Relationship between the Viral Load in Patients with Different COVID-19 Severities and SARS-CoV-2 Variants
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Identification of Positive Cases
- -
- Sequencing of the SARS-CoV-2 genome (CoViGen-Mex)
2.3. Quantification of SARS-CoV-2 Viral Load
- -
- Validation of the endogenous control
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int/ (accessed on 10 January 2022).
- Li, Q.; Wu, J.; Nie, J.; Zhang, L.; Hao, H.; Liu, S.; Zhao, C.; Zhang, Q.; Liu, H.; Nie, L.; et al. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell 2020, 182, 1284–1294.e9. [Google Scholar] [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Taboada, D.B.; Rosales, M. Programa de Vigilancia Genómica del SARS-CoV-2 Realizado por el CoViGen-Mex; UNAM: Mexico City, Mexico, 2022. [Google Scholar]
- Cherian, S.; Potdar, V.; Jadhav, S.; Yadav, P.; Gupta, N.; Das, M.; Rakshit, P.; Singh, S.; Abraham, P.; Panda, S.; et al. SARS-CoV-2 Spike Mutations, L452r, T478k, E484q and P681r, in the Second Wave of COVID-19 in Maharashtra, India. Microorganisms 2021, 9, 1542. [Google Scholar] [CrossRef]
- Riediker, M.; Briceno-Ayala, L.; Ichihara, G.; Albani, D.; Poffet, D.; Tsai, D.H.; Iff, S.; Monn, C. Higher Viral Load and Infectivity Increase Risk of Aerosol Transmission for Delta and Omicron Variants of SARS-CoV-2. Swiss Med. Wkly. 2022, 152, w30133. [Google Scholar] [CrossRef] [PubMed]
- Teyssou, E.; Delagrèverie, H.; Visseaux, B.; Lambert-Niclot, S.; Brichler, S.; Ferre, V.; Marot, S.; Jary, A.; Todesco, E.; Schnuriger, A.; et al. The Delta SARS-CoV-2 Variant Has a Higher Viral Load than the Beta and the Historical Variants in Nasopharyngeal Samples from Newly Diagnosed COVID-19 Patients. J. Infect. 2021, 83, e1–e3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, R.; Hu, F.; Lan, Y.; Yang, Z.; Zhan, C.; Shi, J.; Deng, X.; Jiang, M.; Zhong, S.; et al. Transmission, Viral Kinetics and Clinical Characteristics of the Emergent SARS-CoV-2 Delta VOC in Guangzhou, China. EClinicalMedicine 2021, 40, 101129. [Google Scholar] [CrossRef]
- Li, B.; Deng, A.; Li, K.; Hu, Y.; Li, Z.; Shi, Y.; Xiong, Q.; Liu, Z.; Guo, Q.; Zou, L.; et al. Viral Infection and Transmission in a Large, Well-Traced Outbreak Caused by the SARS-CoV-2 Delta Variant. Nat. Commun. 2022, 13, 460. [Google Scholar] [CrossRef]
- Lee, M.J. Quantifying SARS-CoV-2 Viral Load: Current Status and Future Prospects. Expert Rev. Mol. Diagn. 2021, 21, 1017–1023. [Google Scholar] [CrossRef] [PubMed]
- van Kampen, J.J.A.; van de Vijver, D.A.M.C.; Fraaij, P.L.A.; Haagmans, B.L.; Lamers, M.M.; Okba, N.; van den Akker, J.P.C.; Endeman, H.; Gommers, D.A.M.P.J.; Cornelissen, J.J.; et al. Duration and Key Determinants of Infectious Virus Shedding in Hospitalized Patients with Coronavirus Disease-2019 (COVID-19). Nat. Commun. 2021, 12, 267. [Google Scholar] [CrossRef] [PubMed]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 Viral Load Is Associated with Increased Disease Severity and Mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Sang, L.; Ye, F.; Ruan, S.; Zhong, B.; Song, T.; Alshukairi, A.N.; Chen, R.; Zhang, Z.; et al. Kinetics of Viral Load and Antibody Response in Relation to COVID-19 Severity. J. Clin. Investig. 2020, 130, 5235–5244. [Google Scholar] [CrossRef]
- Zacharioudakis, I.M.; Prasad, P.J.; Zervou, F.N.; Basu, A.; Inglima, K.; Weisenberg, S.A.; Aguero-Rosenfeld, M.E. Association of SARS-CoV-2 Genomic Load with Outcomes in Patients with COVID-19. Ann. Am. Thorac. Soc. 2021, 18, 900–903. [Google Scholar] [CrossRef]
- Dadras, O.; Afsahi, A.M.; Pashaei, Z.; Mojdeganlou, H.; Karimi, A.; Habibi, P.; Barzegary, A.; Fakhfouri, A.; Mirzapour, P.; Janfaza, N.; et al. The Relationship between COVID-19 Viral Load and Disease Severity: A Systematic Review. Immun. Inflamm. Dis. 2022, 10, e580. [Google Scholar] [CrossRef]
- Killingley, B.; Mann, A.J.; Kalinova, M.; Boyers, A.; Goonawardane, N.; Zhou, J.; Lindsell, K.; Hare, S.S.; Brown, J.; Frise, R.; et al. Safety, Tolerability and Viral Kinetics during SARS-CoV-2 Human Challenge in Young Adults. Nat. Med. 2022, 28, 1031–1041. [Google Scholar] [CrossRef] [PubMed]
- Wölfel, R.; Corman, V.M.; Guggemos, W.; Seilmaier, M.; Zange, S.; Müller, M.A.; Niemeyer, D.; Jones, T.C.; Vollmar, P.; Rothe, C.; et al. Virological Assessment of Hospitalized Patients with COVID-2019. Nature 2020, 581, 465–469. [Google Scholar] [CrossRef]
- Zou, L.; Ruan, F.; Huang, M.; Liang, L.; Huang, H.; Hong, Z.; Yu, J.; Kang, M.; Song, Y.; Xia, J.; et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N. Engl. J. Med. 2020, 382, 1177–1179. [Google Scholar] [CrossRef]
- MexCoV2. Consorcio Mexicano de Vigilancia Genómica (CoViGen-Mex). Available online: http://mexcov2.ibt.unam.mx:8080/COVID-TRACKER/somos (accessed on 16 February 2024).
- New England Biolabs. Nucleic Acid Data. Available online: https://www.neb.cn/zh-cn/tools-and-resources/usage-guidelines/nucleic-acid-data (accessed on 16 February 2024).
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV Viral Load Dynamics, Duration of Viral Shedding, and Infectiousness: A Systematic Review and Meta-Analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef] [PubMed]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 Viral Load and Shedding Kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef]
- Vetter, P.; Eberhardt, C.S.; Meyer, B.; Martinez Murillo, P.A.; Torriani, G.; Pigny, F.; Lemeille, S.; Cordey, S.; Laubscher, F.; Vu, D.-L.; et al. Daily Viral Kinetics and Innate and Adaptive Immune Response Assessment in COVID-19: A Case Series. mSphere 2020, 5, e00827-20. [Google Scholar] [CrossRef] [PubMed]
- Aranha, C.; Patel, V.; Bhor, V.; Gogoi, D. Cycle Threshold Values in RT-PCR to Determine Dynamics of SARS-CoV-2 Viral Load: An Approach to Reduce the Isolation Period for COVID-19 Patients. J. Med. Virol. 2021, 93, 6794–6797. [Google Scholar] [CrossRef] [PubMed]
- Yuasa, S.; Nakajima, J.; Takatsuki, Y.; Takahashi, Y.; Tani-Sassa, C.; Iwasaki, Y.; Nagano, K.; Sonobe, K.; Yoshimoto, T.; Nukui, Y.; et al. Viral Load of SARS-CoV-2 Omicron Is Not High despite Its High Infectivity. J. Med. Virol. 2022, 94, 5543–5546. [Google Scholar] [CrossRef] [PubMed]
- Do, T.V.; Manabe, T.; Vu, G.V.; Nong, V.M.; Fujikura, Y.; Phan, D.; Pham, T.T.; Do, C.D.; Doan, T.T.; Nguyen, N.T.; et al. Clinical Characteristics and Mortality Risk among Critically Ill Patients with COVID-19 Owing to the B.1.617.2 (Delta) Variant in Vietnam: A Retrospective Observational Study. PLoS ONE 2023, 18, e0279713. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Pourakbari, B.; Benvari, S.; Hosseinpour Sadeghi, R.; Abdolsalehi, M.R.; Shahbabaie, M.A.; Jalali, F.; Safari, F.; Navaeian, A.; Mamishi, S. Clinical and Laboratory Features of SARS-CoV-2 Variants across Multiple Rounds of Pandemic Waves in Hospitalized Children in an Iranian Referral Hospital. BMC Pediatr. 2023, 23, 241. [Google Scholar] [CrossRef]
- Strasser, Z.H.; Greifer, N.; Hadavand, A.; Murphy, S.N.; Estiri, H. Estimates of SARS-CoV-2 Omicron BA.2 Subvariant Severity in New England. JAMA Netw. Open 2022, 5, E2238354. [Google Scholar] [CrossRef]
- Park, C.; Tavakoli-Tabasi, S.; Sharafkhaneh, A.; Seligman, B.J.; Hicken, B.; Amos, C.I.; Chou, A.; Razjouyan, J. Inflammatory Biomarkers Differ among Hospitalized Veterans Infected with Alpha, Delta, and Omicron SARS-CoV-2 Variants. Int. J. Environ. Res. Public Health 2023, 20, 2987. [Google Scholar] [CrossRef]
- Angeles-Garay, U.; Hernández-Herrera, J.A.; Procopio-Velázquez, J.; Alvarado-Yaah, J.E.; Ramírez-Gonzalez, E.; González-Cruz, I.L. Etiología de Neumonías Bacterianas en Pacientes con COVID-19. Rev. Méd. Inst. Mex. Seguro Soc. 2022, 60, 632–639. [Google Scholar]
- Jacot, D.; Greub, G.; Jaton, K.; Opota, O. Viral Load of SARS-CoV-2 across Patients and Compared to Other Respiratory Viruses. Microbes Infect. 2020, 22, 617–621. [Google Scholar] [CrossRef]
- Jones, T.C.; Mühlemann, B.; Veith, T.; Biele, G.; Zuchowski, M.; Hofmann, J.; Stein, A.; Edelmann, A.; Max Corman, V.; Drosten, C.; et al. An Analysis of SARS-CoV-2 Viral Load by Patient Age. medRxiv 2020. [Google Scholar] [CrossRef]
- Kleiboeker, S.; Cowden, S.; Grantham, J.; Nutt, J.; Tyler, A.; Berg, A.; Altrich, M. SARS-CoV-2 Viral Load Assessment in Respiratory Samples. J. Clin. Virol. 2020, 129, 104439. [Google Scholar] [CrossRef]
- Tsukagoshi, H.; Shinoda, D.; Saito, M.; Okayama, K.; Sada, M.; Kimura, H.; Saruki, N. Relationships between Viral Load and the Clinical Course of COVID-19. Viruses 2021, 13, 304. [Google Scholar] [CrossRef]
- Owusu, M.; Sylverken, A.A.; Ankrah, S.T.; El-Duah, P.; Ayisi-Boateng, N.K.; Yeboah, R.; Gorman, R.; Asamoah, J.; Binger, T.; Acheampong, G.; et al. Epidemiological Profile of SARS-CoV-2 among Selected Regions in Ghana: A Cross-Sectional Retrospective Study. PLoS ONE 2020, 15, e0243711. [Google Scholar] [CrossRef] [PubMed]
- Euser, S.; Aronson, S.; Manders, I.; Van Lelyveld, S.; Herpers, B.; Sinnige, J.; Kalpoe, J.; Van Gemeren, C.; Snijders, D.; Jansen, R.; et al. SARS-CoV-2 Viral-Load Distribution Reveals That Viral Loads Increase with Age: A Retrospective Cross-Sectional Cohort Study. Int. J. Epidemiol. 2021, 50, 1795–1803. [Google Scholar] [CrossRef] [PubMed]
- Levine-Tiefenbrun, M.; Yelin, I.; Alapi, H.; Katz, R.; Herzel, E.; Kuint, J.; Chodick, G.; Gazit, S.; Patalon, T.; Kishony, R. Viral Loads of Delta-Variant SARS-CoV-2 Breakthrough Infections after Vaccination and Booster with BNT162b2. Nat. Med. 2021, 27, 2108–2110. [Google Scholar] [CrossRef] [PubMed]
- Singanayagam, A.; Hakki, S.; Dunning, J.; Madon, K.J.; Crone, M.A.; Koycheva, A.; Derqui-Fernandez, N.; Barnett, J.L.; Whitfield, M.G.; Varro, R.; et al. Community Transmission and Viral Load Kinetics of the SARS-CoV-2 Delta (B.1.617.2) Variant in Vaccinated and Unvaccinated Individuals in the UK: A Prospective, Longitudinal, Cohort Study. Lancet Infect. Dis. 2022, 22, 183–195. [Google Scholar] [CrossRef]
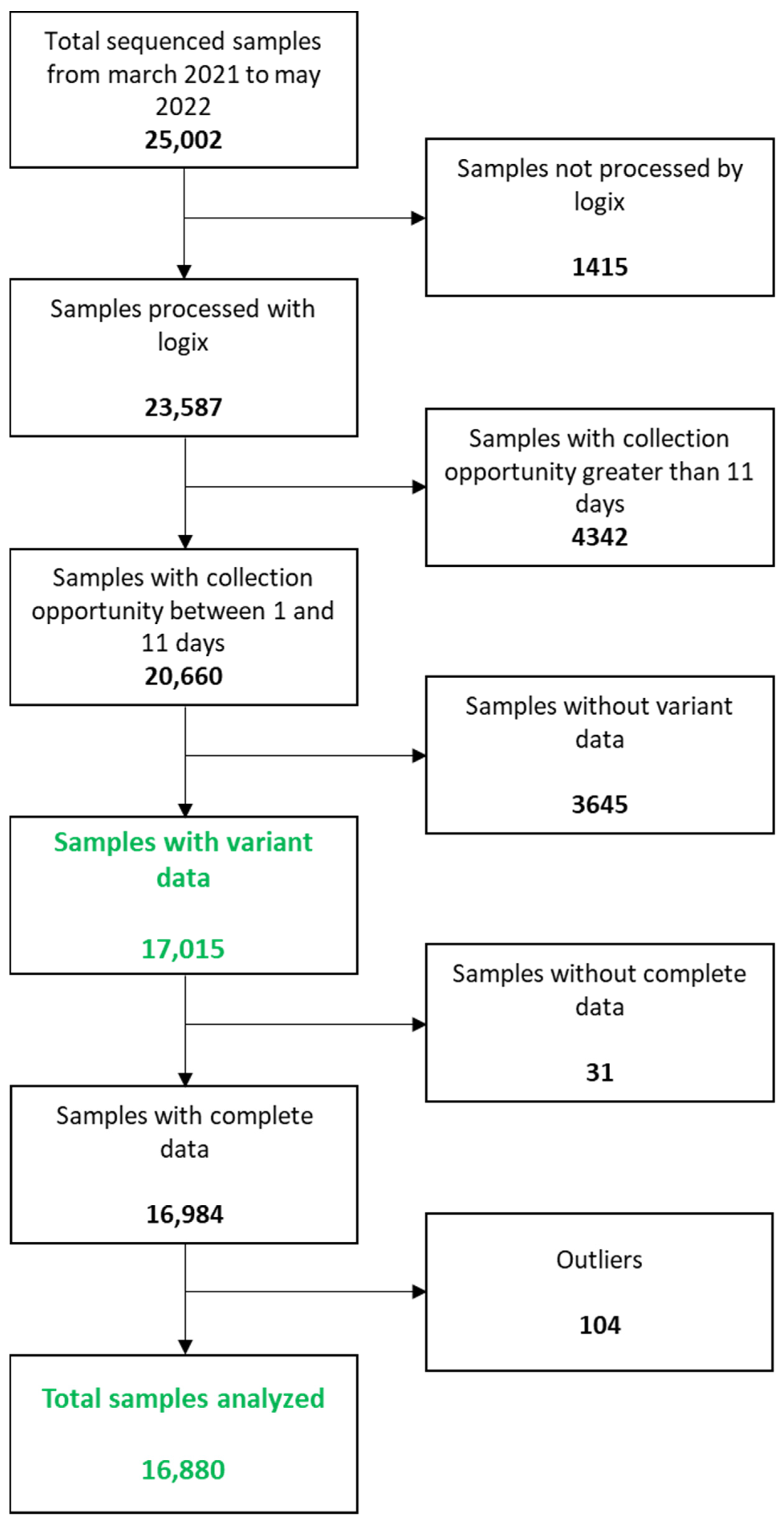
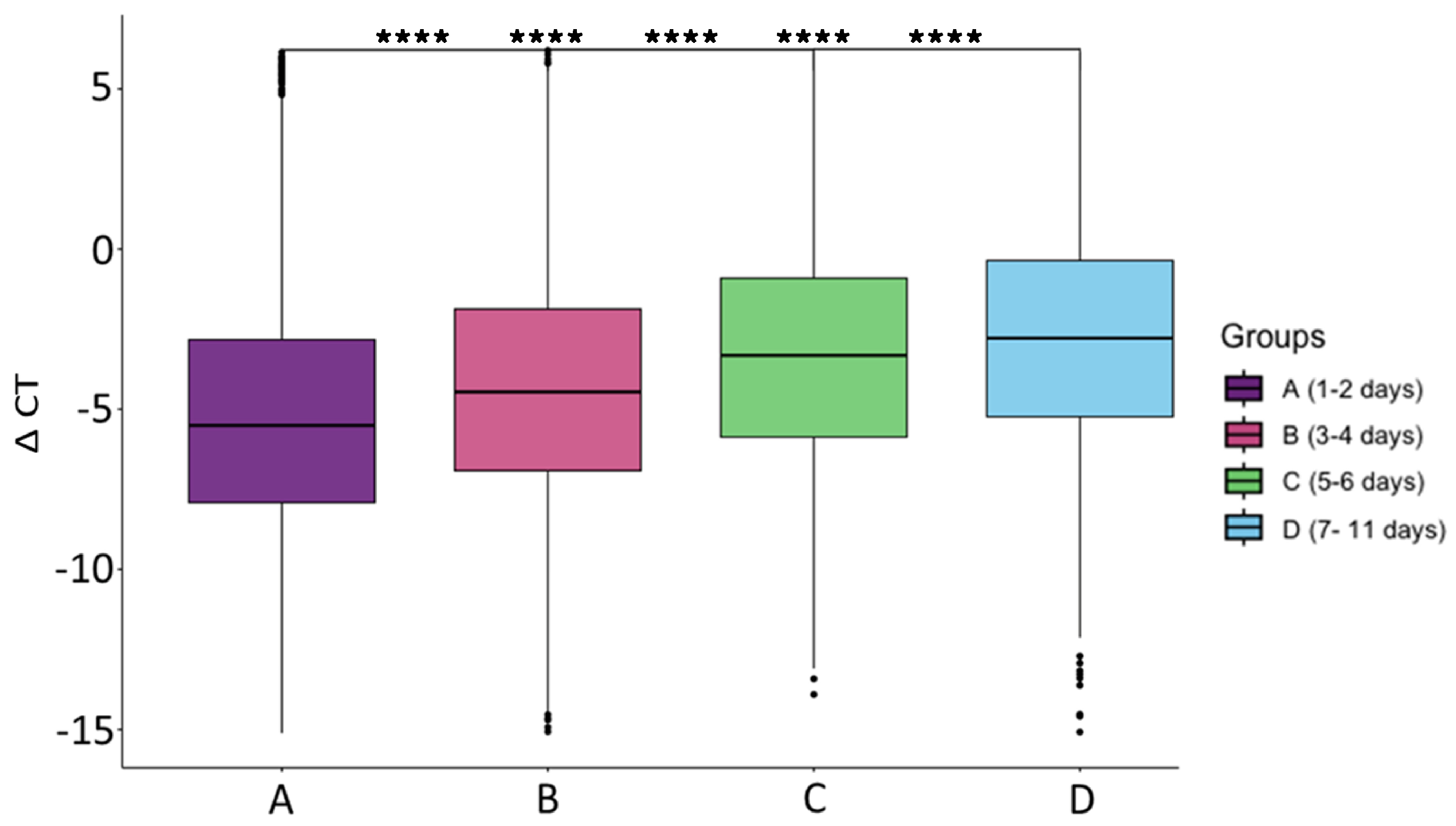
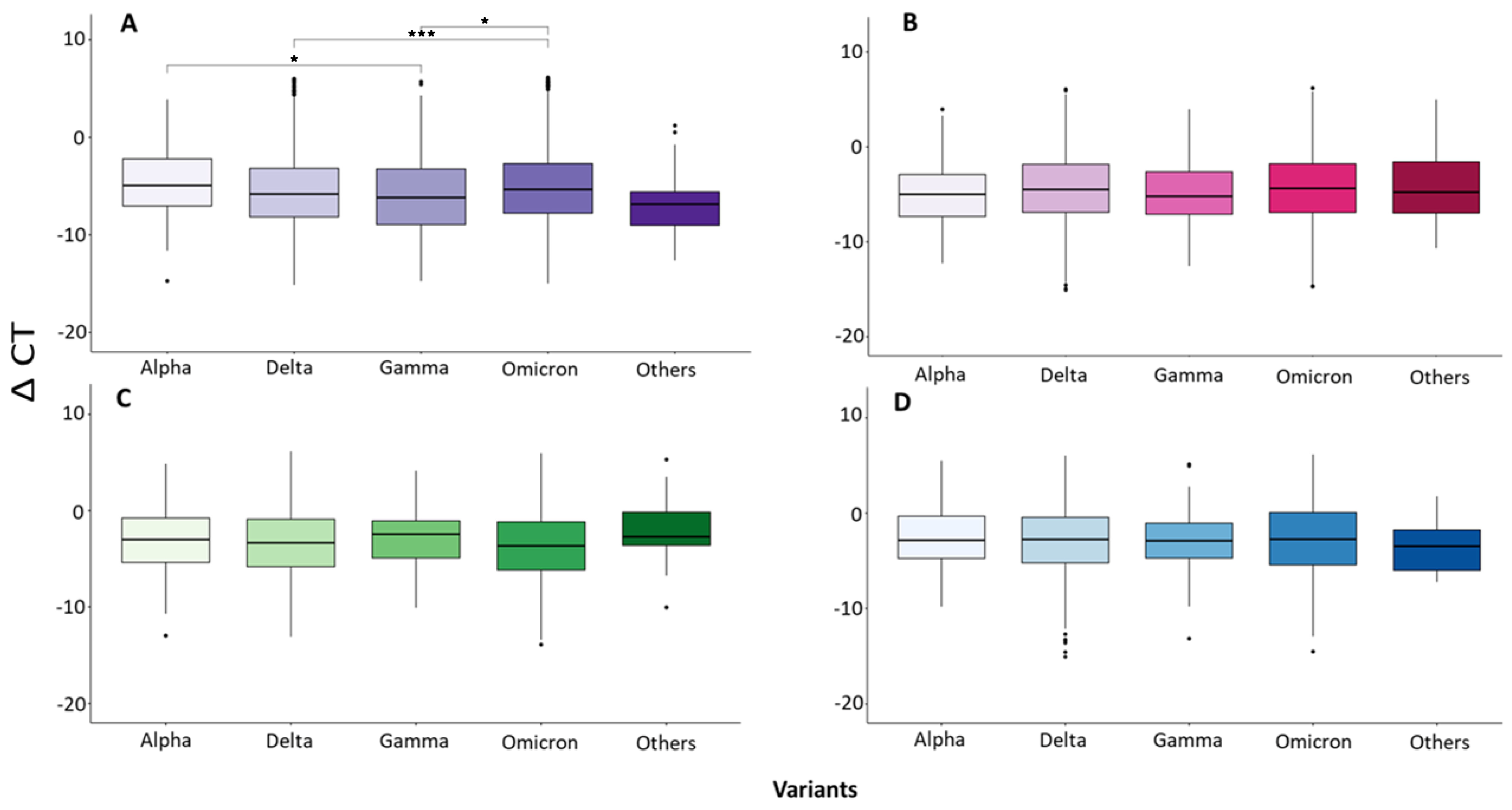
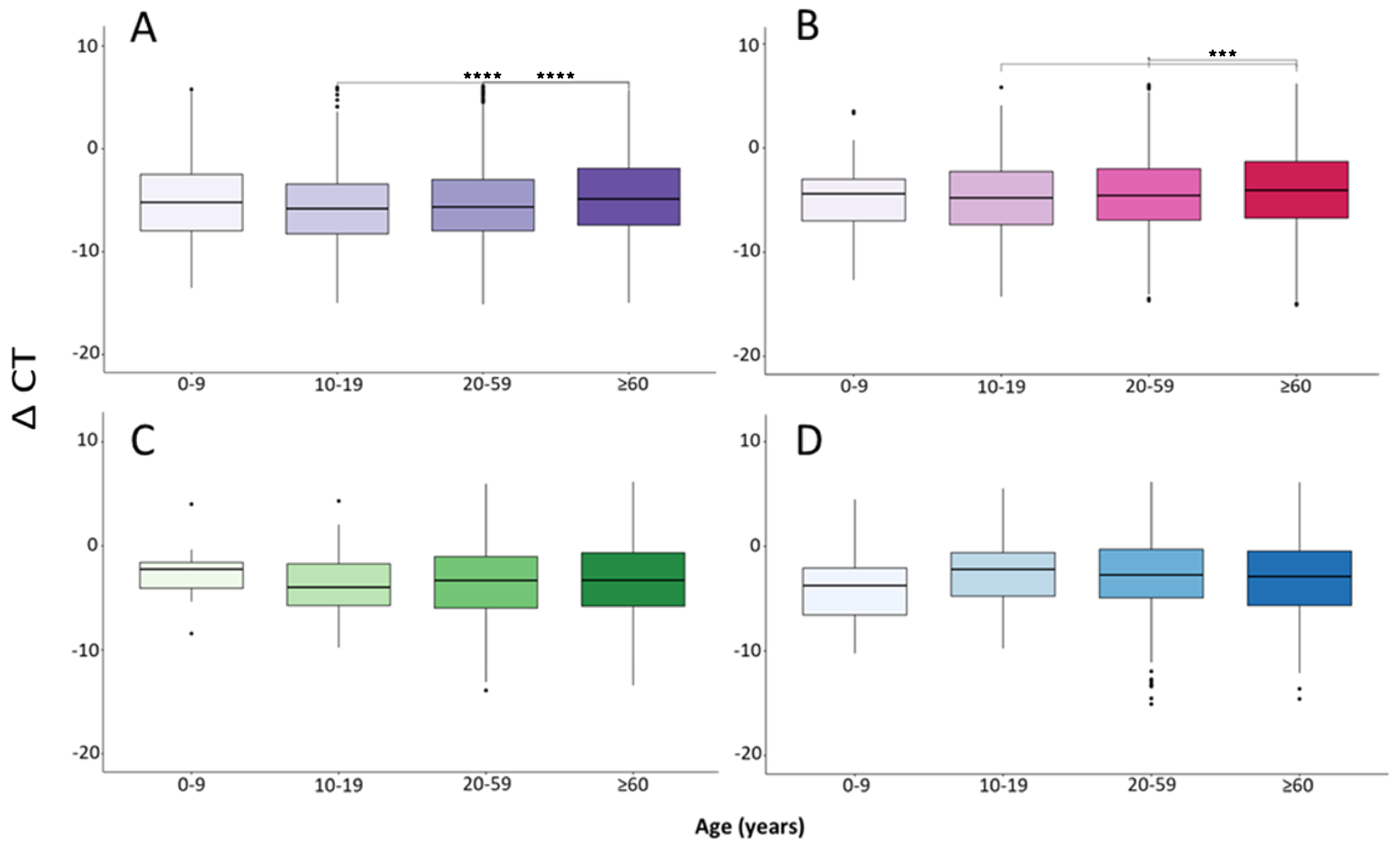
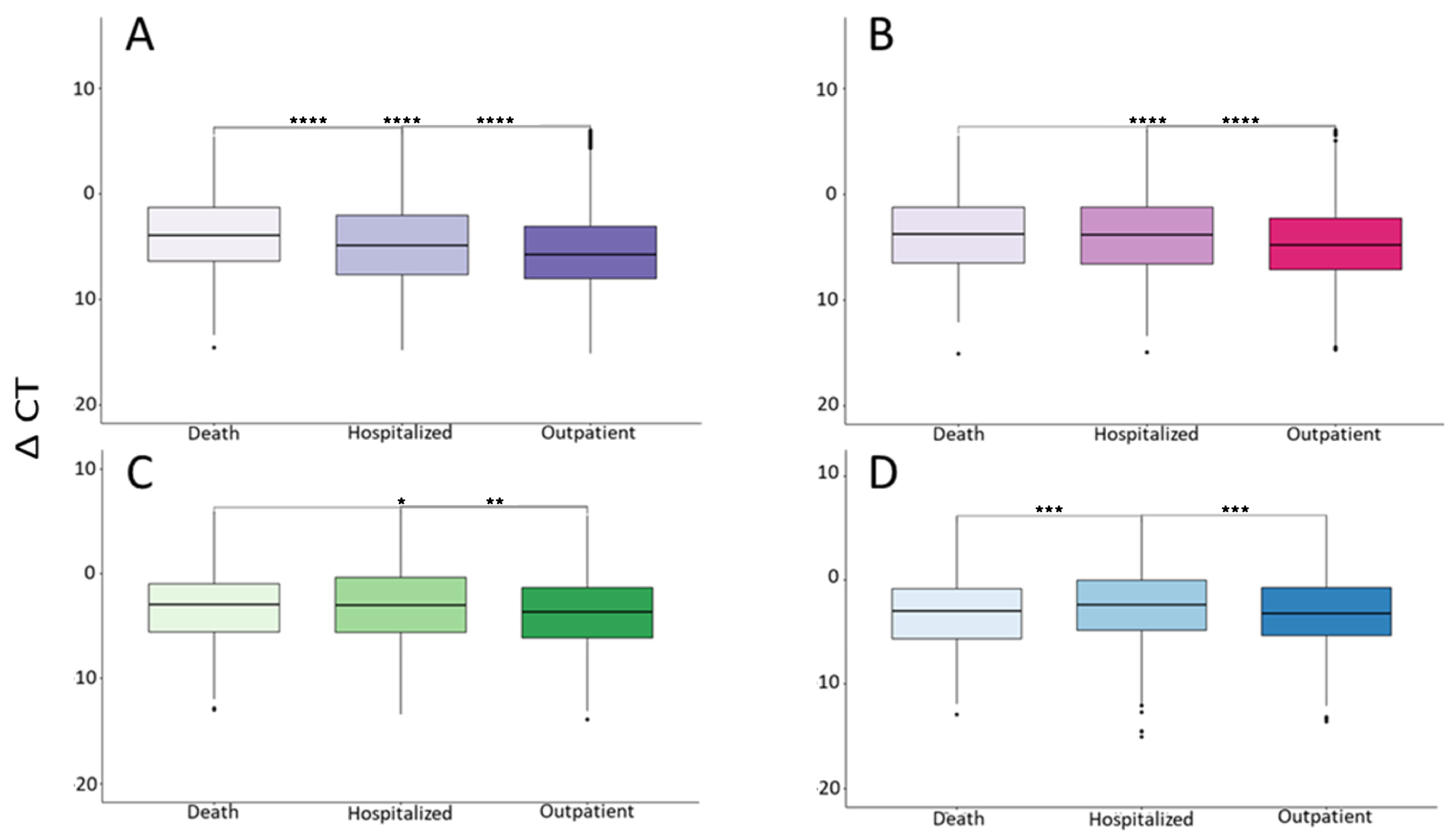
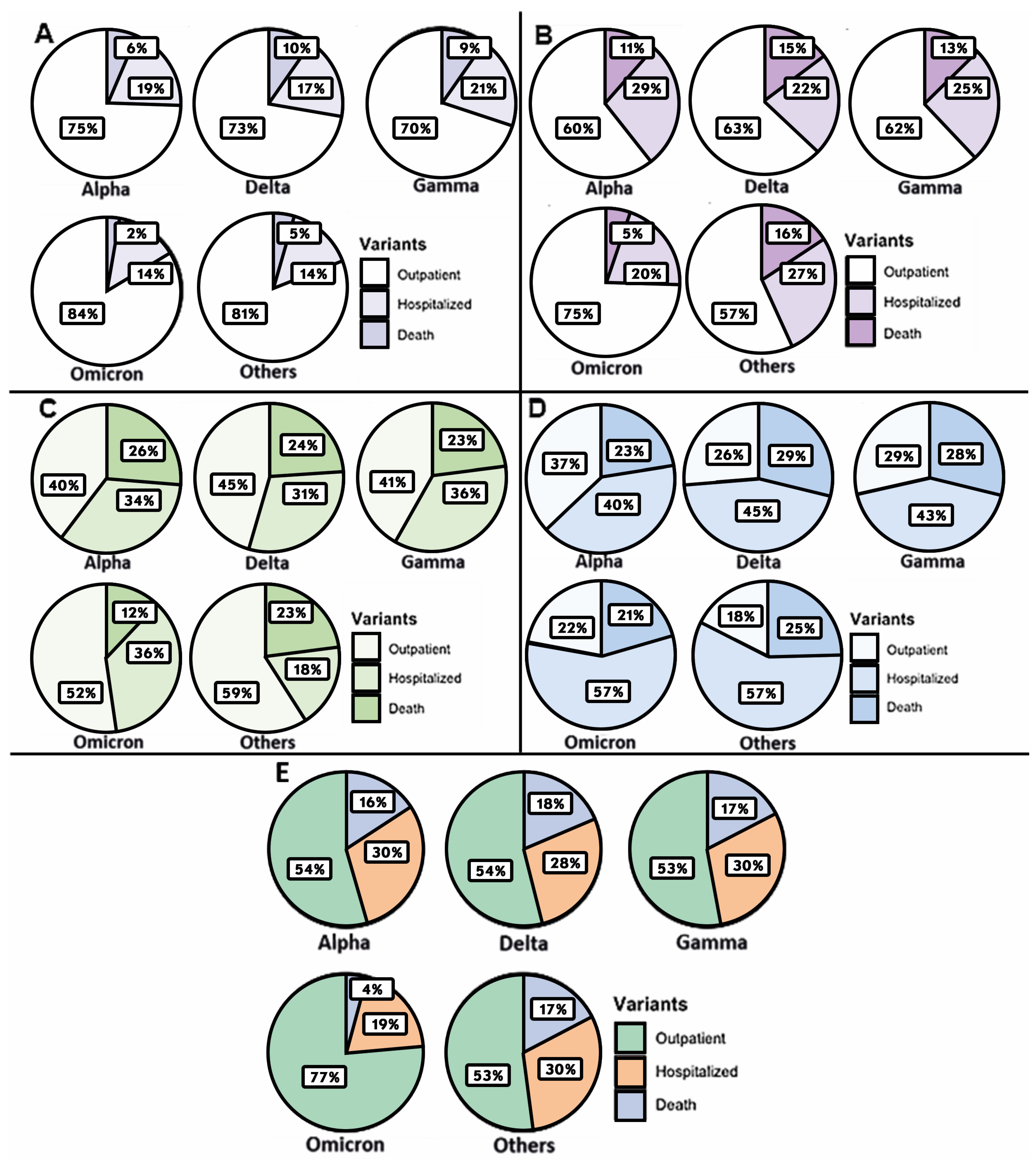
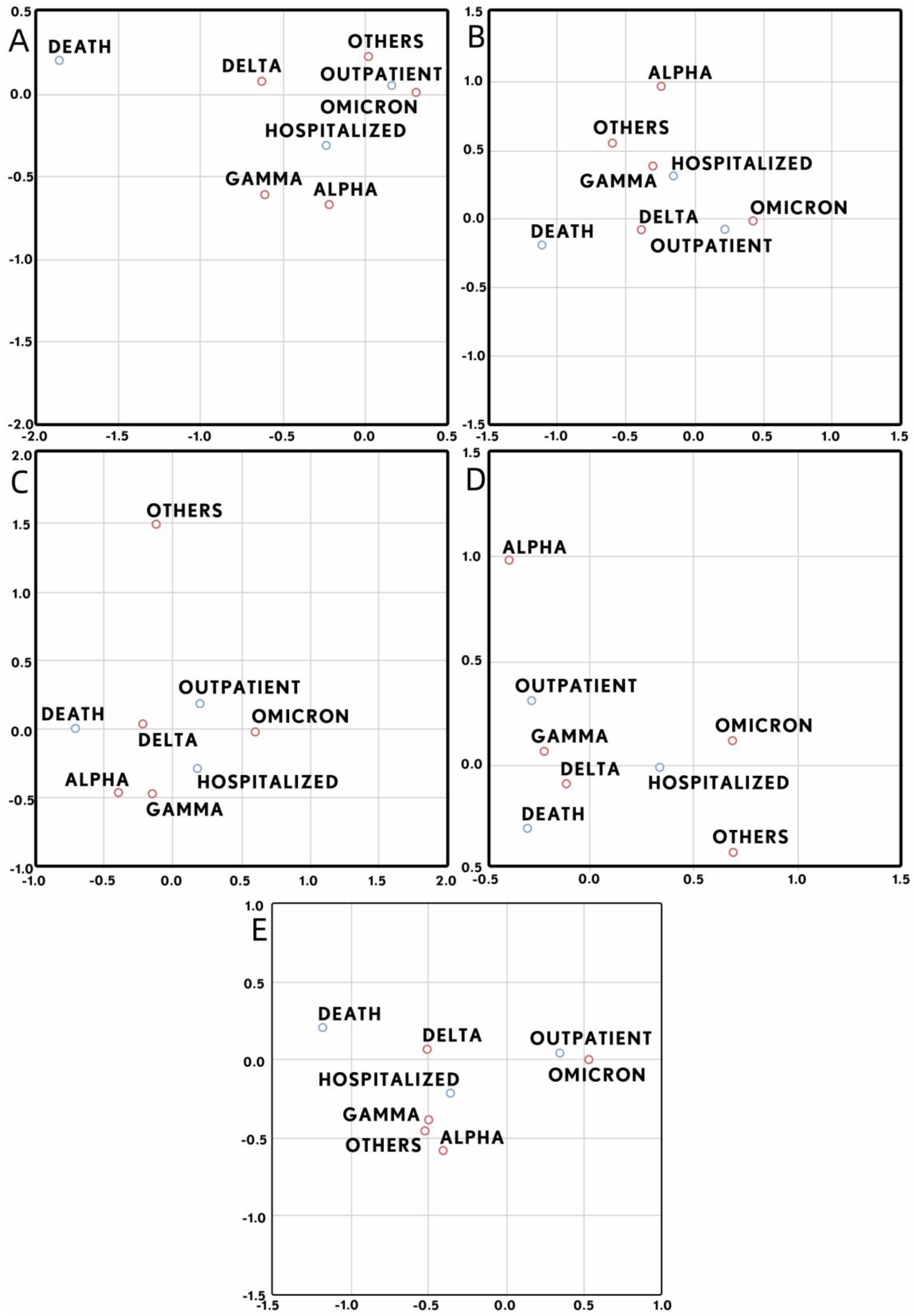
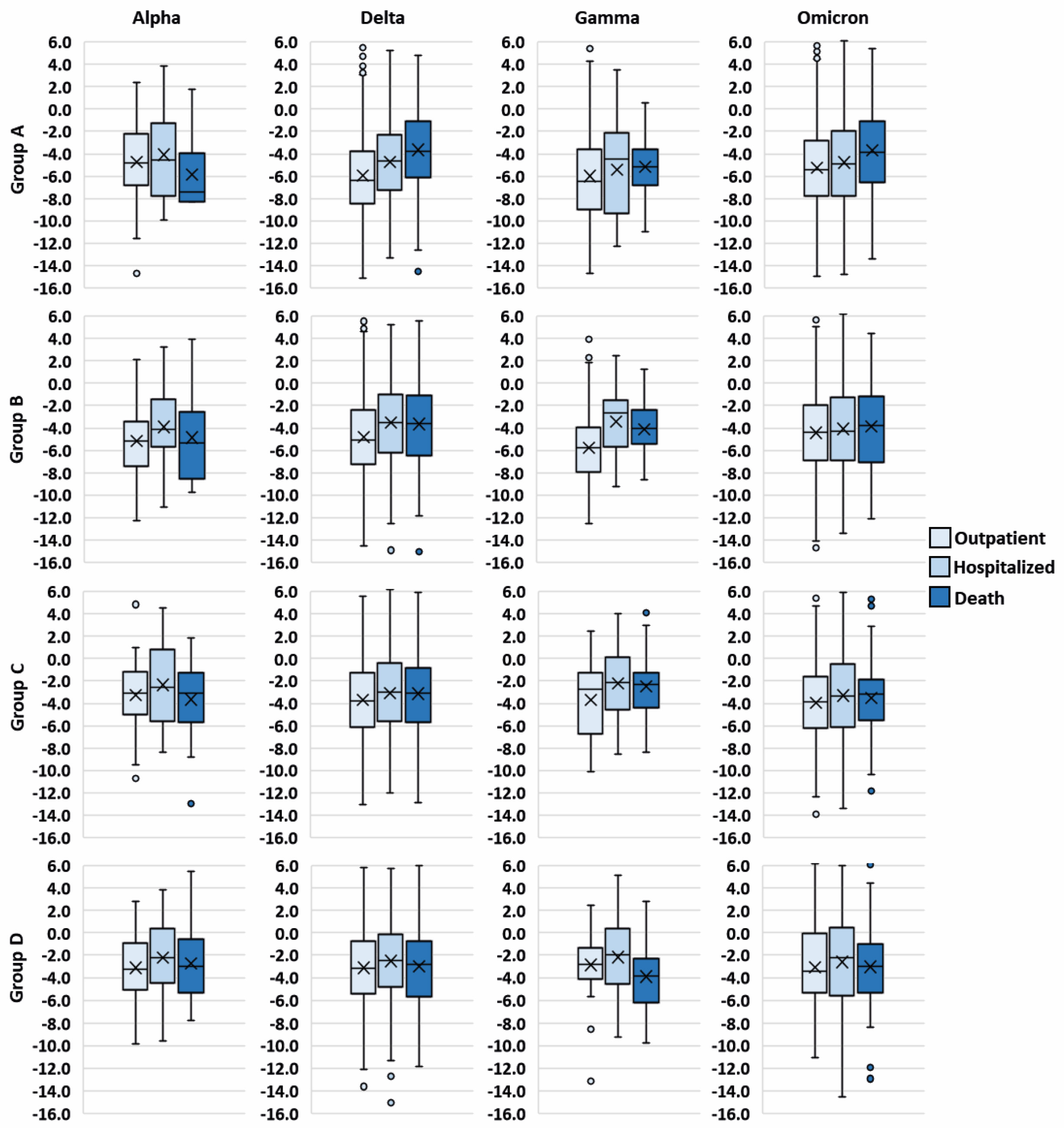
| Total | Group A (1–2 Days) | Group B (3–4 Days) | Group C (5–6 Days) | Group D (7–11 Days) | |
|---|---|---|---|---|---|
| Variants | |||||
| Alpha | 386 | 118 | 90 | 76 | 102 |
| Delta | 7463 | 2224 | 2137 | 1312 | 1790 |
| Gamma | 642 | 213 | 163 | 101 | 165 |
| Omicron | 8274 | 5094 | 2242 | 557 | 381 |
| Others * | 115 | 21 | 44 | 22 | 28 |
| Age group (years) | |||||
| 0–9 | 457 | 304 | 107 | 21 | 25 |
| 10–19 | 753 | 399 | 233 | 64 | 57 |
| 20–59 | 11,609 | 5768 | 3239 | 1258 | 1344 |
| >60 | 4061 | 1199 | 1097 | 725 | 1040 |
| Vaccines | |||||
| No | 10,911 | 4770 | 2981 | 1382 | 1778 |
| Yes (1 shot) | 1651 | 653 | 498 | 233 | 267 |
| Yes (2 shots) | 4318 | 2247 | 1197 | 453 | 421 |
| Sex | |||||
| Female | 9204 | 4439 | 2594 | 1021 | 1150 |
| Male | 7676 | 3231 | 2082 | 1047 | 1316 |
| Patient type | |||||
| Outpatient | 10,947 | 6128 | 3198 | 973 | 648 |
| Hospitalized | 4000 | 1184 | 1003 | 668 | 1145 |
| Death | 1931 | 358 | 475 | 427 | 671 |
| Age Groups (Years) | Outpatient (%) | Hospitalized (%) | Death (%) | Total |
|---|---|---|---|---|
| 0–9 | 296 (64.8) | 156 (34.1) | 5 (1.1) | 457 |
| 10–19 | 626 (83.1) | 123 (16.4) | 4 (0.5) | 753 |
| 20–59 | 9075 (78.2) | 1876 (16.1) | 658 (5.7) | 11,609 |
| >60 | 950 (23.4) | 1845 (45.4) | 1266 (31.2) | 4061 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos Coy-Arechavaleta, A.; Alvarado-Yaah, J.E.; Uribe-Noguez, L.A.; Guerra-Castillo, F.X.; Santacruz-Tinoco, C.E.; Ramón-Gallegos, E.; Muñoz-Medina, J.E.; Fernandes-Matano, L. Relationship between the Viral Load in Patients with Different COVID-19 Severities and SARS-CoV-2 Variants. Microorganisms 2024, 12, 428. https://doi.org/10.3390/microorganisms12030428
Santos Coy-Arechavaleta A, Alvarado-Yaah JE, Uribe-Noguez LA, Guerra-Castillo FX, Santacruz-Tinoco CE, Ramón-Gallegos E, Muñoz-Medina JE, Fernandes-Matano L. Relationship between the Viral Load in Patients with Different COVID-19 Severities and SARS-CoV-2 Variants. Microorganisms. 2024; 12(3):428. https://doi.org/10.3390/microorganisms12030428
Chicago/Turabian StyleSantos Coy-Arechavaleta, Andrea, Julio Elias Alvarado-Yaah, Luis Antonio Uribe-Noguez, Francisco Xavier Guerra-Castillo, Clara Esperanza Santacruz-Tinoco, Eva Ramón-Gallegos, José Esteban Muñoz-Medina, and Larissa Fernandes-Matano. 2024. "Relationship between the Viral Load in Patients with Different COVID-19 Severities and SARS-CoV-2 Variants" Microorganisms 12, no. 3: 428. https://doi.org/10.3390/microorganisms12030428
APA StyleSantos Coy-Arechavaleta, A., Alvarado-Yaah, J. E., Uribe-Noguez, L. A., Guerra-Castillo, F. X., Santacruz-Tinoco, C. E., Ramón-Gallegos, E., Muñoz-Medina, J. E., & Fernandes-Matano, L. (2024). Relationship between the Viral Load in Patients with Different COVID-19 Severities and SARS-CoV-2 Variants. Microorganisms, 12(3), 428. https://doi.org/10.3390/microorganisms12030428






