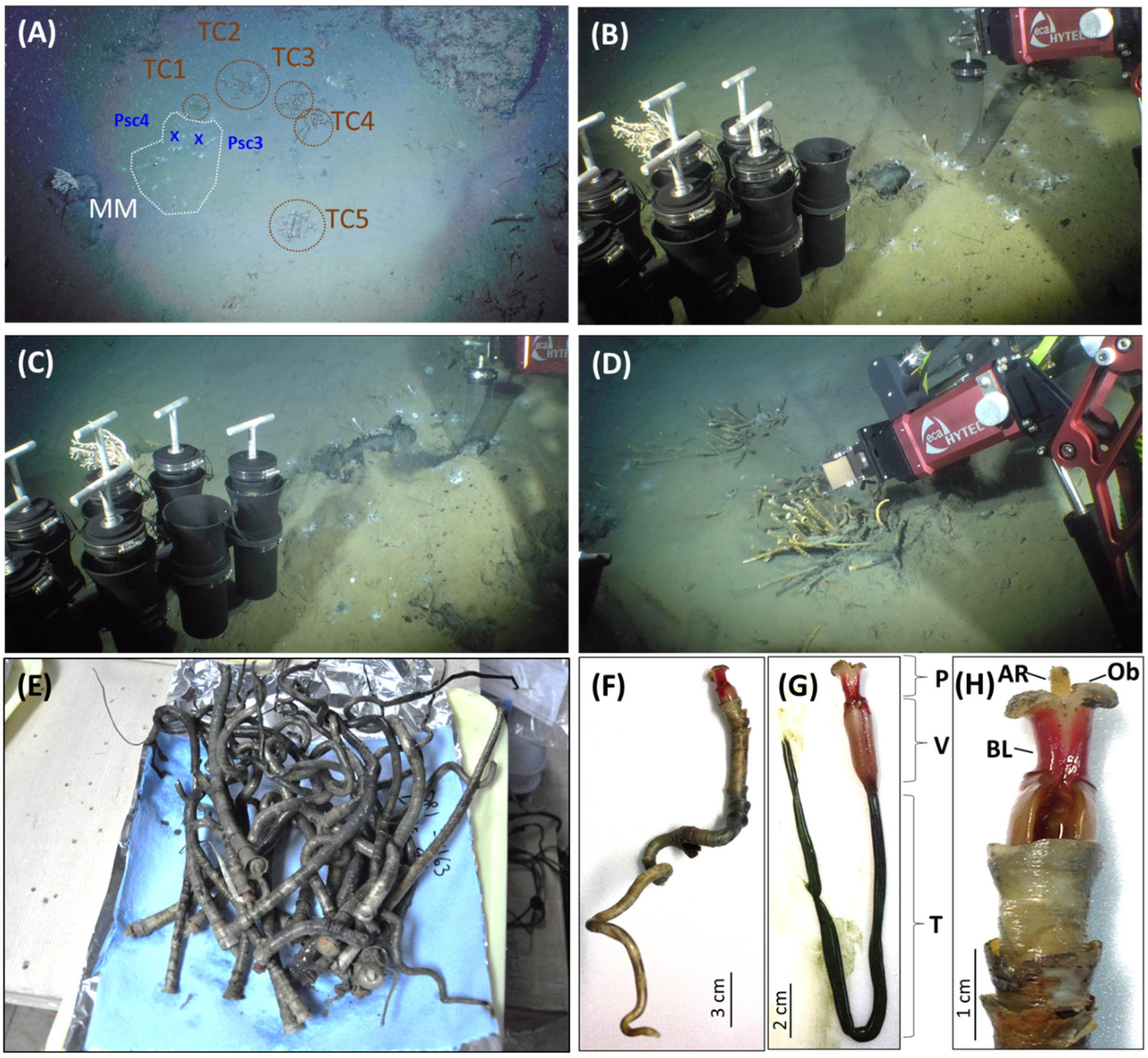
2.1. Study Area, Sample Collection and Geochemical Analysis
2.2. Cloning, Sequencing and Phylogenetic Analysis
2.3. PacBio 16S rRNA Gene Amplicon Sequencing and Analysis of Microbial Communities
3. Results and Discussion
3.1. The Tubeworm ORI-1163B-Tw1 Was Proposed as Paraescarpia Formosa
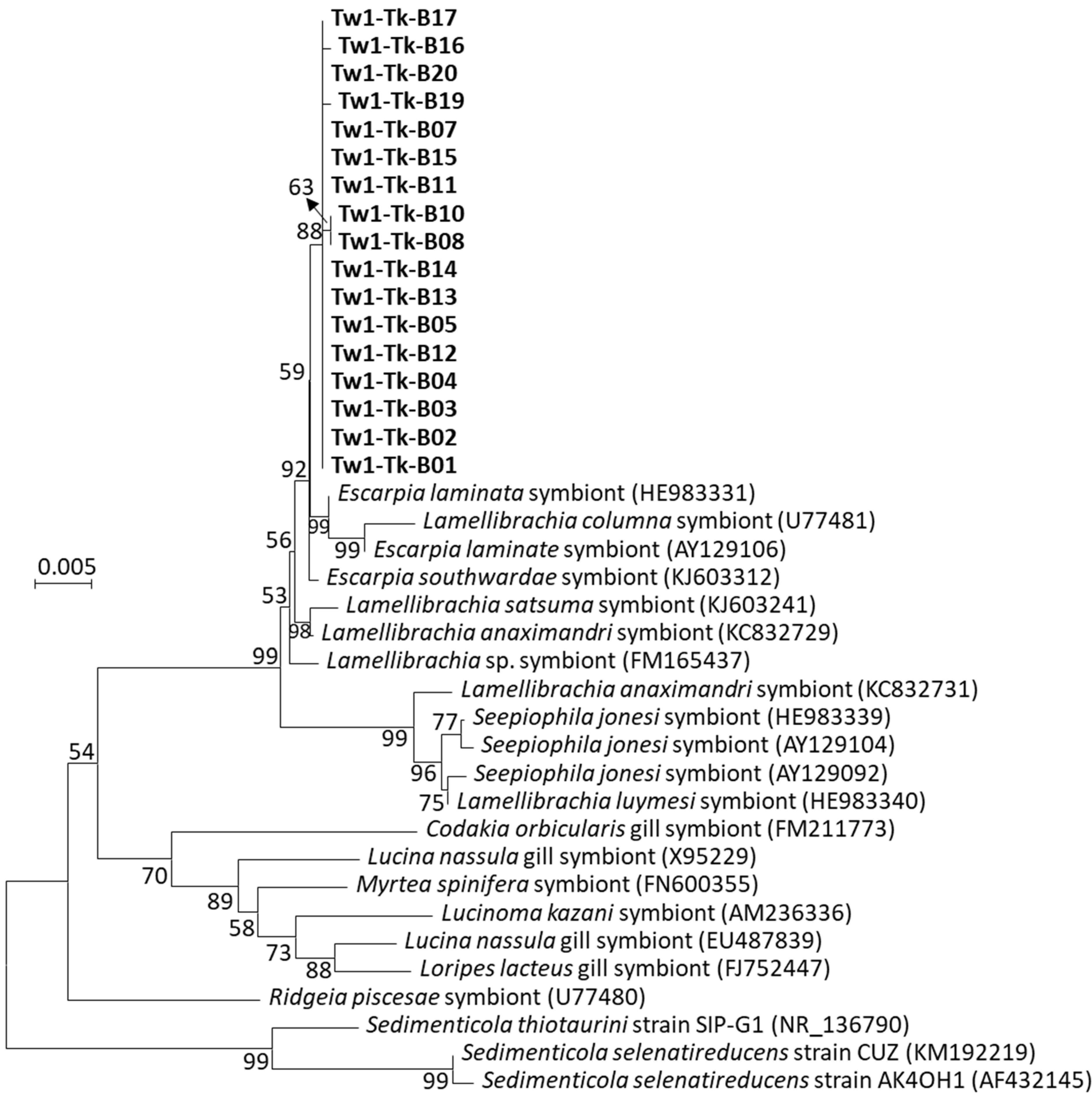
3.2. The Sedimenticolaceae Phylotype Was Identified as a Sulfur-Oxidizing Endosymbiont in the Trunk
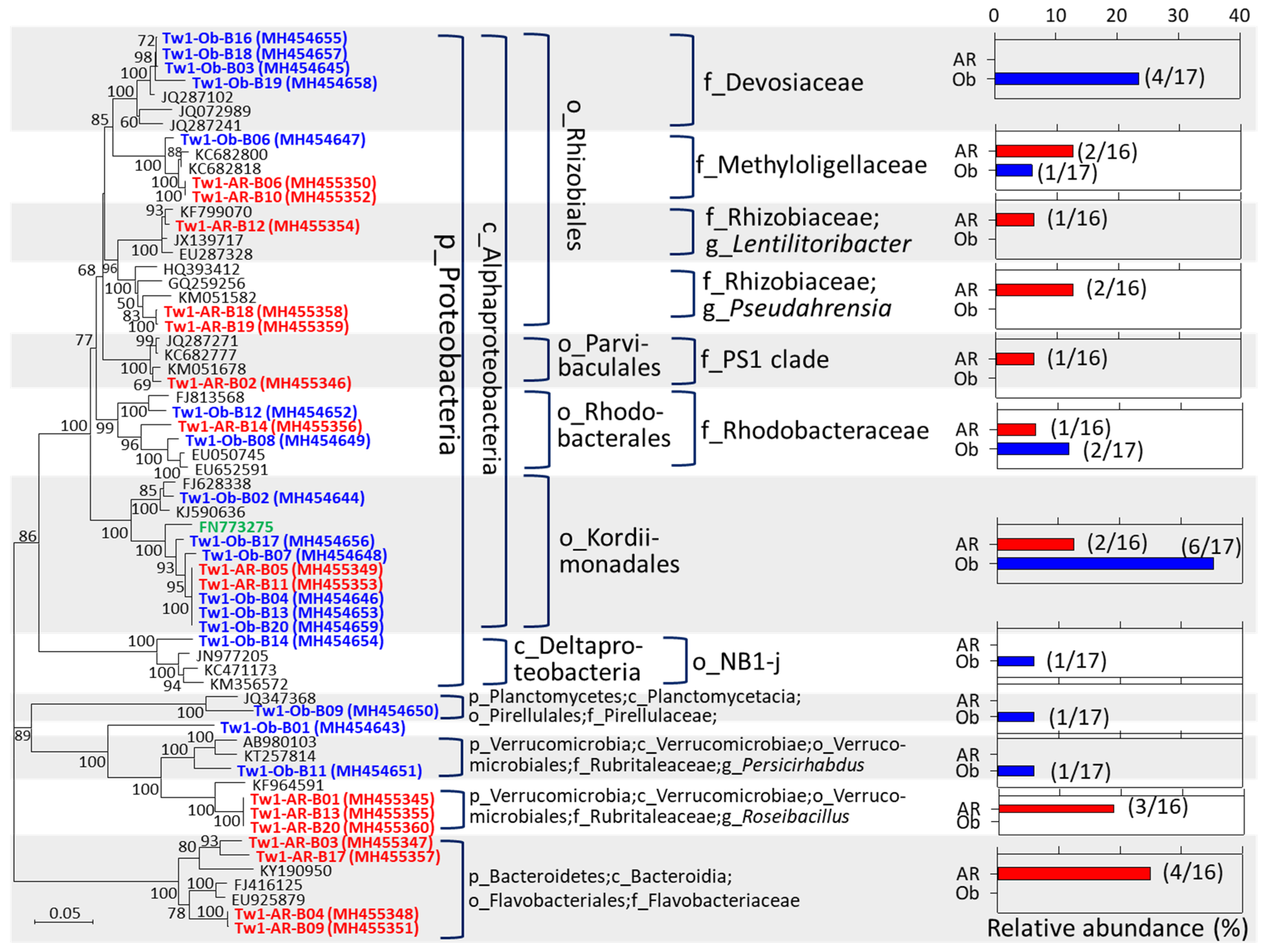
3.3. Potential Sulfur-/Methane-Oxidizing Symbionts Were Found in the Axial Rod and Obturaculum
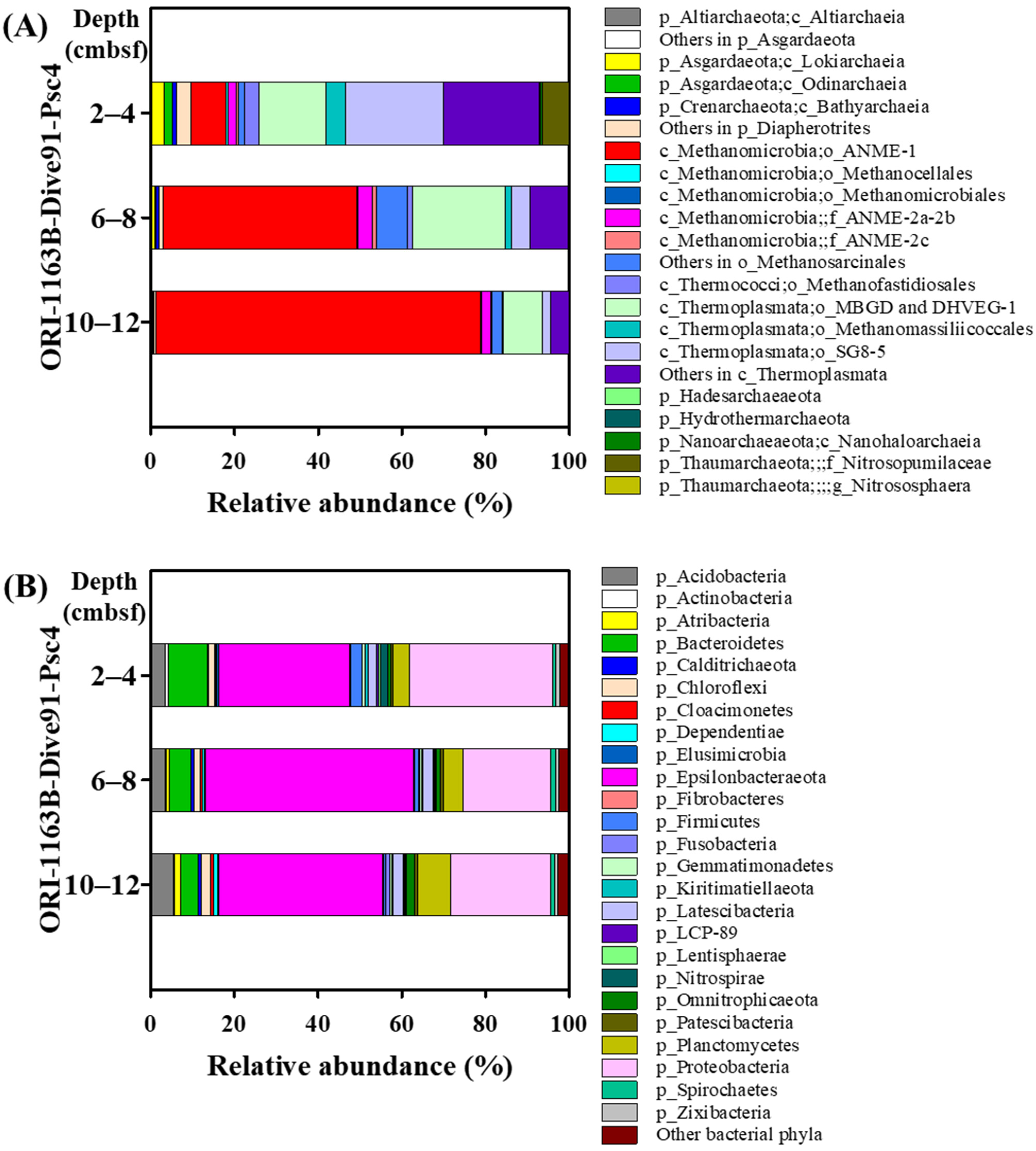
3.4. ANME-1 Was Dominant in AOM Consortia at the SYER Cold Seep
3.5. Various Methanogenic Archaea Were Found at the SYER Cold Seep
3.6. Both Genera Sulfurovum and Sulfurimonas Were Dominant at the SYER Cold Seep
3.7. Diverse Sulfate-Reducing Bacteria Were Found at the SYER Cold Seep
3.8. The Prokaryotic Community of the SYER Cold Seep Is Distinct from Other SCS Cold Seeps
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aharon, P.; Fu, B.S. Microbial sulfate reduction rates and sulfur and oxygen isotope fractionations at oil and gas seeps in deepwater Gulf of Mexico. Geochim. Cosmochim. Acta 2000, 64, 233–246. [Google Scholar] [CrossRef]
- Hinrichs, K.U.; Summons, R.E.; Orphan, V.; Sylva, S.P.; Hayes, J.M. Molecular and isotopic analysis of anaerobic methane-oxidizing communities in marine sediments. Org. Geochem. 2000, 31, 1685–1701. [Google Scholar] [CrossRef]
- Inagaki, F.; Tsunogai, U.; Suzuki, M.; Kosaka, A.; Machiyama, H.; Takai, K.; Nunoura, T.; Nealson, K.H.; Horikoshi, K. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 2004, 70, 7445–7455. [Google Scholar] [CrossRef] [PubMed]
- Niemann, H.; Losekann, T.; de Beer, D.; Elvert, M.; Nadalig, T.; Knittel, K.; Amann, R.; Sauter, E.J.; Schluter, M.; Klages, M.; et al. Novel microbial communities of the Haakon Mosby mud volcano and their role as a methane sink. Nature 2006, 443, 854–858. [Google Scholar] [CrossRef]
- Tunnicliffe, V.; Juniper, S.K.; Sibuet, M. Reducing environments of the deep-sea floor. In Ecosystems of the World; Elsevier: Amsterdam, The Netherlands, 2003; pp. 81–110. [Google Scholar]
- Knittel, K.; Boetius, A. Anaerobic Oxidation of Methane: Progress with an Unknown Process. Annu. Rev. Microbiol. 2009, 63, 311–334. [Google Scholar] [CrossRef]
- Vigneron, A.; Cruaud, P.; Pignet, P.; Caprais, J.-C.; Cambon-Bonavita, M.-A.; Godfroy, A.; Toffin, L. Archaeal and anaerobic methane oxidizer communities in the Sonora Margin cold seeps, Guaymas Basin (Gulf of California). ISME J. 2013, 7, 1595. [Google Scholar] [CrossRef]
- Holler, T.; Widdel, F.; Knittel, K.; Amann, R.; Kellermann, M.Y.; Hinrichs, K.-U.; Teske, A.; Boetius, A.; Wegener, G. Thermophilic anaerobic oxidation of methane by marine microbial consortia. ISME J. 2011, 5, 1946–1956. [Google Scholar] [CrossRef]
- Kleindienst, S.; Ramette, A.; Amann, R.; Knittel, K. Distribution and in situ abundance of sulfate-reducing bacteria in diverse marine hydrocarbon seep sediments. Environ. Microbiol. 2012, 14, 2689–2710. [Google Scholar] [CrossRef]
- Krukenberg, V.; Harding, K.; Richter, M.; Glöckner, F.O.; Gruber-Vodicka, H.R.; Adam, B.; Berg, J.S.; Knittel, K.; Tegetmeyer, H.E.; Boetius, A. Candidatus Desulfofervidus auxilii, a hydrogenotrophic sulfate-reducing bacterium involved in the thermophilic anaerobic oxidation of methane. Environ. Microbiol. 2016, 18, 3073–3091. [Google Scholar] [CrossRef]
- Pernthaler, A.; Dekas, A.E.; Brown, C.T.; Goffredi, S.K.; Embaye, T.; Orphan, V.J. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc. Natl. Acad. Sci. USA 2008, 105, 7052–7057. [Google Scholar] [CrossRef]
- Schreiber, L.; Holler, T.; Knittel, K.; Meyerdierks, A.; Amann, R. Identification of the dominant sulfate-reducing bacterial partner of anaerobic methanotrophs of the ANME-2 clade. Environ. Microbiol. 2010, 12, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Cruaud, P.; Pignet, P.; Caprais, J.C.; Gayet, N.; Cambon-Bonavita, M.A.; Godfroy, A.; Toffin, L. Bacterial communities and syntrophic associations involved in anaerobic oxidation of methane process of the Sonora Margin cold seeps, Guaymas Basin. Environ. Microbiol. 2014, 16, 2777–2790. [Google Scholar] [CrossRef] [PubMed]
- Ruff, S.E.; Biddle, J.F.; Teske, A.P.; Knittel, K.; Boetius, A.; Ramette, A. Global dispersion and local diversification of the methane seep microbiome. Proc. Natl. Acad. Sci. USA 2015, 112, 4015–4020. [Google Scholar] [CrossRef] [PubMed]
- Boetius, A. Microfauna-macrofauna interaction in the seafloor: Lessons from the tubeworm. PLoS. Biol. 2005, 3, 375–378. [Google Scholar] [CrossRef]
- Fisher, C.R. Chemoautotrophic and methanotrophic symbioses in marine invertebrates. CRC Crit. Rev. Aquat. Sci. 1990, 2, 399–436. [Google Scholar]
- Hilario, A.; Capa, M.; Dahlgren, T.G.; Halanych, K.M.; Little, C.T.S.; Thornhill, D.J.; Verna, C.; Glover, A.G. New Perspectives on the ecology and evolution of Siboglinid tubeworms. PLoS ONE 2011, 6, e16309. [Google Scholar] [CrossRef]
- Jones, M.L. Riftia pachyptila Jones: Observations on the vestimentiferan worm from the Galapagos Rift. Science 1981, 213, 333–336. [Google Scholar] [CrossRef]
- Dubilier, N.; Bergin, C.; Lott, C. Symbiotic diversity in marine animals: The art of harnessing chemosynthesis. Nat. Rev. Microbiol. 2008, 6, 725–740. [Google Scholar] [CrossRef]
- Edwards, D.B.; Nelson, D.C. DNA-DNA solution hybridization studies of the bacterial symbionts of hydrothermal vent tube worms (Riftia pachyptila and Tevnia jerichonana). Appl. Environ. Microbiol. 1991, 57, 1082–1088. [Google Scholar] [CrossRef]
- Feldman, R.A.; Black, M.B.; Cary, C.S.; Lutz, R.A.; Vrijenhoek, R.C. Molecular phylogenetics of bacterial endosymbionts and their vestimentiferan hosts. Mol. Mar. Biol. Biotechnol. 1997, 6, 268–277. [Google Scholar]
- Elsaied, H.; Kimura, H.; Naganuma, T. Molecular characterization and endosymbiotic localization of the gene encoding D-ribulose 1, 5-bisphosphate carboxylase–oxygenase (RuBisCO) form II in the deep-sea vestimentiferan trophosome. Microbiology 2002, 148, 1947–1957. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Higashide, Y.; Naganuma, T. Endosymbiotic microflora of the vestimentiferan tubeworm (Lamellibrachia sp.) from a bathyal cold seep. Mar. Biotechnol. 2003, 5, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Kato, C.; Hirayama, H.; Moriyama, N.; Hashimoto, J.; Horikoshi, K.; Shiomoto, A.; Asami, H.; Hama, T.; Shin, K. Intracellular occurrence of ε-Proteobacterial 16S rDNA Sequences in the Vestimentiferan Trophosome. J. Oceanogr. 1997, 53, 193–197. [Google Scholar]
- Zimmermann, J.; Lott, C.; Weber, M.; Ramette, A.; Bright, M.; Dubilier, N.; Petersen, J.M. Dual symbiosis with co-occurring sulfur-oxidizing symbionts in vestimentiferan tubeworms from a Mediterranean hydrothermal vent. Environ. Microbiol. 2014, 16, 3638–3656. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.; Hayes, D.E. Origin and history of the South China Sea basin. Geophys. Monogr. Ser. 1983, 27, 23–56. [Google Scholar]
- Feng, D.; Qiu, J.W.; Hu, Y.; Peckmann, J.; Guan, H.X.; Tong, H.P.; Chen, C.; Chen, J.X.; Gong, S.G.; Li, N.; et al. Cold seep systems in the South China Sea: An overview. J. Asian Earth Sci. 2018, 168, 3–16. [Google Scholar] [CrossRef]
- Zhang, Y.; Su, X.; Chen, F.; Wang, Y.Y.; Jiao, L.; Dong, H.L.; Huang, Y.Y.; Jiang, H.C. Microbial diversity in cold seep sediments from the northern South China Sea. Geosci. Front. 2012, 3, 301–316. [Google Scholar] [CrossRef]
- Niu, M.Y.; Fan, X.B.; Zhuang, G.C.; Liang, Q.Y.; Wang, F.P. Methane-metabolizing microbial communities in sediments of the Haima cold seep area, northwest slope of the South China Sea. FEMS Microbiol. Ecol. 2017, 93, fix101. [Google Scholar] [CrossRef]
- Wu, Y.Z.; Qiu, J.W.; Qian, P.Y.; Wang, Y. The vertical distribution of prokaryotes in the surface sediment of Jiaolong cold seep at the northern South China Sea. Extremophiles 2018, 22, 499–510. [Google Scholar] [CrossRef]
- Cui, H.P.; Su, X.; Chen, F.; Holland, M.; Yang, S.X.; Liang, J.Q.; Dong, H.L.; Hou, W.G. Microbial diversity of two cold seep systems in gas hydrate-bearing sediments in the South China Sea. Mar. Environ. Res. 2019, 144, 230–239. [Google Scholar] [CrossRef]
- Singer, E.; Bushnell, B.; Coleman-Derr, D.; Bowman, B.; Bowers, R.M.; Levy, A.; Gies, E.A.; Cheng, J.F.; Copeland, A.; Klenk, H.P.; et al. High-resolution phylogenetic microbial community profiling. ISME J. 2016, 10, 2020–2032. [Google Scholar] [CrossRef] [PubMed]
- Earl, J.P.; Adappa, N.D.; Krol, J.; Bhat, A.S.; Balashov, S.; Ehrlich, R.L.; Palmer, J.N.; Workman, A.D.; Blasetti, M.; Sen, B.; et al. Species-level bacterial community profiling of the healthy sinonasal microbiome using Pacific Biosciences sequencing of full-length 16S rRNA genes. Microbiome 2018, 6, 190. [Google Scholar] [CrossRef] [PubMed]
- Chuang, P.C.; Yang, T.F.; Hong, W.L.; Lin, S.; Sun, C.H.; Lin, A.T.S.; Chen, J.C.; Wang, Y.; Chung, S.H. Estimation of methane flux offshore SW Taiwan and the influence of tectonics on gas hydrate accumulation. Geofluids 2010, 10, 497–510. [Google Scholar] [CrossRef]
- Cline, J.D. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 1969, 14, 454–458. [Google Scholar] [CrossRef]
- Lin, S.; Huang, K.M.; Chen, S.K. Sulfate reduction and iron sulfide mineral formation in the southern East China Sea continental slope sediment. Deep-Sea Res. Part I-Oceanogr. Res. Pap. 2002, 49, 1837–1852. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Glockner, F.O.; Yilmaz, P.; Quast, C.; Gerken, J.; Beccati, A.; Ciuprina, A.; Bruns, G.; Yarza, P.; Peplies, J.; Westram, R.; et al. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol. 2017, 261, 169–176. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glockner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Southward, E.C.; Schulze, A.; Tunnicliffe, V. Vestimentiferans (Pogonophora) in the Pacific and Indian Oceans: A new genus from Lihir Island (Papua New Guinea) and the Java Trench, with the first report of Arcovestia ivanovi from the North Fiji Basin. J. Nat. Hist. 2002, 36, 1179–1197. [Google Scholar] [CrossRef]
- Gardiner, S.L.; McMullin, E.; Fisher, C.R. Seepiophila jonesi, a new genus and species of vestimentiferan tube worm (Annelida: Pogonophora) from hydrocarbon seep communities in the Gulf of Mexico. Proc. Biol. Soc. Wash. 2001, 114, 694–707. [Google Scholar]
- Andersen, A.C.; Hourdez, S.; Marie, B.; Jollivet, D.; Lallier, F.H.; Sibuet, M. Escarpia southwardae sp nov., a new species of vestimentiferan tubeworm (Annelida, Siboglinidae) from West African cold seeps. Can. J. Zool.-Rev. Can. Zool. 2004, 82, 980–999. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, Q.; Sun, J.; Yang, Y.; Tao, J.; Liang, J.; Feng, D.; Qiu, J.-W.; Qian, P.-Y. The mitochondrial genome of the deep-sea tubeworm Paraescarpia echinospica (Siboglinidae, Annelida) and its phylogenetic implications. Mitochondrial DNA B Resour. 2018, 3, 131–132. [Google Scholar] [CrossRef]
- Kojima, S.; Ohta, S.; Yamamoto, T.; Miura, T.; Fujiwara, Y.; Fujikura, K.; Hashimoto, J. Molecular taxonomy of vestimentiferans of the western Pacific and their phylogenetic relationship to species of the eastern Pacific. Mar. Biol. 2002, 141, 57–64. [Google Scholar]
- Liang, Q.; Hu, Y.; Feng, D.; Peckmann, J.; Chen, L.; Yang, S.; Liang, J.; Tao, J.; Chen, D. Authigenic carbonates from newly discovered active cold seeps on the northwestern slope of the South China Sea: Constraints on fluid sources, formation environments, and seepage dynamics. Deep-Sea Res. I Oceanogr. Res. Pap. 2017, 124, 31–41. [Google Scholar] [CrossRef]
- Flood, B.E.; Jones, D.S.; Bailey, J.V. Sedimenticola thiotaurini sp nov., a sulfur-oxidizing bacterium isolated from salt marsh sediments, and emended descriptions of the genus Sedimenticola and Sedimenticola selenatireducens. Int. J. Syst. Evol. Microbiol. 2015, 65, 2522–2530. [Google Scholar] [CrossRef]
- Carlstrom, C.I.; Loutey, D.E.; Wang, O.W.; Engelbrektson, A.; Clark, I.; Lucas, L.N.; Somasekhar, P.Y.; Coates, J.D. Phenotypic and genotypic description of Sedimenticola selenatireducens strain CUZ, a marine (per) chlorate-respiring gammaproteobacterium, and its close relative the chlorate-respiring Sedimenticola strain NSS. Appl. Environ. Microbiol. 2015, 81, 2717–2726. [Google Scholar] [CrossRef] [PubMed]
- Harmer, T.L.; Rotjan, R.D.; Nussbaumer, A.D.; Bright, M.; Ng, A.W.; DeChaine, E.G.; Cavanaugh, C.M. Free-living tube worm endosymbionts found at deep-sea vents. Appl. Environ. Microbiol. 2008, 74, 3895–3898. [Google Scholar] [CrossRef] [PubMed]
- Morrow, K.M.; Tedford, A.R.; Pankey, M.S.; Lesser, M.P. A member of the Roseobacter clade, Octadecabacter sp., is the dominant symbiont in the brittle star Amphipholis squamata. FEMS Microbiol. Ecol. 2018, 94, fiy030. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.H.; Pratte, Z.A.; Kerwin, A.H.; Rotjan, R.D.; Stewart, F.J. Season, but not symbiont state, drives microbiome structure in the temperate coral Astrangia poculata. Microbiome 2017, 5, 120. [Google Scholar] [CrossRef] [PubMed]
- Elifantz, H.; Horn, G.; Ayon, M.; Cohen, Y.; Minz, D. Rhodobacteraceae are the key members of the microbial community of the initial biofilm formed in Eastern Mediterranean coastal seawater. FEMS Microbiol. Ecol. 2013, 85, 348–357. [Google Scholar] [CrossRef]
- Kwon, K.K.; Lee, H.S.; Yang, S.H.; Kim, S.J. Kordiimonas gwangyangensis gen. nov., sp nov., a marine bacterium isolated from marine sediments that forms a distinct phyletic lineage (Kordiimonadales ord. nov.) in the ‘Alphaproteobacteria’. Int. J. Syst. Evol. Microbiol. 2005, 55, 2033–2037. [Google Scholar] [CrossRef]
- Verna, C.; Ramette, A.; Wiklund, H.; Dahlgren, T.G.; Glover, A.G.; Gaill, F.; Dubilier, N. High symbiont diversity in the bone-eating worm Osedax mucofloris from shallow whale-falls in the North Atlantic. Environ. Microbiol. 2010, 12, 2355–2370. [Google Scholar] [CrossRef]
- Yoon, J.; Matsuo, Y.; Matsuda, S.; Adachi, K.; Kasai, H.; Yokota, A. Rubritalea spongiae sp nov and Rubritalea tangerina sp nov., two carotenoid- and squalene-producing marine bacteria of the family Verrucomicrobiaceae within the phylum ‘Verrucomicrobia’, isolated from marine animals. Int. J. Syst. Evol. Microbiol. 2007, 57, 2337–2343. [Google Scholar] [CrossRef]
- Sylvan, J.B.; Toner, B.M.; Edwards, K.J. Life and death of deep-sea vents: Bacterial diversity and ecosystem succession on inactive hydrothermal sulfides. mBio 2012, 3, e00279-11. [Google Scholar] [CrossRef]
- Takeuchi, M.; Katayama, T.; Yamagishi, T.; Hanada, S.; Tamaki, H.; Kamagata, Y.; Oshima, K.; Hattori, M.; Marumo, K.; Nedachi, M.; et al. Methyloceanibacter caenitepidi gen. nov., sp nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2014, 64, 462–468. [Google Scholar] [CrossRef]
- Vekeman, B.; Kerckhof, F.M.; Cremers, G.; de Vos, P.; Vandamme, P.; Boon, N.; Op den Camp, H.J.M.; Heylen, K. New Methyloceanibacter diversity from North Sea sediments includes methanotroph containing solely the soluble methane monooxygenase. Environ. Microbiol. 2016, 18, 4523–4536. [Google Scholar] [CrossRef]
- Doronina, N.V.; Poroshina, M.N.; Kaparullina, E.N.; Ezhov, V.A.; Trotsenko, Y.A. Methyloligella halotolerans gen. nov., sp nov and Methyloligella solikamskensis sp nov., two non-pigmented halotolerant obligately methylotrophic bacteria isolated from the Ural saline environments. Syst. Appl. Microbiol. 2013, 36, 148–154. [Google Scholar] [CrossRef]
- Judd, A.; Hovland, M. Seabed Fluid Flow: The Impact on Geology, Biology and the Marine Environment; Cambridge University Press: Cambridge, UK, 2009. [Google Scholar]
- Zhou, Z.C.; Pan, J.; Wang, F.P.; Gu, J.D.; Li, M. Bathyarchaeota: Globally distributed metabolic generalists in anoxic environments. FEMS Microbiol. Rev. 2018, 42, 639–655. [Google Scholar] [CrossRef]
- Evans, P.N.; Parks, D.H.; Chadwick, G.L.; Robbins, S.J.; Orphan, V.J.; Golding, S.D.; Tyson, G.W. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 2015, 350, 434–438. [Google Scholar] [CrossRef]
- Zhang, W.P.; Ding, W.; Yang, B.; Tian, R.M.; Gu, S.; Luo, H.W.; Qian, P.Y. Genomic and transcriptomic evidence for carbohydrate consumption among microorganisms in a cold seep brine pool. Front. Microbiol. 2016, 7, 1825. [Google Scholar] [CrossRef]
- He, Y.; Li, M.; Perumal, V.; Feng, X.; Fang, J.; Xie, J.; Sievert, S.M.; Wang, F. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 2016, 1, 16035. [Google Scholar] [CrossRef]
- Lazar, C.S.; Baker, B.J.; Seitz, K.; Hyde, A.S.; Dick, G.J.; Hinrichs, K.U.; Teske, A.P. Genomic evidence for distinct carbon substrate preferences and ecological niches of Bathyarchaeota in estuarine sediments. Environ. Microbiol. 2016, 18, 1200–1211. [Google Scholar] [CrossRef]
- Kubo, K.; Lloyd, K.G.; Biddle, J.F.; Amann, R.; Teske, A.; Knittel, K. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 2012, 6, 1949–1965. [Google Scholar] [CrossRef]
- Lazar, C.S.; Biddle, J.F.; Meador, T.B.; Blair, N.; Hinrichs, K.U.; Teske, A.P. Environmental controls on intragroup diversity of the uncultured benthic archaea of the miscellaneous Crenarchaeotal group lineage naturally enriched in anoxic sediments of the White Oak River estuary (North Carolina, USA). Environ. Microbiol. 2015, 17, 2228–2238. [Google Scholar] [CrossRef]
- Fillol, M.; Auguet, J.C.; Casamayor, E.O.; Borrego, C.M. Insights in the ecology and evolutionary history of the Miscellaneous Crenarchaeotic Group lineage. ISME J. 2016, 10, 665–677. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, R.C.; Wang, H.M.; Gong, L.F.; Man, B.Y.; Xu, Y. Distribution of Bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci. Rep. 2017, 7, 45028. [Google Scholar] [CrossRef]
- Singh, N.; Kendall, M.M.; Liu, Y.T.; Boone, D.R. Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: Description of Methanococcoides alaskense sp nov., and emended description of Methanosarcina baltica. Int. J. Syst. Evol. Microbiol. 2005, 55, 2531–2538. [Google Scholar] [CrossRef]
- Waite, D.W.; Vanwonterghem, I.; Rinke, C.; Parks, D.H.; Zhang, Y.; Takai, K.; Sievert, S.M.; Simon, J.; Campbell, B.J.; Hanson, T.E.; et al. Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 2017, 8, 682. [Google Scholar] [CrossRef]
- Giovannelli, D.; Chung, M.; Staley, J.; Starovoytov, V.; Le Bris, N.; Vetriani, C. Sulfurovum riftiae sp nov., a mesophilic, thiosulfate-oxidizing, nitrate-reducing chemolithoautotrophic epsilonproteobacterium isolated from the tube of the deep-sea hydrothermal vent polychaete Riftia pachyptila. Int. J. Syst. Evol. Microbiol. 2016, 66, 2697–2701. [Google Scholar] [CrossRef]
- Inagaki, F.; Takai, K.; Nealson, K.H.; Horikoshi, K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 2004, 54, 1477–1482. [Google Scholar] [CrossRef]
- Mino, S.; Kudo, H.; Arai, T.; Sawabe, T.; Takai, K.; Nakagawa, S. Sulfurovum aggregans sp nov., a hydrogenoxidizing, thiosulfate-reducing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent chimney, and an emended description of the genus Sulfurovum. Int. J. Syst. Evol. Microbiol. 2014, 64, 3195–3201. [Google Scholar] [CrossRef]
- Mori, K.; Yamaguchi, K.; Hanada, S. Sulfurovum denitrificans sp nov., an obligately chemolithoautotrophic sulfur-oxidizing epsilonproteobacterium isolated from a hydrothermal field. Int. J. Syst. Evol. Microbiol. 2018, 68, 2183–2187. [Google Scholar] [CrossRef]
- Cai, L.; Shao, M.F.; Zhang, T. Non-contiguous finished genome sequence and description of Sulfurimonas hongkongensis sp nov., a strictly anaerobic denitrifying, hydrogen-and sulfur-oxidizing chemolithoautotroph isolated from marine sediment. Stand. Genom. Sci. 2014, 9, 1302–1310. [Google Scholar] [CrossRef]
- Inagaki, F.; Takai, K.; Hideki, K.I.; Nealson, K.H.; Horikishi, K. Sulfurimonas autotrophica gen. nov., sp nov., a novel sulfur-oxidizing epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 2003, 53, 1801–1805. [Google Scholar] [CrossRef]
- Labrenz, M.; Grote, J.; Mammitzsch, K.; Boschker, H.T.S.; Laue, M.; Jost, G.; Glaubitz, S.; Jurgens, K. Sulfurimonas gotlandica sp nov., a chemoautotrophic and psychrotolerant epsilonproteobacterium isolated from a pelagic redoxcline, and an emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2013, 63, 4141–4148. [Google Scholar] [CrossRef]
- Takai, K.; Suzuki, M.; Nakagawa, S.; Miyazaki, M.; Suzuki, Y.; Inagaki, F.; Horikoshi, K. Sulfurimonas paralvinellae sp nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteo-bacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov and emended description of the genus Sulfurimonas. Int. J. Syst. Evol. Microbiol. 2006, 56, 1725–1733. [Google Scholar]
- Robbins, S.J.; Evans, P.N.; Parks, D.H.; Golding, S.D.; Tyson, G.W. Genome-centric analysis of microbial populations enriched by hydraulic fracture fluid additives in a coal bed methane production well. Front. Microbiol. 2016, 7, 731. [Google Scholar] [CrossRef]
- Inagaki, F.; Nunoura, T.; Nakagawa, S.; Teske, A.; Lever, M.; Lauer, A.; Suzuki, M.; Takai, K.; Delwiche, M.; Colwell, F.S.; et al. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments, on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 2006, 103, 2815–2820. [Google Scholar] [CrossRef]
- Vigneron, A.; Alsop, E.B.; Cruaud, P.; Philibert, G.; King, B.; Baksmaty, L.; Lavallee, D.; Lomans, B.P.; Kyrpides, N.C.; Head, I.M.; et al. Comparative metagenomics of hydrocarbon and methane seeps of the Gulf of Mexico. Sci. Rep. 2017, 7, 16015. [Google Scholar] [CrossRef]
- Wegener, G.; Krukenberg, V.; Riedel, D.; Tegetmeyer, H.E.; Boetius, A. Intercellular wiring enables electron transfer between methanotrophic archaea and bacteria. Nature 2015, 526, 587–590. [Google Scholar] [CrossRef]
- Chung, S.H.; Lin, T.S.; Lin, C.C.; Liu, C.S.; Chen, S.C.; Wang, Y.S.; Wei, C.Y.; Chen, P.C. Geological investigation of gas hydrate resource potential in the offshore areas of south-western Taiwan. Spec. Publ. Cent. Geol. Surv. 2016, 30, 1–42. [Google Scholar]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar]
- Gray, J.P.; Herwig, R.P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl. Environ. Microbiol. 1996, 62, 4049–4059. [Google Scholar] [CrossRef]
- Katayama, T.; Nishioka, M.; Yamamoto, M. Phylogenetic relationships among turbellarian orders inferred from 18S rDNA sequences. Zoolog Sci. 1996, 13, 747–756. [Google Scholar] [CrossRef]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Reysenbach, A.L.; Wickham, G.S.; Pace, N.R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 1994, 60, 2113–2119. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Ye, Z.; Lai, M.-C.; Liu, C.-S.; Paull, C.K.; Lin, S.; Lai, S.-J.; You, Y.-T.; Wu, S.-Y.; Hung, C.-C.; et al. Microbial Communities in and Around the Siboglinid Tubeworms from the South Yungan East Ridge Cold Seep Offshore Southwestern Taiwan at the Northern South China Sea. Microorganisms 2024, 12, 2452. https://doi.org/10.3390/microorganisms12122452
Li Y, Ye Z, Lai M-C, Liu C-S, Paull CK, Lin S, Lai S-J, You Y-T, Wu S-Y, Hung C-C, et al. Microbial Communities in and Around the Siboglinid Tubeworms from the South Yungan East Ridge Cold Seep Offshore Southwestern Taiwan at the Northern South China Sea. Microorganisms. 2024; 12(12):2452. https://doi.org/10.3390/microorganisms12122452
Chicago/Turabian StyleLi, Yin, Zhiwei Ye, Mei-Chin Lai, Char-Shine Liu, Charles K. Paull, Saulwood Lin, Shu-Jung Lai, Yi-Ting You, Sue-Yao Wu, Chuan-Chuan Hung, and et al. 2024. "Microbial Communities in and Around the Siboglinid Tubeworms from the South Yungan East Ridge Cold Seep Offshore Southwestern Taiwan at the Northern South China Sea" Microorganisms 12, no. 12: 2452. https://doi.org/10.3390/microorganisms12122452
APA StyleLi, Y., Ye, Z., Lai, M.-C., Liu, C.-S., Paull, C. K., Lin, S., Lai, S.-J., You, Y.-T., Wu, S.-Y., Hung, C.-C., Ding, J.-Y., Shih, C.-J., Wu, Y.-C., Zhao, J., Xiao, W., Wu, C.-H., Dong, G., Zhang, H., Qiu, W., ... Chen, S.-C. (2024). Microbial Communities in and Around the Siboglinid Tubeworms from the South Yungan East Ridge Cold Seep Offshore Southwestern Taiwan at the Northern South China Sea. Microorganisms, 12(12), 2452. https://doi.org/10.3390/microorganisms12122452




