Microbial and Monosaccharide Composition of Biofilms Developing on Sandy Loams from an Aquifer Contaminated with Liquid Radioactive Waste
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Object of the Study
2.2. Medium Composition and Cultivation Conditions
2.3. Analytical Methods
2.4. Microscopic Methods
2.5. DNA Extraction from Pure Cultures and Biofilms
2.6. High-Throughput Sequencing of V4 Fragment of the 16S rRNA Genes
2.7. Identification of Pure Cultures
2.8. Bioinformatics Analysis
2.9. Nucleotide Sequence Accession Numbers
3. Results
3.1. The Composition of Biofilms on Sandy Loams
3.2. The Dynamics of Biofilms’ Formation on Sandy Loams
3.3. The Monosaccharides’ Composition in the Biofilms
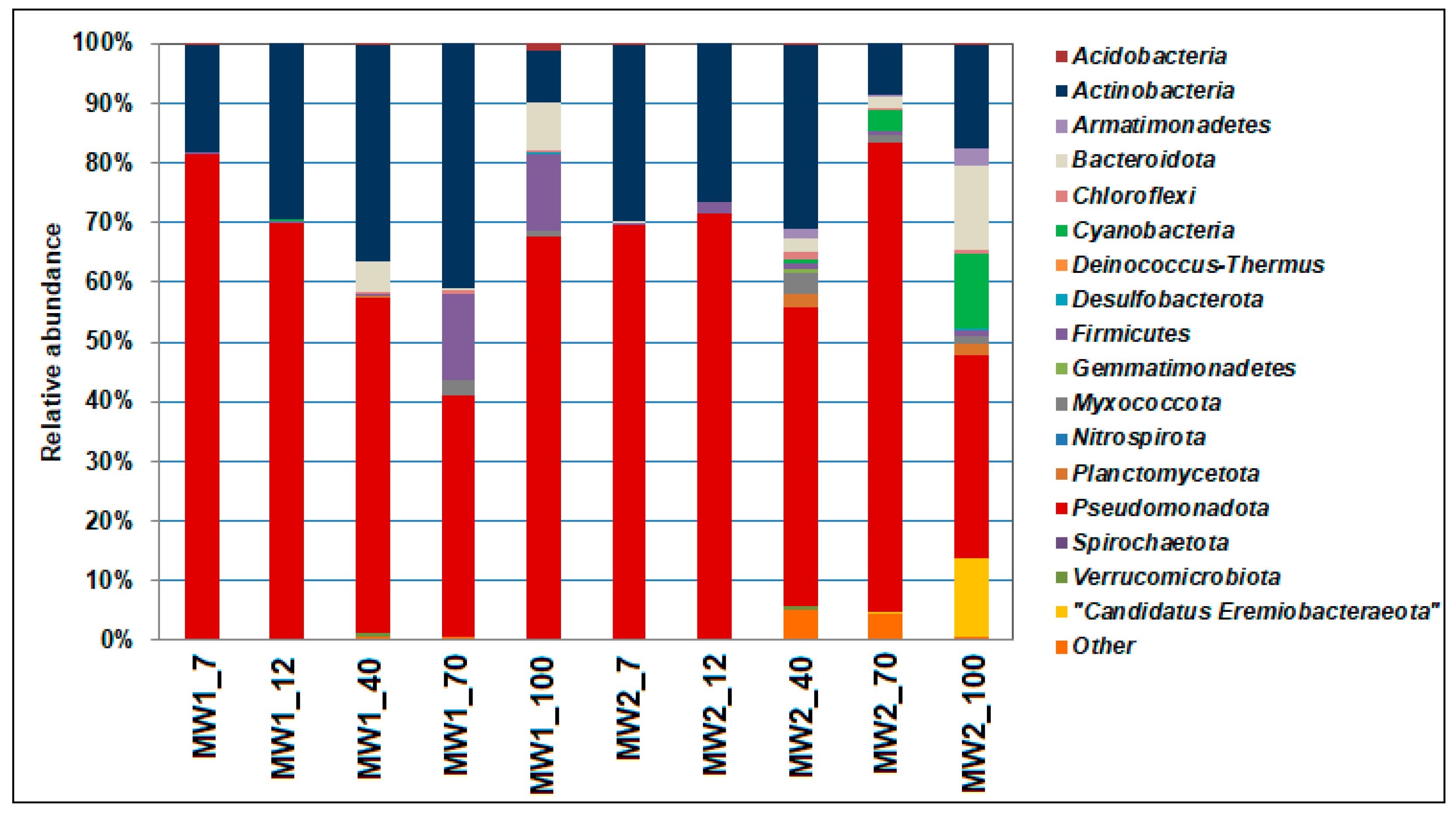
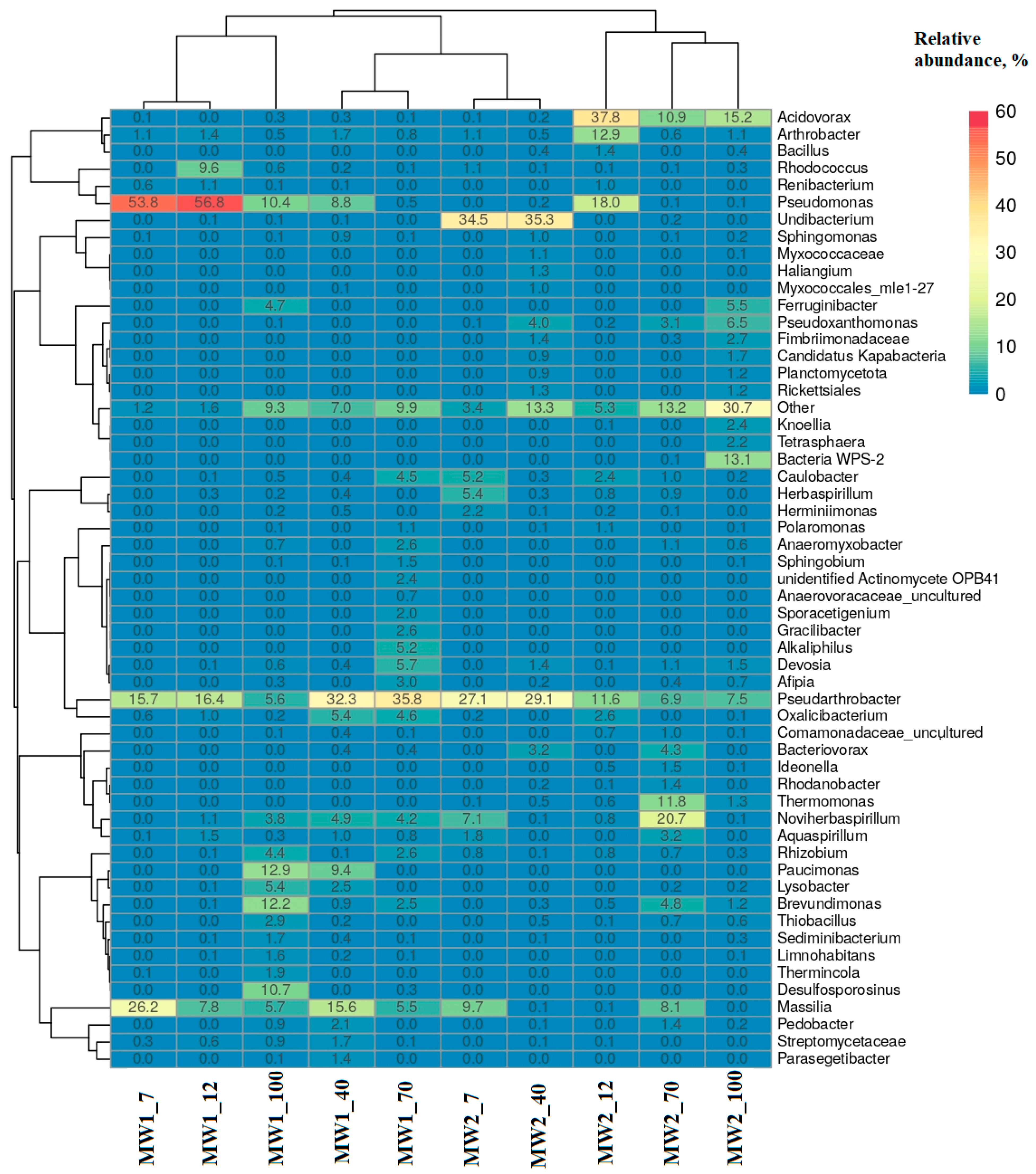
3.4. Phylogenetic Diversity of Bacteria in the Biofilms
3.5. Pure Cultures of Bacteria Isolated from Biofilms
3.6. Reduction of Nitrate Ions and Heavy Metals by the Biofilm Microbial Community and Pure Bacterial Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Poinssot, C.; Bourg, S.; Ouvrier, N.; Combernoux, N.; Rostaing, C.; Vargas-Gonzalez, M.; Bruno, J. Assessment of the environmental footprint of nuclear energy systems. Comparison between closed and open fuel cycles. Energy 2014, 69, 199–211. [Google Scholar] [CrossRef]
- Rybal’chenko, A.I.; Pimenov, M.K.; Kostin, P.P.; Balukova, V.D.; Nosukhin, A.V.; Mikerin, E.I.; Egorov, N.N.; Kaimin, E.P.; Kosareva, I.M.; Kurochkin, V.M. Deep Injection Disposal of Liquid Radioactive Waste in Russia; Battelle Press: Columbus, OH, USA, 1998; p. 206. [Google Scholar]
- Lloyd, J.R.; Renshaw, J.C. Bioremediation of radioactive waste: Radionuclide–microbe interactions in laboratory and field-scale studies. Curr. Opin. Biotechnol. 2005, 16, 254–260. [Google Scholar] [CrossRef]
- Zachara, J.M.; Long, P.E.; Bargar, J.; Davis, J.A.; Fox, P.; Fredrickson, J.K.; Freshley, M.D.; Konopka, A.E.; Liu, C.; McKinley, J.P.; et al. Persistence of uranium groundwater plumes: Contrasting mechanisms at two DOE sites in the groundwater–river interaction zone. J. Contam. Hydrol. 2013, 147, 45–72. [Google Scholar] [CrossRef] [PubMed]
- Safonov, A.V.; Boguslavsky, A.E.; Gaskova, O.L.; Boldyrev, K.A.; Shvartseva, O.S.; Khvashchevskaya, A.A.; Popova, N.M. Biogeochemical modelling of uranium immobilization and aquifer remediation strategies near NCCP sludge storage facilities. Appl. Sci. 2021, 11, 2875. [Google Scholar] [CrossRef]
- Safonov, A.; Popova, N.; Boldyrev, K.; Lavrinovich, E.; Boeva, N.; Artemiev, G.; Kuzovkina, E.; Emelyanov, A.; Myasnikov, I.; Zakharova, E.; et al. The microbial impact on U, Pu, Np, and Am immobilization on aquifer sandy rocks, collected at the deep LRW injection site. J. Geochem. Explor. 2022, 240, 107052. [Google Scholar] [CrossRef]
- Ma, R.; Zheng, C.; Liu, C. Groundwater impacts of radioactive wastes and associated environmental modeling assessment. In Encyclopedia of Sustainability Science and Technology; Meyers, R., Ed.; Springer: New York, NY, USA, 2018. [Google Scholar] [CrossRef]
- Berk, W.; Fu, Y. Redox roll-front mobilization of geogenic uranium by nitrate input into aquifers: Risks for groundwater resources. Environ. Sci. Technol. 2017, 51, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Meena, A.H.; Arai, Y. Environmental geochemistry of technetium. Environ. Chem. Lett. 2017, 15, 241–263. [Google Scholar] [CrossRef]
- Lorah, M.M.; Cozzarelli, I.M.; Böhlke, J.K. Biogeochemistry at a wetland sediment-alluvial aquifer interface in a landfill leachate plume. J. Contam. Hydrol. 2009, 105, 99–117. [Google Scholar] [CrossRef]
- Nazina, T.N.; Luk’yanova, E.A.; Zakharova, E.V.; Ivoilov, V.S.; Poltaraus, A.B.; Kalmykov, S.N.; Belyaev, S.S.; Zubkov, A.A. Distribution and activity of microorganisms in the deep repository for liquid radioactive waste at the Siberian Chemical Combine. Microbiology 2006, 75, 727–738. [Google Scholar] [CrossRef]
- Artemiev, G.; Safonov, A. Authigenic mineral formation in aquifers near the uranium sludge storage facility of Chepetsky Mechanical Plant during the formation of a biogeochemical barrier in a laboratory and field experiment. Minerals 2023, 13, 1319. [Google Scholar] [CrossRef]
- Townsend, L.T.; Morris, K.; Lloyd, J.R. Chapter 11—Microbial transformations of radionuclides in geodisposal systems. In The Microbiology of Nuclear Waste Disposal; Elsevier: Amsterdam, The Netherlands, 2021; pp. 245–265. [Google Scholar] [CrossRef]
- Simonoff, M.; Sergeant, C.; Poulain, S.; Pravikoff, M.S. Microorganisms and migration of radionuclides in environment. Comptes Rendus Chim. 2007, 10, 1092–1107. [Google Scholar] [CrossRef]
- Berridge, M.V.; Herst, P.M.; Tan, A.S. Tetrazolium dyes as tools in cell biology: New insights into their cellular reduction. Biotechnol. Annu. Rev. 2005, 11, 127–152. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.M.; Carley, J.; Fienen, M.; Mehlhorn, T.; Lowe, K.; Nyman, J.; Luo, J.; Gentile, M.E.; Rajan, R.; Wagner, D.; et al. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ. Sci. Technol. 2006, 40, 3978–3985. [Google Scholar] [CrossRef] [PubMed]
- Babich, T.L.; Safonov, A.V.; Grouzdev, D.S.; Andryuschenko, N.D.; Zakharova, E.V.; Nazina, T.N. Bacteria of the genus Shewanella from radionuclide-contaminated groundwater. Microbiology 2019, 88, 613–623. [Google Scholar] [CrossRef]
- Vettese, G.F.; Morris, K.; White-Pettigrew, M.; Townsend, L.T.; Shaw, S.; Boothman, C.; Lloyd, J.R. In situ (bio) remediation treatment options for U and Sr contaminated land: A comparison of radionuclide retention and remobilization. Environ. Sci. Adv. 2023, 2, 1423–1435. [Google Scholar] [CrossRef]
- You, W.; Peng, W.; Tian, Z.; Zheng, M. Uranium bioremediation with U(VI)-reducing bacteria. Sci. Total Environ. 2021, 798, 149107. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Liu, M.; Shang, M.; Yi, X. Horizontal well test for simulating the in situ remediation of nitrate contaminated groundwater by microbial denitrification. Water Air Soil Pollut. 2019, 230, 189. [Google Scholar] [CrossRef]
- Christensen, B.H.; Characklis, W.G. Physical and chemical properties of biofilms. In Biofilms; Characklis, W.G., Marshall, K.C., Eds.; Wiley: New York, NY, USA, 1990; pp. 93–130. [Google Scholar]
- Newsome, L.; Morris, K.; Lloyd, J.R. The biogeochemistry and bioremediation of uranium and other priority radionuclides. Chem. Geol. 2014, 363, 164–184. [Google Scholar] [CrossRef]
- Safonov, A.V.; Babich, T.L.; Sokolova, D.S.; Grouzdev, D.S.; Tourova, T.P.; Poltaraus, A.B.; Zakharova, E.V.; Merkel, A.Y.; Novikov, A.P.; Nazina, T.N. Microbial community and in situ bioremediation of groundwater by nitrate removal in the zone of a radioactive waste surface repository. Front. Microbiol. 2018, 9, 1985. [Google Scholar] [CrossRef]
- Wufuer, R.; Duo, J.; Li, W.; Fan, J.; Pan, X. Bioremediation of uranium- and nitrate-contaminated groundwater after the in situ leach mining of uranium. Water 2021, 13, 3188. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef]
- Edwards, S.J.; Kjellerup, B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 2013, 97, 9909–9921. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Mayer, C. Physico-chemical properties of biofilms. In Biofilms: Recent Advances in Their Study and Control; Evans, L.V., Ed.; Harwood Academic Publishers: Amsterdam, The Netherlands, 2000; pp. 19–34. [Google Scholar]
- Sharma, P.; Pandey, A.K.; Kim, S.-H.; Singh, S.P.; Chaturvedi, P.; Varjani, S. Critical review on microbial community during in-situ bioremediation of heavy metals from industrial wastewater. Environ. Technol. Innov. 2021, 24, 101826. [Google Scholar] [CrossRef]
- Mishra, S.; Huang, Y.; Li, J.; Wu, X.; Zhou, Z.; Lei, Q.; Bhatt, P.; Chen, S. Biofilm-mediated bioremediation is a powerful tool for the removal of environmental pollutants. Chemosphere 2022, 294, 133609. [Google Scholar] [CrossRef]
- Fletcher, M. How do bacteria attach to solid surfaces? Microbiol. Sci. 1987, 4, 133–136. [Google Scholar]
- Jeffrey, W.H.; Paul, J.H. Activity measurements of planktonic microbial and microfouling communities in a eutrophic estuary. Appl. Environ. Microbiol. 1986, 46, 157–162. [Google Scholar] [CrossRef]
- Lawrence, J.R.; Korber, D.R.; Wolfaardt, G.M. Heterogeneity of natural biofilm communities. Cells Mater. 1996, 6, 19. Available online: https://digitalcommons.usu.edu/cellsandmaterials/vol6/iss1/19 (accessed on 20 February 2023).
- Nwodo, U.U.; Green, E.; Okoh, A.I. Bacterial exopolysaccharides: Functionality and prospects. Int. J. Mol. Sci. 2012, 13, 14002–14015. [Google Scholar] [CrossRef]
- Fletcher, M. Bacterial metabolism in biofilms. In Biofilms—Science and Technology; Melo, L.F., Bott, T.R., Fletcher, M., Capdeville, B., Eds.; NATO ASI Series; Springer: Dordrecht, The Netherlands, 1992; Volume 223, pp. 113–124. [Google Scholar] [CrossRef]
- Maamar, S.B. Groundwater isolation governs chemistry and microbial community structure along hydrologic flowpaths. Front. Microbiol. 2015, 6, 1457. [Google Scholar] [CrossRef]
- Gupta, P.; Diwan, B. Bacterial exopolysaccharide mediated heavy metal removal: A review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef]
- Safonov, A.V.; Perepelov, A.V.; Babich, T.L.; Popova, N.M.; Grouzdev, D.S.; Filatov, A.V.; Shashkov, A.S.; Demina, L.I.; Nazina, T.N. Structure and gene cluster of the O-polysaccharide from Pseudomonas veronii A-6-5 and its uranium bonding. Int. J. Biol. Macromol. 2020, 165, 2197–2204. [Google Scholar] [CrossRef]
- ASTM Standards 04.08:117–127; Standard Test Method for Particle-Size Analysis of Soils D422-63 (2007). American Society for Testing Materials: Philadelphia, PA, USA, 2008.
- Pfennig, N.; Lippert, K.D. Uber das Vitamin B12-Bedfirfnis phototropher Schwefelbakterien. Arch. Mikrobiol. 1966, 55, 245–256. [Google Scholar] [CrossRef]
- Cory, A.H.; Owen, T.C.; Barltrop, J.A.; Cory, J.G. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991, 3, 207–212. [Google Scholar] [CrossRef]
- Plakunov, V.K.; Mart’yanov, S.V.; Teteneva, N.A.; Zhurina, M.V. A universal method for quantitative characterization of growth and metabolic activity of microbial biofilms in static models. Microbiology 2016, 85, 509–513. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Wang, F.; Wei, D.; Wang, X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J. Microbiol. Meth. 2010, 82, 330–333. [Google Scholar] [CrossRef]
- Leontein, K.; Lönngren, J. Determination of the absolute configuration of sugars by gas–liquid chromatography of their acetylated 2-octylglycosides. Methods Carbohydr. Chem. 1993, 9, 87–89. [Google Scholar] [CrossRef]
- Al-Kayssi, M.; Magee, R.J.; Wilson, C.L. Spectrophotometric studies on technetium and rhenium. Talanta 1962, 9, 125–132. [Google Scholar] [CrossRef]
- Onishi, H.; Sekine, K. Spectrophotometric determination of zirconium, uranium, thorium and rare earths with arsenazo III after extractions with thenoyltrifluoroacetone and tri-n-octylamine. Talanta 1972, 19, 473–478. [Google Scholar] [CrossRef]
- Haass-Koffler, C.L.; Naeemuddin, M.; Bartlett, S.E. An analytical tool that quantifies cellular morphology changes from three-dimensional fluorescence images. J. Vis. Exp. 2012, 66, e4233. [Google Scholar] [CrossRef]
- Vishnyakova, A.; Popova, N.; Artemiev, G.; Botchkova, E.; Litti, Y.; Safonov, A. Effect of mineral carriers on biofilm formation and nitrogen removal activity by an indigenous anammox community from cold groundwater ecosystem alone and bioaugmented with biomass from a “warm” anammox reactor. Biology 2022, 11, 1421. [Google Scholar] [CrossRef]
- Allen, R.D.; David, G.B.; Nomarski, G. The zeiss-Nomarski differential interference equipment for transmitted-light microscopy. Mikrosk. Mikrosk. Tech. 1969, 69, 193–221. [Google Scholar]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef]
- Brunk, C.F.; Avaniss-Aghajani, E.; Brunk, C.A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl. Environ. Microbiol. 1996, 61, 872–879. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- V1.3.2. Available online: https://github.com/jstjohn/SeqPrep (accessed on 4 October 2016).
- Version: 1.9.10/1.4.9; SILVA: r138.1. Available online: https://www.arb-silva.de/ngs/ (accessed on 14 March 2023).
- Metsalu, T.; Vilo, J. Clustvis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef]
- Available online: http://bioinfogp.cnb.csic.es/tools/venny/ (accessed on 12 April 2022).
- Schoina, E.; Doulgeraki, A.I.; Miliou, H.; Nychas, G.-J.E. Dynamics of water and biofilm bacterial community composition in a Mediterranean recirculation aquaculture system. Aquac. J. 2022, 2, 164–179. [Google Scholar] [CrossRef]
- Grzegorczyk, M.; Pogorzelski, S.J.; Pospiech, A.; Boniewicz-Szmyt, K. Monitoring of marine biofilm formation dynamics at submerged solid surfaces with multitechnique sensors. Front. Mar. Sci. 2018, 5, 363. [Google Scholar] [CrossRef]
- Xenopoulos, E.; Giannikakis, I.; Chatzifragkou, A.; Koutinas, A.; Papanikolaou, S. Lipid production by yeasts growing on commercial xylose in submerged cultures with process water being partially replaced by olive mill wastewaters. Processes 2020, 8, 819. [Google Scholar] [CrossRef]
- King, J.D.; Kocincova, D.; Westman, E.L.; Lam, J.S. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 2009, 15, 261–312. [Google Scholar] [CrossRef]
- Kasimova, A.A.; Shashkov, A.S.; Perepelov, A.V.; Babich, T.; Demina, L.; Popova, N.; Krivonos, D.; Safonov, A. Structure elucidation and gene cluster of the O-antigen of Shewanella xiamenensis strain DCB-2-1 containing an amide of d-glucuronic acid with d-alanine and its bonding with U, Cr and V. Int. J. Biol. Macromol. 2023, 253, 127546. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Panova, I.A.; Beletsky, A.V.; Avakyan, M.R.; Kadnikov, V.V.; Antsiferov, D.V.; Banks, D.; Frank, Y.A.; Pimenov, N.V.; Ravin, N.V.; et al. Genomic insights into a new acidophilic, copper-resistant Desulfosporosinus isolate from the oxidized tailings area of an abandoned gold mine. FEMS Microbiol. Ecol. 2016, 92, fiw111. [Google Scholar] [CrossRef]
- Panova, I.A.; Ikkert, O.; Avakyan, M.R.; Kopitsyn, D.S.; Mardanov, A.V.; Pimenov, N.V.; Shcherbakova, V.A.; Ravin, N.V.; Karnachuk, O.V. Desulfosporosinus metallidurans sp. nov.; an acidophilic, metal-resistant sulfate-reducing bacterium from acid mine drainage. Int. J. Syst. Evol. Microbiol. 2021, 71, 004876. [Google Scholar] [CrossRef]
- Liu, J.L.; Yao, J.; Zhou, D.L.; Liu, B.; Liu, H.; Li, M.; Zhao, C.; Sunahara, G.; Duran, R. Mining-related multi-resistance genes in sulfate-reducing bacteria treatment of typical karst nonferrous metal(loid) mine tailings in China. Environ. Sci. Pollut. Res. Int. 2023, 30, 104753–104766. [Google Scholar] [CrossRef]
- Ishii, S.; Ashida, N.; Ohno, H.; Segawa, T.; Yabe, S.; Otsuka, S.; Yokota, A.; Senoo, K. Noviherbaspirillum denitrificans sp. nov.; a denitrifying bacterium isolated from rice paddy soil and Noviherbaspirillum autotrophicum sp. nov.; a denitrifying, facultatively autotrophic bacterium isolated from rice paddy soil and proposal to reclassify Herbaspirillum massiliense as Noviherbaspirillum massiliense comb. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 1841–1848. [Google Scholar] [CrossRef]
- Alexander, B.; Priest, F.G. Bacillus glucanolyticus, a new species that degrades a variety of [beta]-glucans. Int. J. Syst. Bacteriol. 1989, 39, 112–115. [Google Scholar] [CrossRef]
- Velazquez, E.; de Miguel, T.; Poza, M.; Rivas, R.; Rossello-Mora, R.; Villa, T.G. Paenibacillus favisporus sp. nov.; a xylanolytic bacterium isolated from cow faeces. Int. J. Syst. Evol. Microbiol. 2004, 54, 59–64. [Google Scholar] [CrossRef]
- Takeuchi, M.; Hatano, K. Union of the genera Microbacterium Orla-Jensen and Aureobacterium Collins et al. in a redefined genus Microbacterium. Int. J. Syst. Bacteriol. 1998, 48, 739–747. [Google Scholar] [CrossRef]
- Mahto, K.U.; Vandana; Priyadarshanee, M.; Samantaray, D.P.; Das, S. Bacterial biofilm and extracellular polymeric substances in the treatment of environmental pollutants: Beyond the protective role in survivability. J. Clean. Prod. 2022, 379, 134759. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef]
- Lewandowski, Z. MIC and biofilm heterogeneity. Proc. Corros. 2000, 400, 1–7. [Google Scholar]
- Anderson, R.T.; Vrionis, H.A.; Ortiz-Bernad, I.; Resch, C.T.; Long, P.E.; Dayvault, R.; Karp, K.; Marutzky, S.; Metzler, D.R.; Peacock, A.; et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl. Environ. Microbiol. 2003, 69, 5884–5891. [Google Scholar] [CrossRef]
- Senko, J.M.; Istok, J.D.; Suflita, J.M.; Krumholz, L.R. In-situ evidence for uranium immobilization and remobilization. Environ. Sci. Technol. 2002, 36, 1491–1496. [Google Scholar] [CrossRef]
- Xu, M.; Wu, W.M.; Wu, L.; He, Z.; Van Nostrand, J.D.; Deng, Y.; Luo, J.; Carley, J.; Ginder-Vogel, M.; Gentry, T.J.; et al. Responses of microbial community functional structures to pilot-scale uranium in situ bioremediation. ISME J. 2010, 4, 1060–1070. [Google Scholar] [CrossRef]
- Wang, G.; Liu, Y.; Wang, J.; Xiang, J.; Zeng, T.; Li, S.; Song, J.; Zhang, Z.; Liu, J. The remediation of uranium-contaminated groundwater via bioreduction coupled to biomineralization with different pH and electron donors. Environ. Sci. Pollut. Res. Int. 2023, 30, 23096–23109. [Google Scholar] [CrossRef]
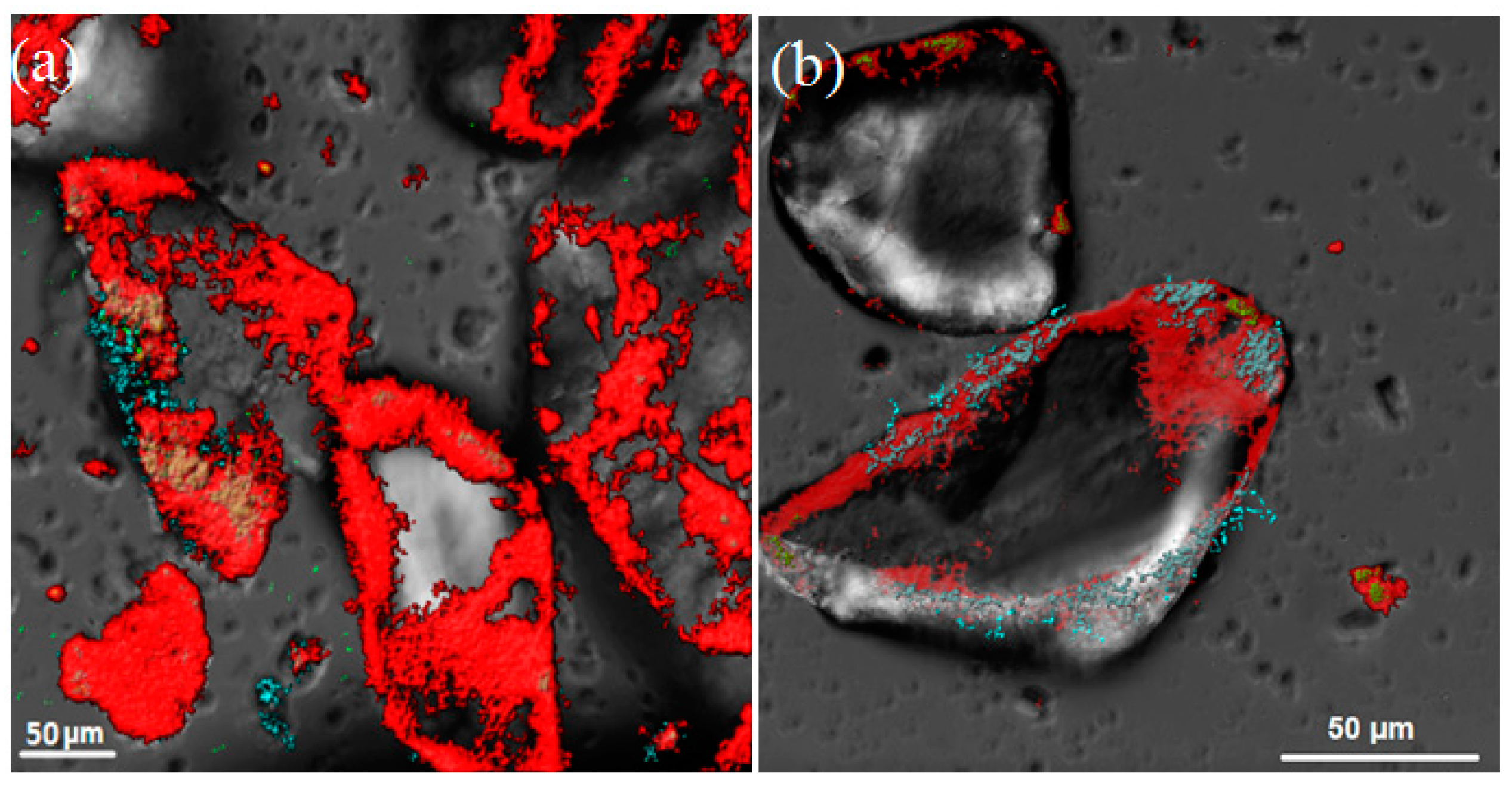
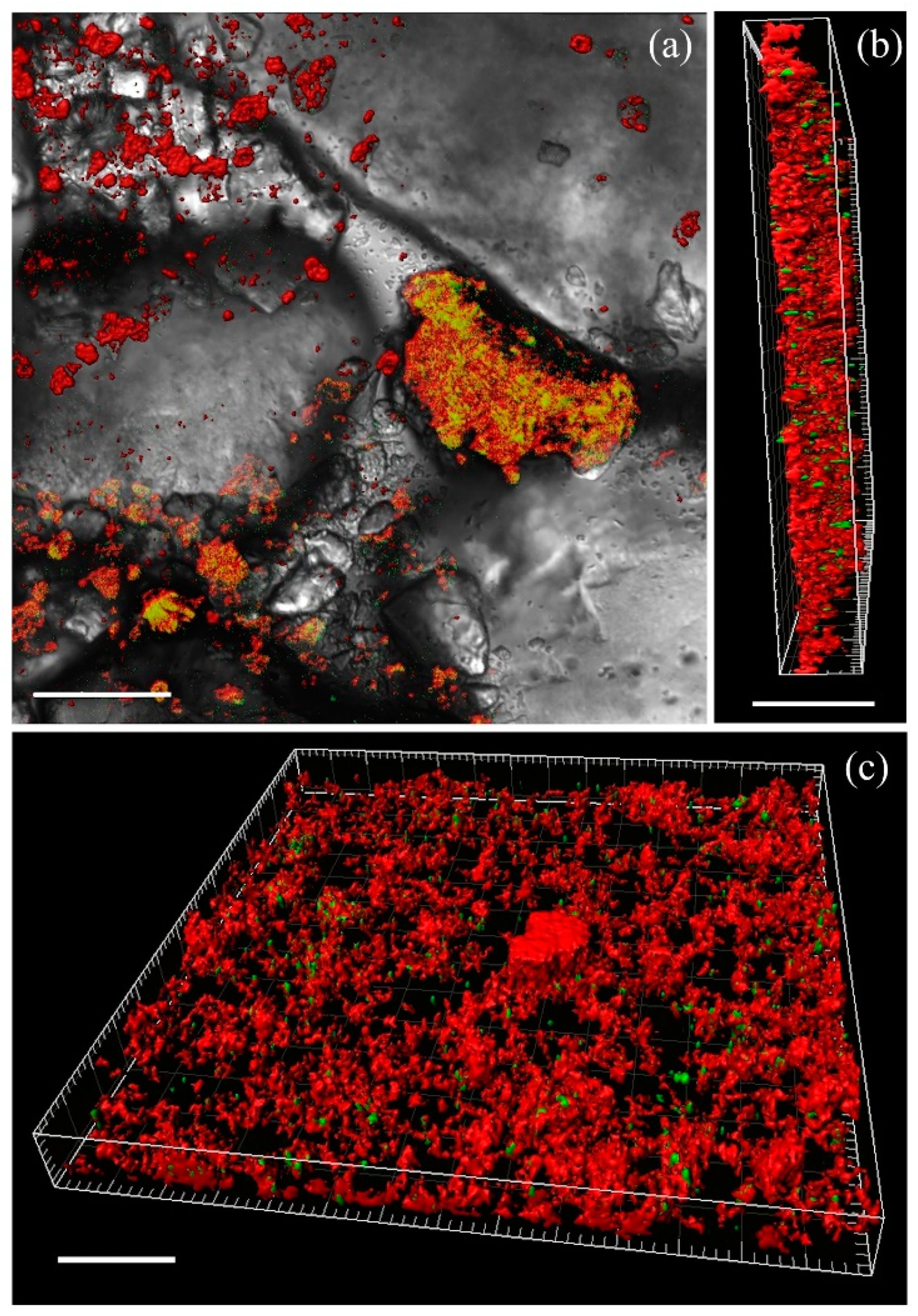
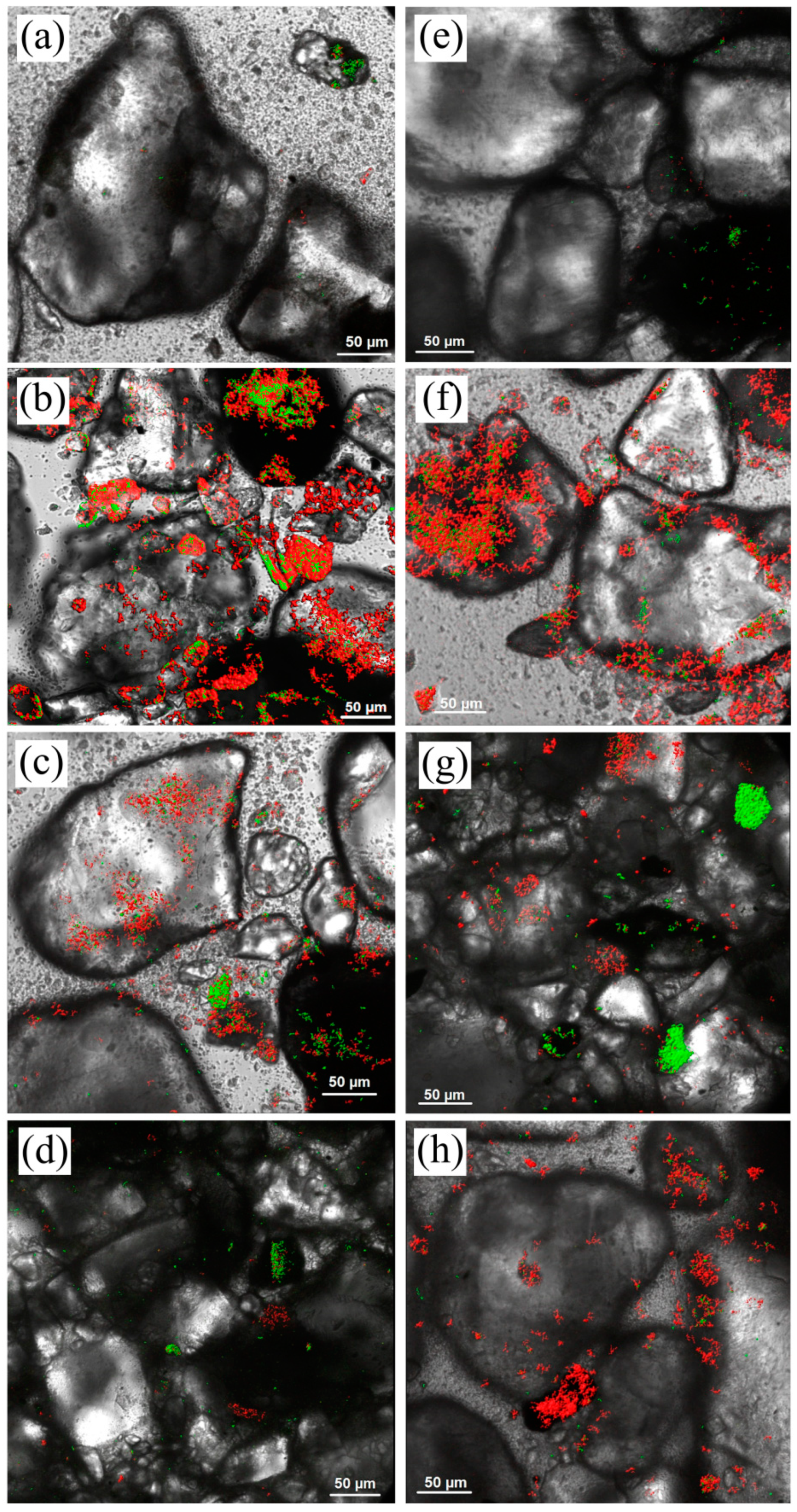



| Sample | Parameters | Incubation Time, Days | |||
|---|---|---|---|---|---|
| 1 | 20 | 40 | 100 | ||
| MW1 | MTT test | 0.4 | 0.8 | 1.2 | 1.5 |
| Proteins, mg/L | 0.8 | 6.4 | 4.2 | 7.3 | |
| Corg, mass. % | 1.6 | 9.0 | 2.4 | 0.2 | |
| Polysaccharides, % | 0.3 | 19.8 | 7.8 | 2.8 | |
| Nucleic acids, % | 0.2 | 4.2 | 2.7 | 1.8 | |
| MW2 | MTT test | 0.8 | 0.9 | 2.1 | 2.5 |
| Proteins, mg/L | 0.9 | 2.8 | 5.6 | 9.9 | |
| Corg, mass. % | 1.2 | 8.5 | 1.2 | 0.4 | |
| Polysaccharides, % | 0.3 | 23.6 | 10.9 | 3.3 | |
| Nucleic acids, % | 0.5 | 3.8 | 11.2 | 3.4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babich, T.L.; Popova, N.M.; Sokolova, D.S.; Perepelov, A.V.; Safonov, A.V.; Nazina, T.N. Microbial and Monosaccharide Composition of Biofilms Developing on Sandy Loams from an Aquifer Contaminated with Liquid Radioactive Waste. Microorganisms 2024, 12, 275. https://doi.org/10.3390/microorganisms12020275
Babich TL, Popova NM, Sokolova DS, Perepelov AV, Safonov AV, Nazina TN. Microbial and Monosaccharide Composition of Biofilms Developing on Sandy Loams from an Aquifer Contaminated with Liquid Radioactive Waste. Microorganisms. 2024; 12(2):275. https://doi.org/10.3390/microorganisms12020275
Chicago/Turabian StyleBabich, Tamara L., Nadezhda M. Popova, Diyana S. Sokolova, Andrei V. Perepelov, Alexey V. Safonov, and Tamara N. Nazina. 2024. "Microbial and Monosaccharide Composition of Biofilms Developing on Sandy Loams from an Aquifer Contaminated with Liquid Radioactive Waste" Microorganisms 12, no. 2: 275. https://doi.org/10.3390/microorganisms12020275
APA StyleBabich, T. L., Popova, N. M., Sokolova, D. S., Perepelov, A. V., Safonov, A. V., & Nazina, T. N. (2024). Microbial and Monosaccharide Composition of Biofilms Developing on Sandy Loams from an Aquifer Contaminated with Liquid Radioactive Waste. Microorganisms, 12(2), 275. https://doi.org/10.3390/microorganisms12020275









