Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Determination of the Transformation and Mutation Frequencies
2.3. Bacterial Strain Genomic Sequences
2.4. Determination of cts Operon, CJIEs and Associated DNase Genes
2.5. DNase Detection
2.6. Determination of Phylogeny
3. Results
3.1. Transformation Frequencies of C. jejuni and C. coli
3.2. Multiple Mechanisms for Non-Transformability among ST-50 Strains
3.3. Global Examination of the ST-50 Non-Transformability Genotype
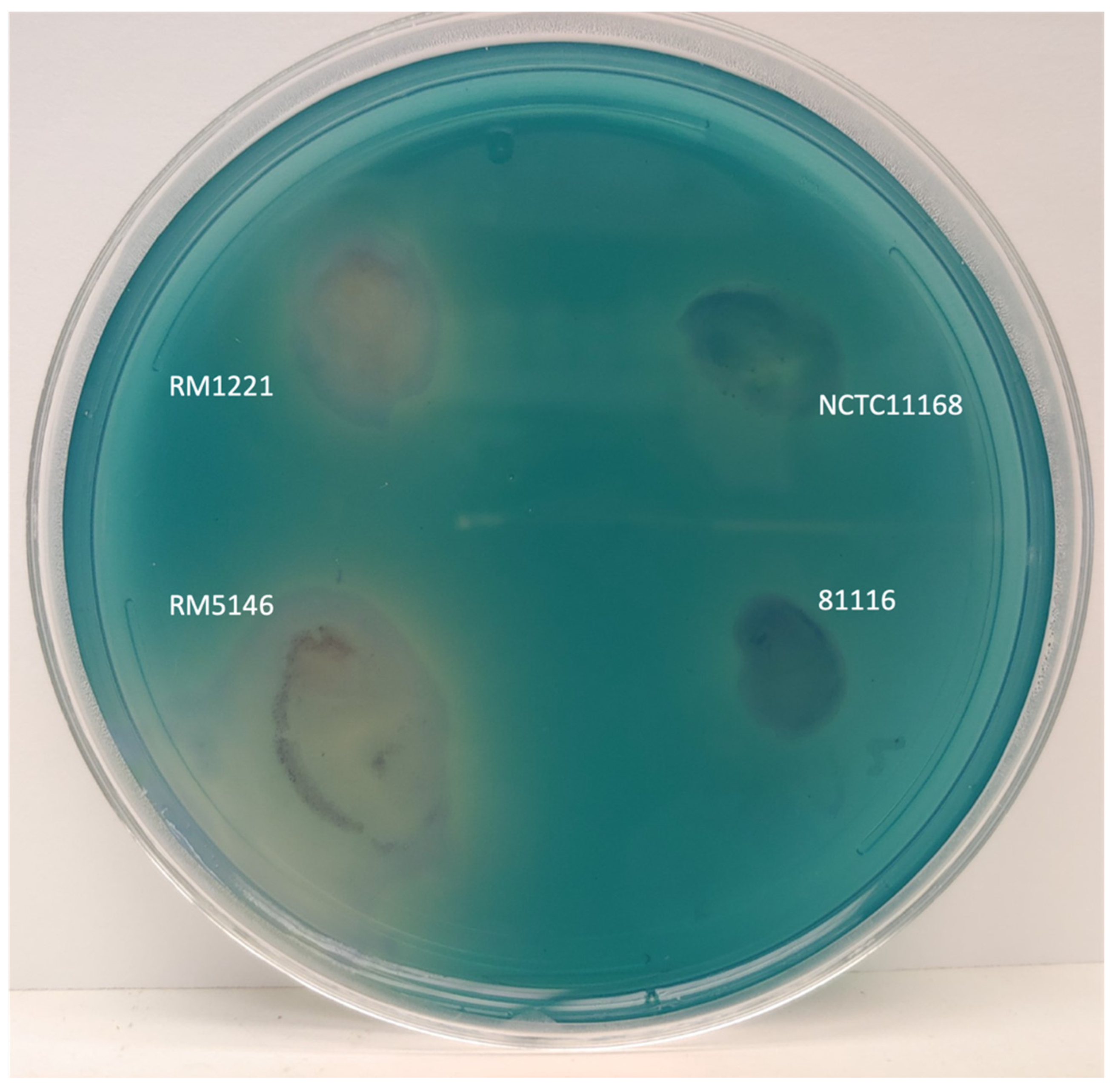
3.4. Evidence of Extracellular DNase from dns Positive Strains
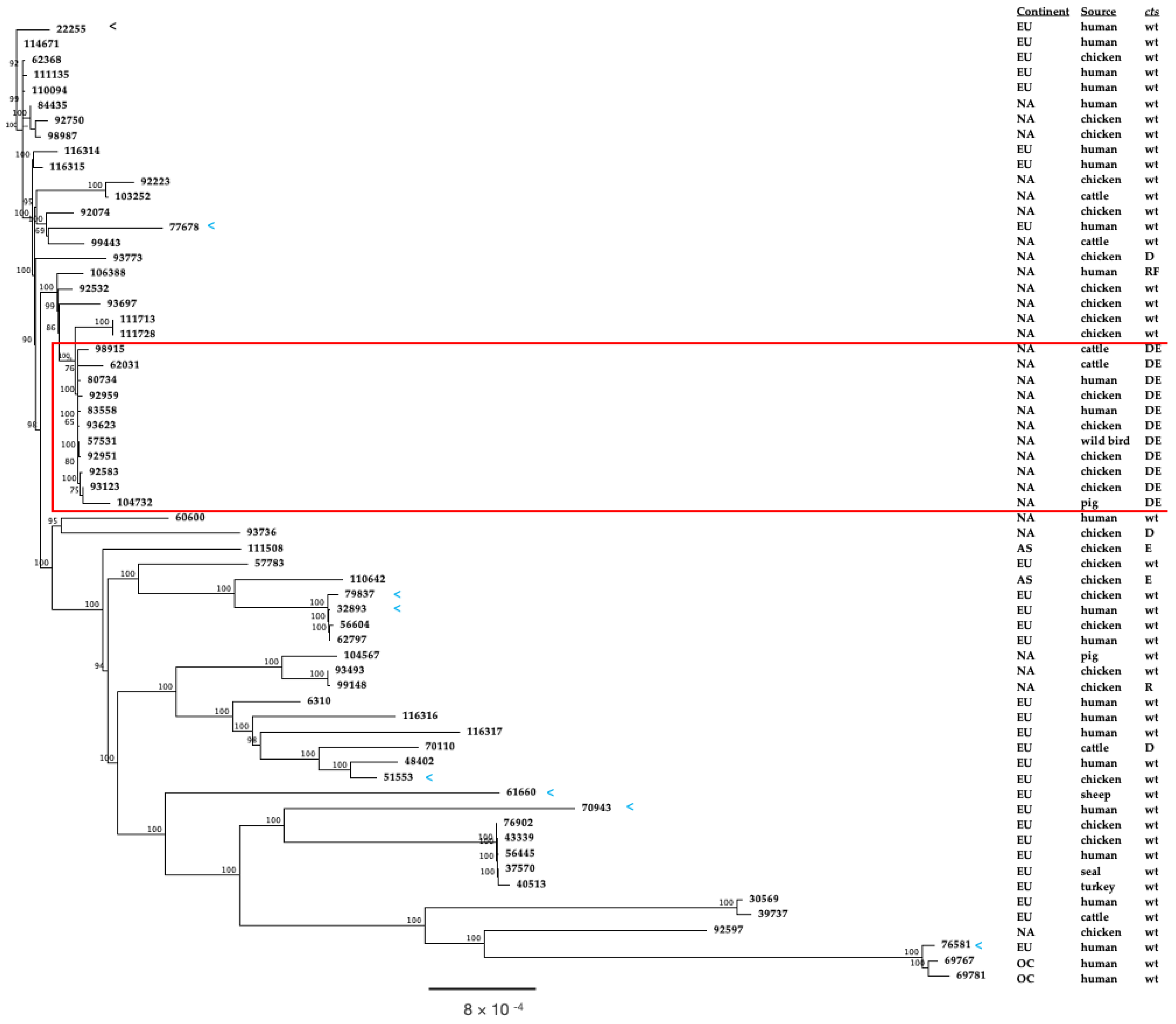
3.5. Phylogenetic Comparison of North American and European ST-50 Strains
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Man, S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States—Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Nachamkin, I.; Allos, B.M.; Ho, T. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 1998, 11, 555–567. [Google Scholar] [CrossRef]
- Scallan Walter, E.J.; Crim, S.M.; Bruce, B.B.; Griffin, P.M. Incidence of Campylobacter-Associated Guillain- Barré syndrome estimated from health insurance data. Foodborne Pathog. Dis. 2020, 17, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Antibiotic resistance threats in the United States. 2019. Available online: https://stacks.cdc.gov/view/cdc/82532 (accessed on 2 January 2024).
- Bolinger, H.; Kathariou, S. The current state of macrolide resistance in Campylobacter spp.: Trends and impacts of resistance mechanisms. Appl. Environ. Microbiol. 2017, 83, e00416-17. [Google Scholar] [CrossRef] [PubMed]
- Sproston, E.L.; Wimalarathna, H.M.L.; Sheppard, S.K. Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 2018, 4, e000198. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Dingle, K.E.; Colles, F.M.; Wareing, D.R.; Ure, R.; Fox, A.J.; Bolton, F.E.; Bootsma, H.J.; Willems, R.J.; Urwin, R.; Maiden, M.C. Multilocus sequence typing system for Campylobacter jejuni. J. Clin. Microbiol. 2001, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Cody, A.J.; McCarthy, N.D.; Bray, J.E.; Wimalarathna, H.M.; Colles, F.M.; Jansen van Rensburg, M.J.; Dingle, K.E.; Waldenstrom, J.; Maiden, M.C. Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ. Microbiol. Rep. 2015, 7, 782–788. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Dallas, J.F.; Strachan, N.J.; MacRae, M.; McCarthy, N.D.; Wilson, D.J.; Gormley, F.J.; Falush, D.; Ogden, I.D.; Maiden, M.C.; et al. Campylobacter genotyping to determine the source of human infection. Clin. Infect. Dis. 2009, 48, 1072–1078. [Google Scholar] [CrossRef]
- Golz, J.C.; Stingl, K. Natural competence and horizontal gene transfer in Campylobacter. Curr. Top. Microbiol. Immunol. 2021, 431, 265–292. [Google Scholar] [CrossRef]
- Golz, J.C.; Stingl, K. “Take It or Leave It”-Factors regulating competence development and DNA uptake in Campylobacter jejuni. Int. J. Mol. Sci. 2021, 22, 10169. [Google Scholar] [CrossRef]
- Shen, Z.; Wang, Y.; Zhang, Q.; Shen, J. Antimicrobial Resistance in Campylobacter spp. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Taylor, D.E. Genetics of Campylobacter and Helicobacter. Annu. Rev. Microbiol. 1992, 46, 35–64. [Google Scholar] [CrossRef]
- Wang, Y.; Taylor, D.E. Natural transformation in Campylobacter species. J. Bacteriol. 1990, 172, 949–955. [Google Scholar] [CrossRef]
- Gaasbeek, E.J.; Wagenaar, J.A.; Guilhabert, M.R.; Wosten, M.M.; van Putten, J.P.; van der Graaf-van Bloois, L.; Parker, C.T.; van der Wal, F.J. A DNase encoded by integrated element CJIE1 inhibits natural transformation of Campylobacter jejuni. J. Bacteriol. 2009, 191, 2296–2306. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.L.; Bell, J.A.; Young, V.B.; Wilder, S.R.; Mansfield, L.S.; Linz, J.E. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 2003, 149, 3603–3615. [Google Scholar] [CrossRef] [PubMed]
- Gaasbeek, E.J.; Wagenaar, J.A.; Guilhabert, M.R.; van Putten, J.P.; Parker, C.T.; van der Wal, F.J. Nucleases encoded by the integrated elements CJIE2 and CJIE4 inhibit natural transformation of Campylobacter jejuni. J. Bacteriol. 2010, 192, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Carver, D.K.; Kathariou, S. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 2006, 72, 1316–1321. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, R.S.; Hendrixson, D.R.; DiRita, V.J. Natural transformation of Campylobacter jejuni requires components of a type II secretion system. J. Bacteriol. 2003, 185, 5408–5418. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, J.M.; Erfurt, R.S.; DiRita, V.J. Characterization and localization of the Campylobacter jejuni transformation system proteins CtsE, CtsP, and CtsX. J. Bacteriol. 2015, 197, 636–645. [Google Scholar] [CrossRef]
- Beauchamp, J.M.; Leveque, R.M.; Dawid, S.; DiRita, V.J. Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni. Proc. Natl. Acad. Sci. USA 2017, 114, E8053–E8061. [Google Scholar] [CrossRef]
- Hanafy, Z.; Osborne, J.A.; Miller, W.G.; Parker, C.T.; Olson, J.W.; Jackson, J.H., 3rd; Kathariou, S. Differences in the propensity of different antimicrobial resistance determinants to be disseminated via transformation in Campylobacter jejuni and Campylobacter coli. Microorganisms 2022, 10, 1194. [Google Scholar] [CrossRef]
- Miller, W.G.; Huynh, S.; Parker, C.T.; Niedermeyer, J.A.; Kathariou, S. Complete genome sequences of multidrug-resistant Campylobacter jejuni strain 14980A (turkey feces) and Campylobacter coli Strain 14983A (housefly from a turkey farm), harboring a novel gentamicin resistance mobile element. Genome Announc. 2016, 4, e01175-16. [Google Scholar] [CrossRef]
- Miller, W.G.; On, S.L.; Wang, G.; Fontanoz, S.; Lastovica, A.J.; Mandrell, R.E. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 2005, 43, 2315–2329. [Google Scholar] [CrossRef]
- Fouts, D.E.; Mongodin, E.F.; Mandrell, R.E.; Miller, W.G.; Rasko, D.A.; Ravel, J.; Brinkac, L.M.; DeBoy, R.T.; Parker, C.T.; Daugherty, S.C.; et al. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005, 3, e15. [Google Scholar] [CrossRef]
- Wallace, R.L.; Cribb, D.M.; Bulach, D.M.; Ingle, D.J.; Joensen, K.G.; Nielsen, E.M.; Leekitcharoenphon, P.; Stingl, K.; Kirk, M.D. Campylobacter jejuni ST50, a pathogen of global importance: A comparative genomic analysis of isolates from Australia, Europe and North America. Zoonoses Public Health 2021, 68, 638–649. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Suerbaum, S.; Lohrengel, M.; Sonnevend, A.; Ruberg, F.; Kist, M. Allelic diversity and recombination in Campylobacter jejuni. J. Bacteriol. 2001, 183, 2553–2559. [Google Scholar] [CrossRef] [PubMed]
- de Boer, P.; Wagenaar, J.A.; Achterberg, R.P.; van Putten, J.P.; Schouls, L.M.; Duim, B. Generation of Campylobacter jejuni genetic diversity in vivo. Mol. Microbiol. 2002, 44, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Guernier-Cambert, V.; Trachsel, J.; Maki, J.; Qi, J.; Sylte, M.J.; Hanafy, Z.; Kathariou, S.; Looft, T. Natural horizontal gene transfer of antimicrobial resistance genes in Campylobacter spp. from turkeys and swine. Front. Microbiol. 2021, 12, 732969. [Google Scholar] [CrossRef]
| C. jejuni Strains | ST | Average Transform. Frequency | TF SD | Average Mutation Frequency | MF SD | Competence of Recipient | TF/MF Log Diff. | AMR 1 |
|---|---|---|---|---|---|---|---|---|
| FSIS11812945 | 10398 | 1.98 × 10−7 | 1.72 × 10−7 | 2.32 × 10−8 | 3.03 × 10−8 | No | 0–1 | - |
| FSIS11812081 | 2132 | 1.06 × 10−6 | 4.60 × 10−7 | 3.09 × 10−9 | 2.95 × 10−9 | Yes | ≥3 | - |
| FSIS11810577 | 353 | 6.14 × 10−3 | 5.03 × 10−3 | 1.19 × 10−8 | 1.10 × 10−8 | Yes | ≥3 | - |
| FSIS11706266 | 464 | 3.49 × 10−4 | 3.98 × 10−4 | 3.96 × 10−8 | 1.98 × 10−8 | Yes | ≥3 | - |
| FSIS11811270 | 50 | 1.92 × 10−7 | 4.10 × 10−9 | 1.02 × 10−7 | 1.03 × 10−7 | No | 0–1 | - |
| FSIS11812063 | 50 | 8.68 × 10−8 | 2.93 × 10−9 | 1.16 × 10−7 | 9.51 × 10−8 | No | 0–1 | - |
| FSIS12028218 | 50 | 1.51 × 10−7 | 8.81 × 10−8 | 2.68 × 10−8 | 2.98 × 10−8 | No | 0–1 | T |
| FSIS12028305 | 50 | 6.76 × 10−8 | 2.79 × 10−8 | 1.77 × 10−8 | 8.26 × 10−9 | No | 0–1 | T |
| FSIS12029464 | 50 | 1.09 × 10−6 | 1.32 × 10−6 | 1.23 × 10−7 | 2.56 × 10−8 | No | 0–1 | - |
| FSIS12029904 | 50 | 4.42 × 10−8 | 4.81 × 10−8 | 3.54 × 10−8 | 3.89 × 10−8 | No | 0–1 | - |
| FSIS12030287 | 50 | 1.53 × 10−7 | 1.16 × 10−7 | 7.59 × 10−8 | 6.56 × 10−8 | No | 0–1 | - |
| FSIS12030565 | 50 | 1.03 × 10−7 | 8.59 × 10−8 | 5.66 × 10−8 | 2.71 × 10−8 | No | 0–1 | - |
| FSIS12030692 | 50 | 3.51 × 10−7 | 3.64 × 10−7 | 4.65 × 10−7 | 4.48 × 10−7 | No | 0–1 | TK |
| FSIS12030816 | 50 | 3.58 × 10−8 | 2.94 × 10−8 | 4.47 × 10−8 | 4.99 × 10−8 | No | 0–1 | - |
| FSIS12031002 | 50 | 5.12 × 10−8 | 1.25 × 10−8 | 2.39 × 10−8 | 3.97 × 10−9 | No | 0–1 | - |
| FSIS12031145 | 50 | 7.25 × 10−8 | 5.77 × 10−8 | 5.77 × 10−7 | 7.84 × 10−7 | No | 0–1 | - |
| FSIS22028286 | 50 | 4.50 × 10−7 | 4.75 × 10−7 | 4.36 × 10−8 | 3.80 × 10−8 | No | 0–1 | - |
| FSIS22028636 | 50 | 7.60 × 10−8 | 2.25 × 10−8 | 1.91 × 10−7 | 2.08 × 10−7 | No | 0–1 | - |
| FSIS1607853 | 50 | 8.74 × 10−8 | 7.30 × 10−8 | 1.90 × 10−8 | 1.18 × 10−8 | No | 0–1 | T |
| FSIS1608758 | 50 | 4.49 × 10−8 | 2.72 × 10−8 | 2.17 × 10−7 | 2.73 × 10−7 | No | 0–1 | - |
| FSIS1609357 | 50 | 4.14 × 10−8 | 9.19 × 10−10 | 1.40 × 10−8 | 1.38 × 10−8 | No | 0–1 | - |
| FSIS1609374 | 50 | 6.58 × 10−8 | 4.50 × 10−8 | 1.10 × 10−8 | 4.52 × 10−9 | No | 0–1 | - |
| FSIS1709833 | 50 | 2.33 × 10−7 | 2.61 × 10−7 | 8.34 × 10−9 | 1.18 × 10−9 | No | ~2 | T |
| FSIS1710700 | 50 | 4.10 × 10−8 | 2.52 × 10−8 | 1.26 × 10−8 | 4.30 × 10−9 | No | 0–1 | - |
| FSIS1710996 | 50 | 2.45 × 10−8 | 1.10 × 10−8 | 2.45 × 10−9 | 1.10 × 10−9 | No | 0–1 | QT |
| FSIS1701236 | 50 | 6.57 × 10−7 | 1.44 × 10−7 | 6.43 × 10−8 | 7.03 × 10−8 | No | 0–1 | T |
| FSIS1702913 | 50 | 3.14 × 10−7 | 1.02 × 10−7 | 2.09 × 10−7 | 2.74 × 10−7 | No | 0–1 | - |
| FSIS1703025 | 50 | 9.48 × 10−8 | 1.09 × 10−7 | 5.89 × 10−8 | 4.72 × 10−8 | No | 0–1 | QT |
| FSIS11705500 | 50 | 5.22 × 10−8 | 5.23 × 10−8 | 4.35 × 10−10 | 0 | No | ~2 | QT |
| FSIS21720655 | 50 | 5.26 × 10−8 | 2.77 × 10−8 | 7.66 × 10−9 | 7.69 × 10−9 | No | 0–1 | QT |
| FSIS21720686 | 50 | 1.40 × 10−6 | 1.69 × 10−6 | 3.25 × 10−8 | 2.38 × 10−8 | No | ~2 | - |
| FSIS21820901 | 50 | 3.20 × 10−6 | 4.32 × 10−6 | 8.85 × 10−8 | 1.11 × 10−7 | No | ~2 | - |
| FSIS1606748 | 50 | 3.80 × 10−8 | 7.57 × 10−9 | 4.31 × 10−8 | 5.69 × 10−8 | No | 0–1 | TK |
| FSIS1607146 | 50 | 2.30 × 10−8 | 1.77 × 10−8 | 8.87 × 10−9 | 7.54 × 10−9 | No | 0–1 | T |
| FSIS1701497 | 50 | 2.70 × 10−8 | 3.25 × 10−8 | 2.51 × 10−9 | 2.27 × 10−9 | No | 0–1 | QT |
| FSIS11812592 | 50 | 5.87 × 10−7 | 4.89 × 10−7 | 1.59 × 10−7 | 5.66 × 10−9 | No | 0–1 | T |
| FSIS11917669 | 50 | 5.16 × 10−8 | 2.69 × 10−8 | 1.16 × 10−8 | 1.24 × 10−8 | No | 0–1 | - |
| FSIS11918239 | 50 | 7.66 × 10−8 | 4.30 × 10−8 | 5.71 × 10−8 | 5.17 × 10−8 | No | 0–1 | - |
| FSIS12028219 | 50 | 2.80 × 10−8 | 2.31 × 10−8 | 6.21 × 10−10 | 4.68 × 10−10 | No | ~2 | - |
| FSIS12032984 | 50 | 1.20 × 10−7 | 1.52 × 10−7 | 7.50 × 10−9 | 3.13 × 10−9 | No | ~2 | T |
| FSIS12033376 | 50 | 4.63 × 10−8 | 2.30 × 10−8 | 5.34 × 10−8 | 6.03 × 10−8 | No | 0–1 | - |
| FSIS12138180 | 50 | 5.01 × 10−9 | 2.04 × 10−9 | 7.88 × 10−10 | 3.71 × 10−10 | No | 0–1 | T |
| FSIS11814023 | 939 | 1.40 × 10−7 | 1.50 × 10−7 | 2.19 × 10−8 | 2.60 × 10−8 | No | 0–1 | - |
| FSIS12028216 | 939 | 9.62 × 10−9 | 1.07 × 10−8 | 2.85 × 10−9 | 1.20 × 10−9 | No | 0–1 | TK |
| FSIS12028439 | 939 | 4.80 × 10−4 | 3.96 × 10−4 | 1.41 × 10−8 | 1.88 × 10−8 | Yes | ≥3 | T |
| FSIS12030679 | 939 | 2.24 × 10−4 | 1.08 × 10−4 | 3.23 × 10−9 | 3.51 × 10−9 | Yes | ≥3 | - |
| FSIS12031025 | 939 | 3.65 × 10−5 | 2.99 × 10−5 | 6.16 × 10−8 | 8.11 × 10−8 | Yes | ≥3 | K |
| FSIS12031178 | 939 | 3.98 × 10−4 | 2.86 × 10−4 | 4.50 × 10−9 | 1.08 × 10−9 | Yes | ≥3 | T |
| FSIS12031661 | 939 | 2.81 × 10−4 | 3.68 × 10−4 | 3.35 × 10−7 | 4.69 × 10−7 | Yes | ≥3 | - |
| FSIS12031779 | 939 | 1.40 × 10−7 | 1.25 × 10−7 | 4.77 × 10−9 | 3.54 × 10−9 | No | ~2 | - |
| FSIS22027247 | 939 | 1.38 × 10−8 | 1.07 × 10−8 | 1.40 × 10−8 | 1.39 × 10−8 | No | 0–1 | - |
| FSIS22027921 | 939 | 3.72 × 10−8 | 3.05 × 10−8 | 4.64 × 10−9 | 5.15 × 10−9 | No | 0–1 | - |
| FSIS22028453 | 939 | 3.71 × 10−7 | 4.85 × 10−7 | 1.33 × 10−7 | 1.83 × 10−7 | No | 0–1 | - |
| FSIS32003146 | 939 | 4.02 × 10−4 | 4.00 × 10−4 | 8.17 × 10−10 | 2.33 × 10−11 | Yes | ≥3 | K |
| RM 3405 | 50 | 1.33 × 10−8 | 1.29 × 10−8 | 1.45 × 10−8 | 1.58 × 10−8 | No | 0–1 | - |
| RM 3412 | 50 | 2.79 × 10−5 | 3.60 × 10−5 | 1.04 × 10−6 | 7.57 × 10−7 | No | 0–1 | T |
| RM 5146 | 50 | 3.80 × 10−8 | 3.61 × 10−8 | 1.47 × 10−9 | 1.46 × 10−9 | No | 0–1 | T |
| RM 5148 | 50 | 8.25 × 10−8 | 8.84 × 10−8 | 1.09 × 10−8 | 7.78 × 10−10 | No | 0–1 | T |
| RM 5149 | 50 | 5.91 × 10−9 | 1.08 × 10−9 | 3.06 × 10−9 | 3.74 × 10−9 | No | 0–1 | TQ |
| RM 5156 | 50 | 1.52 × 10−4 | 9.86 × 10−5 | 6.13 × 10−8 | 6.75 × 10−8 | Yes | ≥3 | T |
| C. coli Strains | ST | Average Transform. Frequency | TF SD | Average Mutation Frequency | MF SD | Competence of recipient | TF/MF Log Diff. | AMR |
| FSIS11813367 | 1050 | 5.76 × 10−3 | 3.69 × 10−3 | 3.48 × 10−8 | 3.75 × 10−8 | Yes | ≥3 | - |
| FSIS1710488 | 7818 | 8.32 × 10−4 | 4.16 × 10−5 | 8.03 × 10−9 | 3.50 × 10−9 | Yes | ≥3 | - |
| FSIS1710329 | 7818 | 1.87 × 10−4 | 2.36 × 10−4 | 1.52 × 10−8 | 1.94 × 10−8 | Yes | ≥3 | - |
| FSIS11811291 | 829 | 8.29 × 10−4 | 4.08 × 10−4 | 1.13 × 10−8 | 1.10 × 10−8 | Yes | ≥3 | E |
| FSIS11813365 | 829 | 4.62 × 10−4 | 2.44 × 10−4 | 2.10 × 10−8 | 1.87 × 10−8 | Yes | ≥3 | E |
| FSIS1607221 | 829 | 4.64 × 10−7 | 2.24 × 10−8 | 3.16 × 10−7 | 4.30 × 10−7 | No | 0–1 | - |
| FSIS21822106 | 829 | 1.25 × 10−6 | 1.49 × 10−6 | 2.31 × 10−7 | 2.42 × 10−7 | No | 0–1 | - |
| FSIS11813852 | 902 | 6.64 × 10−4 | 3.31 × 10−4 | 1.42 × 10−9 | 1.00 × 10−9 | Yes | ≥3 | Q |
| FSIS1710767 | 3262 | 1.89 × 10−8 | 1.05 × 10−8 | 8.17 × 10−10 | 2.33 × 10−11 | No | ~2 | Q |
| FSIS12027778 | 3262 | 1.73 × 10−5 | 1.05 × 10−5 | 2.38 × 10−9 | 1.99 × 10−10 | Yes | ≥3 | Q |
| FSIS12030275 | 3262 | 1.92 × 10−8 | 9.89 × 10−9 | 1.13 × 10−9 | 1.04 × 10−9 | No | 0–1 | Q |
| FSIS12031023 | 3262 | 1.54 × 10−3 | 1.84 × 10−3 | 6.96 × 10−9 | 7.83 × 10−9 | Yes | ≥3 | Q |
| FSIS12031175 | 3262 | 3.02 × 10−7 | 3.41 × 10−7 | 8.95 × 10−7 | 7.15 × 10−7 | No | 0–1 | - |
| FSIS12031458 | 3262 | 1.59 × 10−8 | 1.68 × 10−8 | 2.44 × 10−9 | 6.90 × 10−13 | No | 0–1 | Q |
| FSIS12031835 | 3262 | 1.11 × 10−7 | 8.88 × 10−8 | 1.25 × 10−8 | 3.13 × 10−9 | No | 0–1 | - |
| FSIS12031855 | 3262 | 3.07 × 10−8 | 2.73 × 10−8 | 4.51 × 10−9 | 3.39 × 10−9 | No | 0–1 | Q |
| FSIS22027509 | 3262 | 7.38 × 10−4 | 4.58 × 10−4 | 1.34 × 10−8 | 6.14 × 10−9 | Yes | ≥3 | Q |
| FSIS22028223 | 3262 | 1.82 × 10−7 | 2.17 × 10−7 | 1.31 × 10−8 | 1.02 × 10−9 | No | 0–1 | Q |
| FSIS22028629 | 3262 | 5.18 × 10−8 | 6.10 × 10−8 | 2.36 × 10−8 | 3.26 × 10−8 | No | 0–1 | Q |
| C. jejuni Strains | ST | cts Mutations 1 | dns 2 | dns2 | dns3 | Competence |
|---|---|---|---|---|---|---|
| FSIS11812945 | 10398 | D | N | N | N | No |
| FSIS11812081 | 2132 | none | N | N | Y | Yes |
| FSIS11810577 | 353 | none | Y (FS) | N | N | Yes |
| FSIS11706266 | 464 | none | N | N | N | Yes |
| FSIS11811270 | 50 | none | Y | N | N | No |
| FSIS11812063 | 50 | N.D. | N | Y | N | No |
| FSIS12028218 | 50 | DE | Y | N | N | No |
| FSIS12028305 | 50 | DE | Y | N | N | No |
| FSIS12029464 | 50 | none | Y | N | N | No |
| FSIS12029904 | 50 | none | Y | N | Y | No |
| FSIS12030287 | 50 | DE | Y (FS) | N | N | No |
| FSIS12030565 | 50 | DE | Y (FS) | N | N | No |
| FSIS12030692 | 50 | DE | Y | N | N | No |
| FSIS12030816 | 50 | none | N | Y | N | No |
| FSIS12031002 | 50 | none | N | Y | Y | No |
| FSIS12031145 | 50 | DE | Y (FS) | N | N | No |
| FSIS22028286 | 50 | none | Y | N | N | No |
| FSIS22028636 | 50 | none | N | Y | N | No |
| FSIS1607853 | 50 | none | Y | N | N | No |
| FSIS1608758 | 50 | DE | Y (FS) | N | N | No |
| FSIS1609357 | 50 | DE | Y (FS) | N | N | No |
| FSIS1609374 | 50 | none | N | Y | N | No |
| FSIS1709833 | 50 | DE | N | N | N | No |
| FSIS1710700 | 50 | DE | Y | N | N | No |
| FSIS1710996 | 50 | none | Y | N | N | No |
| FSIS1701236 | 50 | none | Y | N | N | No |
| FSIS1702913 | 50 | DE | N | N | N | No |
| FSIS1703025 | 50 | none | Y | N | N | No |
| FSIS11705500 | 50 | D | N | N | N | No |
| FSIS21720655 | 50 | D | N | N | N | No |
| FSIS21720686 | 50 | none | Y | N | N | No |
| FSIS21820901 | 50 | none | Y | N | N | No |
| FSIS1606748 | 50 | DE | Y | N | N | No |
| FSIS1607146 | 50 | none | Y | N | Y | No |
| FSIS1701497 | 50 | none | Y | N | N | No |
| FSIS11812592 | 50 | DE | Y | N | N | No |
| FSIS11917669 | 50 | DE | N | N | N | No |
| FSIS11918239 | 50 | none | Y | N | N | No |
| FSIS12028219 | 50 | none | N | N | Y | No |
| FSIS12032984 | 50 | N.D. | Y | N | N | No |
| FSIS12033376 | 50 | none | N | Y | N | No |
| FSIS12138180 | 50 | DE | Y | N | N | No |
| FSIS11814023 | 939 | none | N | N | Y | No |
| FSIS12028216 | 939 | none | N | N | Y | No |
| FSIS12028439 | 939 | none | N | N | N | Yes |
| FSIS12030679 | 939 | none | N | N | N | Yes |
| FSIS12031025 | 939 | none | N | N | N | Yes |
| FSIS12031178 | 939 | none | N | N | N | Yes |
| FSIS12031661 | 939 | none | N | N | N | Yes |
| FSIS12031779 | 939 | none | N | N | Y | No |
| FSIS22027247 | 939 | none | N | N | Y | No |
| FSIS22027921 | 939 | none | N | N | Y | No |
| FSIS22028453 | 939 | none | N | N | Y | No |
| FSIS32003146 | 939 | none | N | N | N | Yes |
| RM3405 | 50 | RF | N | N | N | No |
| RM3412 | 50 | none | N | N | N | No |
| RM5146 | 50 | none | Y | N | N | No |
| RM5148 | 50 | none | Y | N | Y | No |
| RM5149 | 50 | none | Y | N | Y | No |
| RM5156 | 50 | none | Y (FS) | N | N | Yes |
| C. coli Strains | ST | cts Mutations 1 | dns | dns2 3 | dns3 | Competence |
| FSIS11813367 | 1050 | none | Y | N | N | Yes |
| FSIS1710488 | 7818 | none | N | N | N | Yes |
| FSIS1710329 | 7818 | none | N | N | N | Yes |
| FSIS11811291 | 829 | none | N | N | N | Yes |
| FSIS11813365 | 829 | none | N | N | N | Yes |
| FSIS1607221 | 829 | none | N | N | N | No |
| FSIS21822106 | 829 | none | Y | N | N | No |
| FSIS11813852 | 902 | none | N | N | N | Yes |
| FSIS1710767 | 3262 | none | N | Y | N | No |
| FSIS12027778 | 3262 | none | N | Y (*) | N | Yes |
| FSIS12030275 | 3262 | none | N | Y | N | No |
| FSIS12031023 | 3262 | none | N | Y (*) | N | Yes |
| FSIS12031175 | 3262 | none | N | Y | N | No |
| FSIS12031458 | 3262 | none | N | Y | N | No |
| FSIS12031835 | 3262 | none | N | Y | N | No |
| FSIS12031855 | 3262 | none | N | Y | N | No |
| FSIS22027509 | 3262 | none | N | Y (*) | N | Yes |
| FSIS22028223 | 3262 | none | N | Y | N | No |
| FSIS22028629 | 3262 | none | N | Y | N | No |
| Region | cts Mutations | Total Strains | % cts Mutations | Mu-dns+ | Total Strains | % Mu-dns+ | dns2 or dns3 | Total Strains | % dns2 or dns3 |
|---|---|---|---|---|---|---|---|---|---|
| Europe | 37 | 1124 | 3.3 | 933 | 1450 | 64.3 | 412 | 1450 | 28.4 |
| N. America | 214 | 566 | 37.8 | 416 | 780 | 53.3 | 237 | 780 | 30.3 |
| Australia | 8 | 23 | 34.7 | 5 | 23 | 21.7 | 12 | 23 | 52.2 |
| ST-8 | 49 | 641 | 7.6 | 62 | 809 | 7.7 | 218 | 809 | 26.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parker, C.T.; Villafuerte, D.A.; Miller, W.G.; Huynh, S.; Chapman, M.H.; Hanafy, Z.; Jackson, J.H., III; Miller, M.A.; Kathariou, S. Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50. Microorganisms 2024, 12, 327. https://doi.org/10.3390/microorganisms12020327
Parker CT, Villafuerte DA, Miller WG, Huynh S, Chapman MH, Hanafy Z, Jackson JH III, Miller MA, Kathariou S. Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50. Microorganisms. 2024; 12(2):327. https://doi.org/10.3390/microorganisms12020327
Chicago/Turabian StyleParker, Craig T., David A. Villafuerte, William G. Miller, Steven Huynh, Mary H. Chapman, Zahra Hanafy, James H. Jackson, III, Morgan A. Miller, and Sophia Kathariou. 2024. "Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50" Microorganisms 12, no. 2: 327. https://doi.org/10.3390/microorganisms12020327
APA StyleParker, C. T., Villafuerte, D. A., Miller, W. G., Huynh, S., Chapman, M. H., Hanafy, Z., Jackson, J. H., III, Miller, M. A., & Kathariou, S. (2024). Genomic Analysis Points to Multiple Genetic Mechanisms for Non-Transformable Campylobacter jejuni ST-50. Microorganisms, 12(2), 327. https://doi.org/10.3390/microorganisms12020327







