An Evaluation of the Antibacterial, Antileishmanial, and Cytotoxic Potential of the Secondary Metabolites of Streptomyces sp. ARH (A3)
Abstract
1. Introduction
2. Materials and Methods
2.1. Soil Collection, Isolation, and Classical Identification of the Microorganism
2.2. Molecular Identification of the Active Isolate
2.2.1. Genome Sequencing
2.2.2. Phylogenetic Tree
2.3. Submerged Fermentation
Liquid–Liquid Extraction
2.4. Screening of Antibacterial Tests
2.4.1. Microorganisms Used
2.4.2. Test in Solid Medium
2.4.3. Agar Diffusion Assay
2.4.4. Determination of Minimum Inhibitory Concentration (MIC)
2.5. In Vitro leishmanicidal Activity
2.6. Cytotoxic Action of Metabolites in Mammalian Cells
2.7. Evaluation of the Hemolysis Index
3. Results
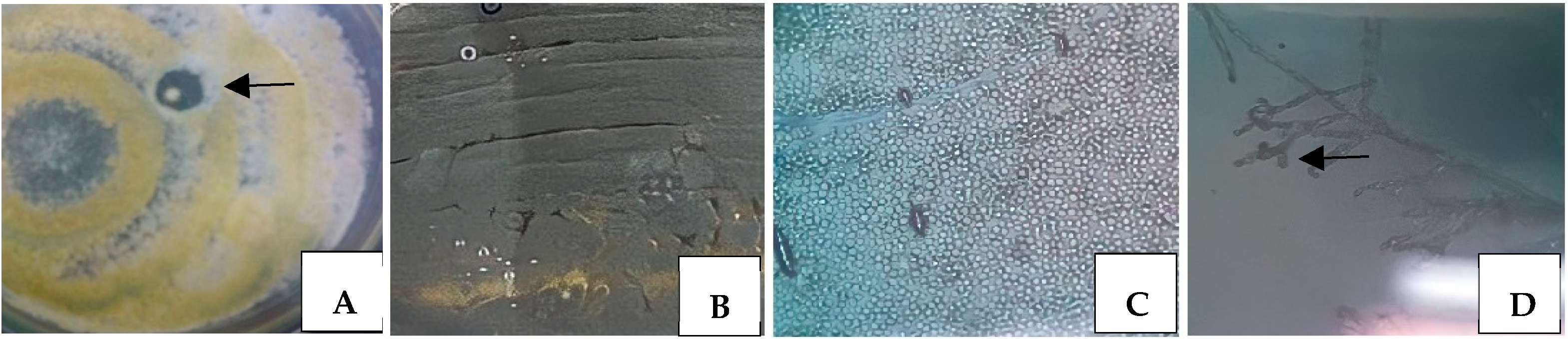
3.1. Isolation and Classical Identification of Microorganisms
3.2. Molecular Identification
3.2.1. Genome and 16S rRNA Gene
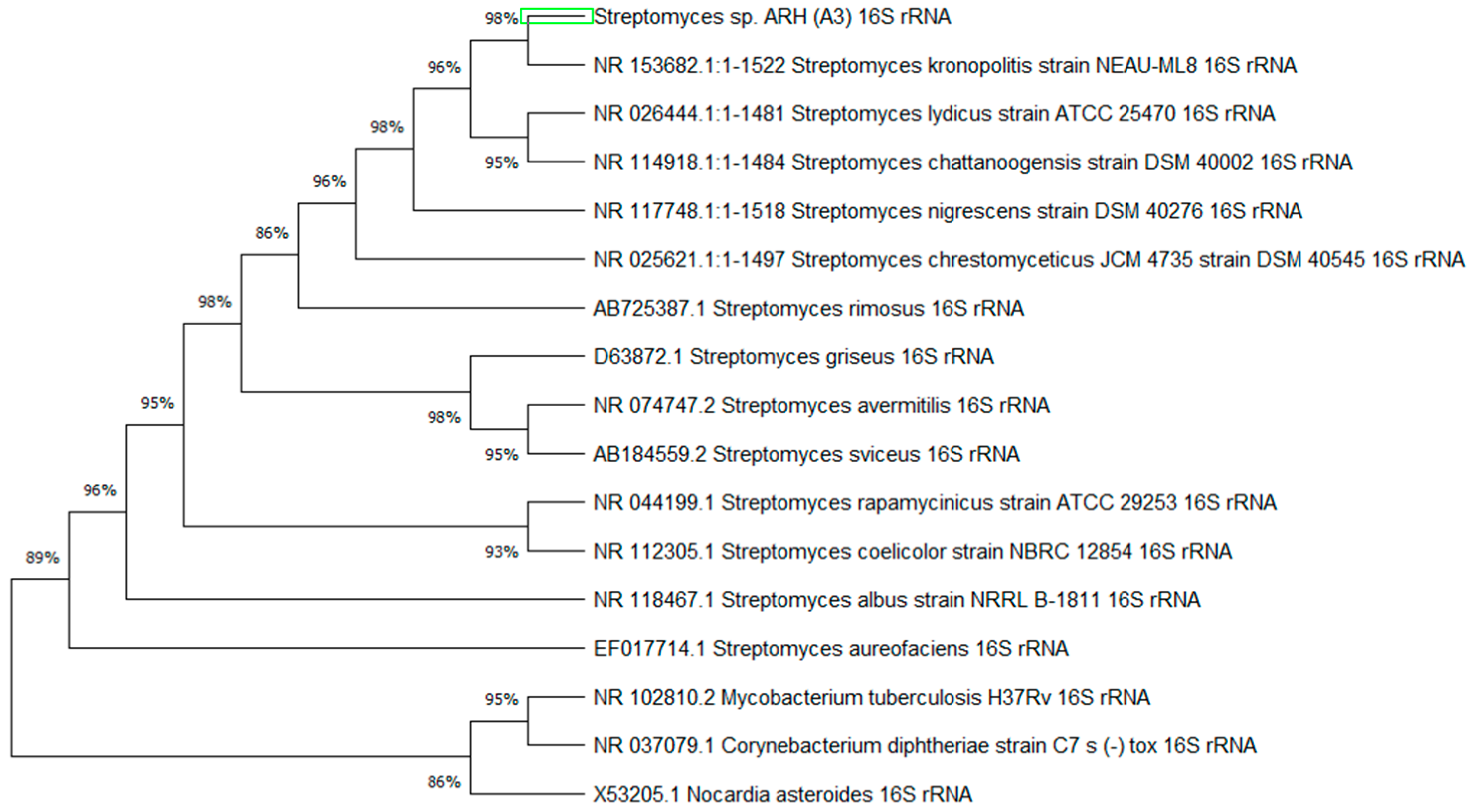
3.2.2. Phylogenetic Tree
3.3. Antibacterial Test Screening
3.3.1. Test in Solid Medium
3.3.2. Agar Diffusion Assay
3.3.3. Determination of Minimum Inhibitory Concentration (MIC)
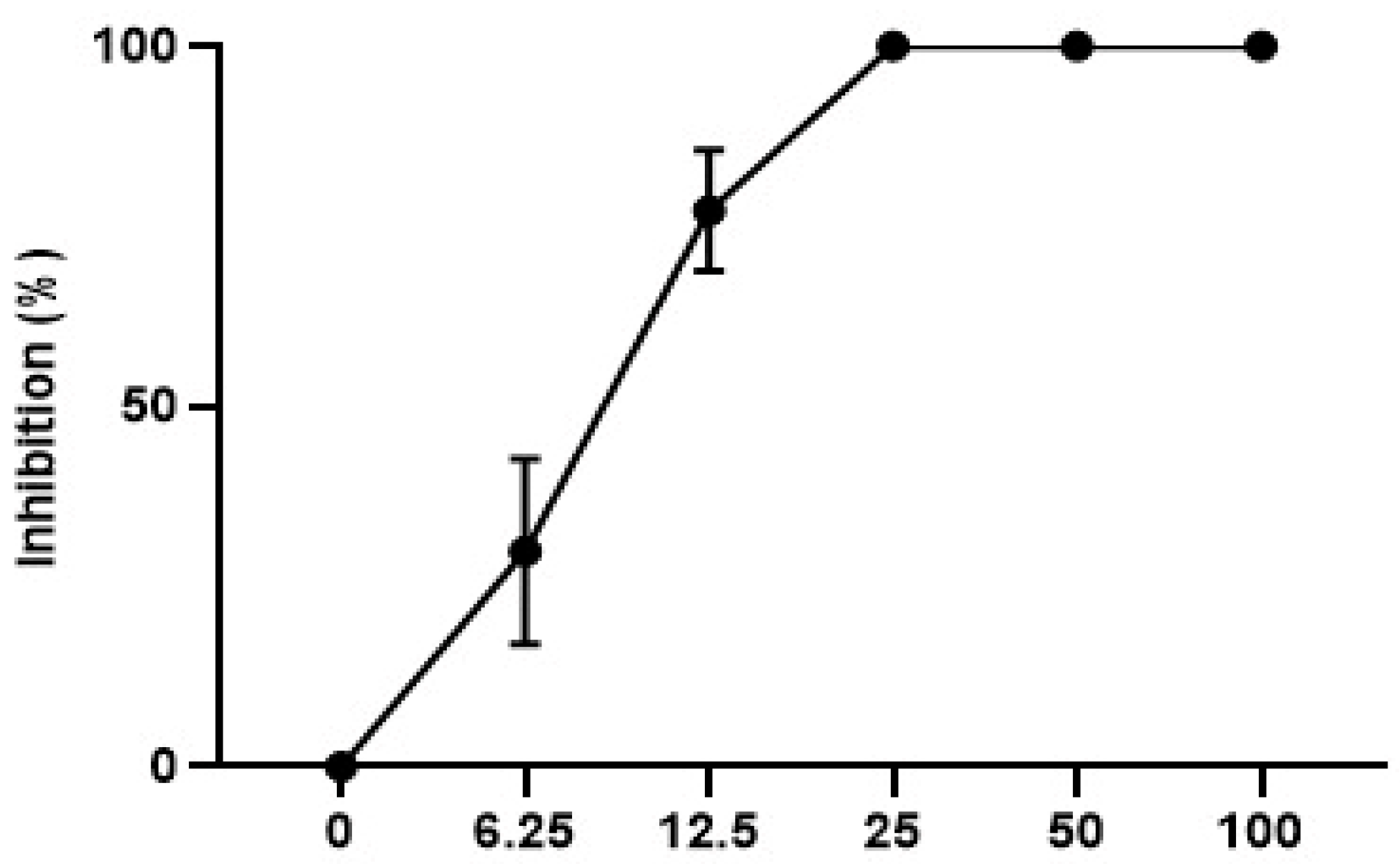
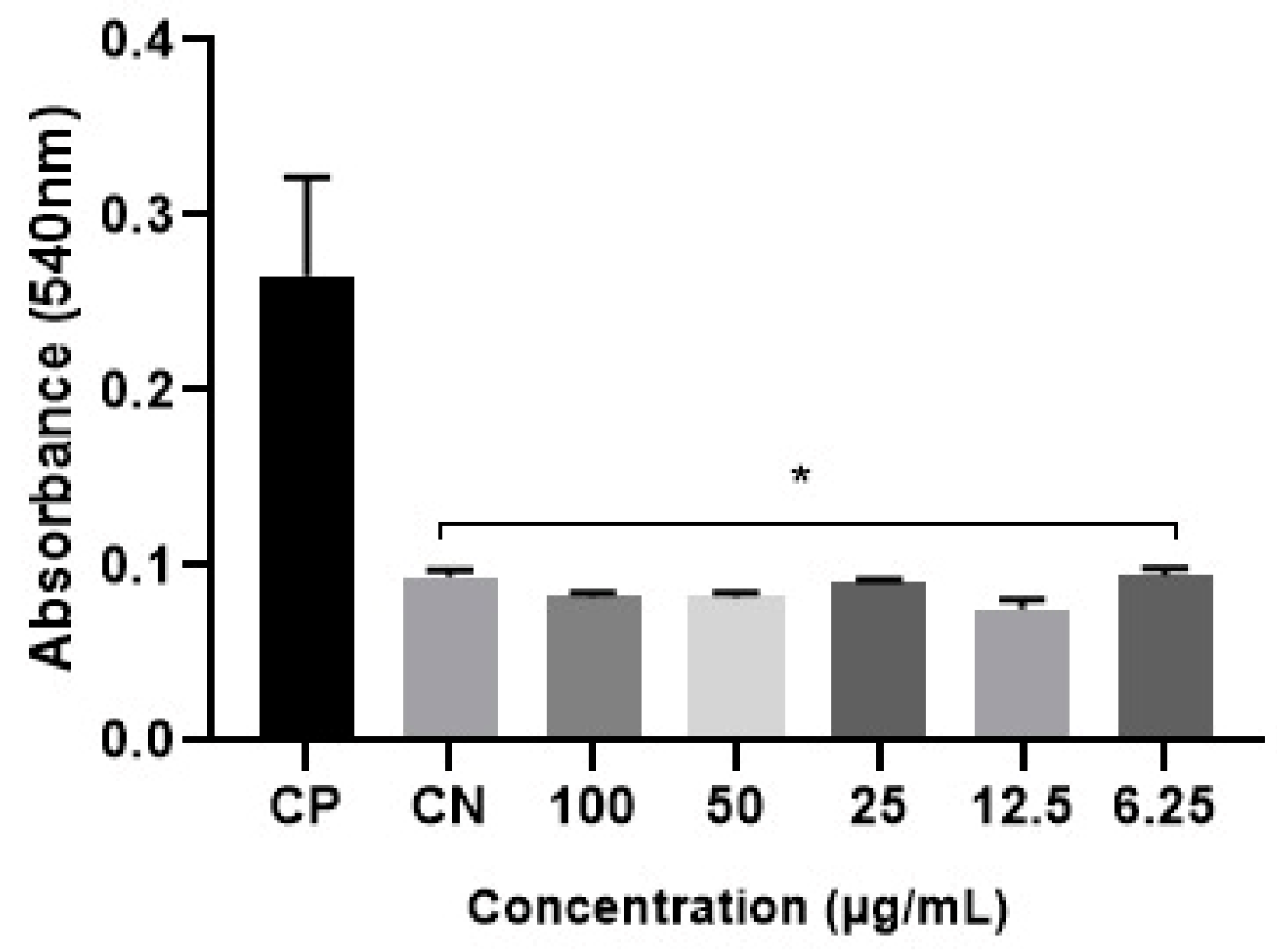
3.3.4. In Vitro leishmanicidal Activity
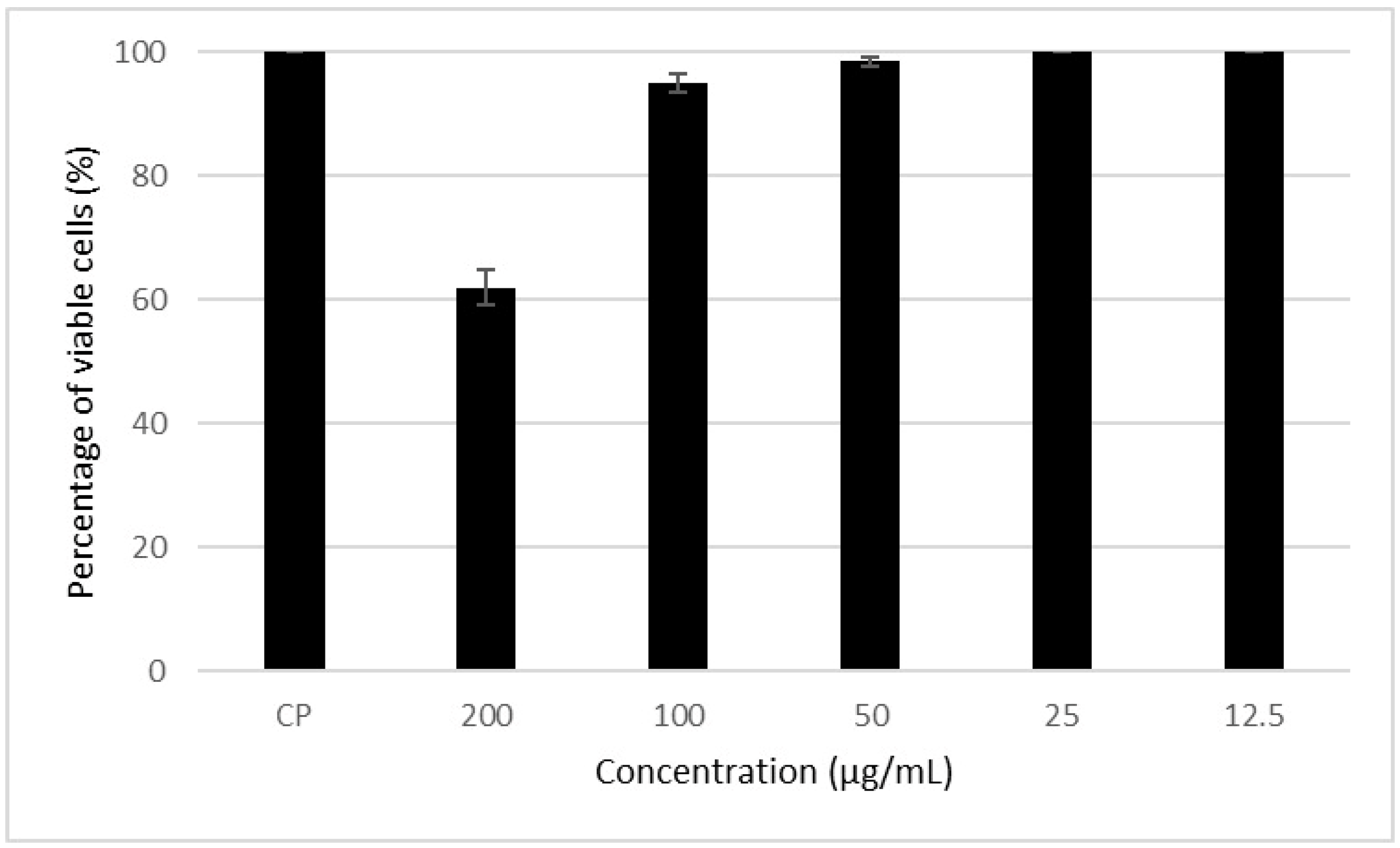
3.4. Cytotoxic Action of the Metabolite in Mammalian Cells
3.5. Evaluation of the Hemolysis Index
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parker, S.S. Buried treasure: Soil biodiversity and conservation. Biodivers. Conserv. 2010, 19, 3743–3756. [Google Scholar] [CrossRef]
- Al-Dhabi, N.A.; Esmail, G.A.; Ghilan, A.K.M.; Arasu, M.V. Isolation and screening of Streptomyces sp. Al-Dhabi-49 from the environment of Saudi Arabia with concomitant production of lipase and protease in submerged fermentation. Saudi J. Biol. Sci. 2020, 27, 474–479. [Google Scholar] [CrossRef]
- Carmin, A.A.; Santiago, P.A.L.; Santiago, S.R.S.S.; Gomes, A.M.S.; Pereira, K.D.E.S. Evaluation of the antimicrobial potential of fungi isolated from the soil of the Municipality of Iranduba. Res. Soc. Dev. 2021. [Google Scholar] [CrossRef]
- Thiele-Bruhn, S.; Schloter, M.; Wilke, B.M.; Beaudette, L.A.; Martin-Laurent, F.; Cheviron, N.; Mougin, C.; Römbke, J. Identification of new microbial functional standards for soil quality assessment. Soil 2020, 6, 17–34. [Google Scholar] [CrossRef]
- Supong, K.; Thawai, C.; Choowong, W.; Kittiwongwattana, C.; Thanaboripat, D.; Laosinwattana, C.; Koohakan, P.; Parinthawong, N.; Pittayakhajonwut, P. Antimicrobial compounds from endophytic Streptomyces sp. BCC72023 isolated from rice (Oryza sativa L.). Res. Microb. 2016, 167, 290–298. [Google Scholar] [CrossRef]
- Amorim, E.A.d.F.; Castro, E.J.M.; da Souza, S.V.; Alves, M.S.; Dias, L.R.L.; Melo, M.H.F.; da Silva, I.M.A.; Villis, P.C.M.; Bonfim, M.R.Q.; Falcai, A.; et al. Antimicrobial Potential of Streptomyces ansochromogenes (PB3) isolated from a plant native to the Amazon against Pseudomonas aeruginosa. Front. Microb. 2020, 11, 574693. [Google Scholar] [CrossRef] [PubMed]
- Elkhayat, E.S.; Goda, A.M. Antifungal and cytotoxic constituents from the endophytic fungus Penicillium sp. Bullet Fac. Pharm. 2017, 55, 85–89. [Google Scholar] [CrossRef]
- Bunbamrung, N.; Intaraudom, C.; Dramae, A. Antibacterial, antitubercular, antimalarial and cytotoxic substances from the endophytic Streptomyces sp. TBRC7642. Phytochemistry 2020, 172, 112275. [Google Scholar] [CrossRef]
- Pagmadulam, B.; Tserendulam, D.; Rentsenkhand, T.; Igarashi, M.; Sawa, R.; Nihei, C.-I.; Nishikawa, Y. Isolation and characterization of antiprotozoal compound-producing Streptomyces species from Mongolian soils. Parasitol. Int. 2020, 74, 101961. [Google Scholar] [CrossRef]
- Rosa, F.C.; de Almeida, I.A.; Mota, A.G.; de Miranda, R.D.C.M. Bioprospecting of secondary metabolites produced by endophytic actinomycete isolated from Aloe vera. Concilium 2023, 23, 227–239. [Google Scholar] [CrossRef]
- Encalada Álvarez, R.C.; Arteaga Sarmiento, S.D. Vigilância epidemiológica do Acinetobacter baumannii multirresistente em nível hospitalar. Vive Rev. Salud 2021, 4, 66–86. [Google Scholar]
- Böger, B.; Surek, M.; Vilhena, O.R.; Fachi, M.M.; Junkert, A.M.; Santos, J.M.; Domingos, E.L.; Cobre, A.d.F.; Momade, D.R.; Pontarolo, R. Occurrence of antibiotics and antibiotic resistant bacteria in subtropical urban rivers in Brazil. J. Hazard. Mater. 2021, 402, a123448. [Google Scholar] [CrossRef]
- Wang, Q.; Mao, C.; Lei, L.; Yan, B.; Yuan, J.; Guo, Y.; Li, T.; Xiong, X.; Cao, X.; Huang, J.; et al. Antibiotic resistance genes and their links with bacteria and environmental factors in three predominant freshwater aquaculture modes. Ecotoxicol. Environ. Saf. 2022, 241, a113832. [Google Scholar] [CrossRef]
- Lopes, R.K.; Carvalho, C.E.G.; Marques, D.D.; Ferreira, A.B.; Almeida, R.M.; Aguilar, M.G. Evaluation of the leishmanicida activity of Symphonia globulifera latex. Braz. J. Dev. 2020, 6, a16705–a16730. [Google Scholar] [CrossRef]
- Pereira, B.A.S. Leishmania (Leishmania) amazonensis: Participação de Fatores do Hospedeiro e do Parasito no Curso da Infecção Experimental em Camundongos. Ph.D. Thesis, Instituto Oswaldo Cruz, Rio de Janerio, Brazil, 2017. [Google Scholar]
- Oddone, R.; Schweynoch, C.; Schönian, G. Development of a multilocus microsatellite typing approach for discriminating strains of Leishmania (Viannia) species. J. Clin. Microb. 2017, 47, 2818–2825. [Google Scholar] [CrossRef] [PubMed]
- Pan American Health Organization; World Health Organization. Manual of Procedures for Leishmaniases Surveillance and Control in the Americas; Pan American Health Organization: Washington, DC, USA, 2019. [Google Scholar]
- Clinical and Laboratory Standards Institute. Quality Control Minimal Inhibitory Concentration (MIC) Limits for Broth Dilution and MIC Interpretative Breakpoints (M27-S2); Clinical and Laboratory Standards Institute Wayne: Wayne, PA, USA, 2006. [Google Scholar]
- Clark, F.E. Agar-plate method for total microbial count. In Methods of Soil Analysis, Part Chemical and Microbiological Properties; Blanc, C.A., Evans, D., White, J.L., Ensminger, L.E., Clark, F.E., Dinauer, R.C., Eds.; Madson Inc.: New York, NY, USA, 1965; pp. 1460–1466. [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Rapper, K.B.; Fennell, D.I. The Genus Aspergillus; Williams & Wilkins: Baltimore, MD, USA, 1965. [Google Scholar]
- Pitt, J.I. The Genus Penicillium and Its Teleomorphic States Eupenicillium and Talaromyces; Academic Press: London, UK, 1979. [Google Scholar]
- Samson, R.A.; Pitt, J.I. Modern Concepts in Penicillium and Aspergillus Classification; Plenum Press: New York, NY, USA, 1990. [Google Scholar]
- Klich, M.A.; Pitt, J.I. Differentiation of Aspergillus flavus from A. parasiticus and other closely related species. Trans. Br. Mycol. Soc. 1988, 91, 99–108. [Google Scholar] [CrossRef]
- Trisuwan, K.; Rukachaisirikul, V.; Sukpondma, Y.; Preedanon, S.; Phongpaichit, S.; Rungjindamai, N.; Sakayaroj, J. Epoxydons and a pyrone from the marine-derived fungus Nigrospora sp. PSU-F5. J. Nat. Prod. 2008, 71, 1323–1326. [Google Scholar] [CrossRef]
- Uchida, K.; Ichikawa, T.; Shimauchi, Y.; Ishikura, T.; Ozaki, A. HIkizimycin, a new antibiotic. J. Antibiot. 1971, 14, 259–262. [Google Scholar] [CrossRef]
- Matsuura, T. Caracterização Taxonômica de Actinomicetos Endofíticos Produtores de Antibióticos Isolados de Cupuaçuzeiro (Theobroma grandiflorum Schum.). Ph.D. Thesis, Universidade Estadual de Campinas, Campinas, Brazil, 2004. [Google Scholar]
- Bauer, A.W.; Kirby, W.M.M.; Sherrys, J.C.; Turk, M. Antibiotic susceptibility testing by a standardized single disc method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement M100-S25; Clinical and Laboratory Standards Institute Wayne: Wayne, PA, USA, 2015. [Google Scholar]
- Zgoda, J.R.; Porter, J.R. A convenient microdilution method for screening natural products against bacteria and fungi. Pharm. Biol. 2001, 39, 221–225. [Google Scholar] [CrossRef]
- Aliança, A.S.S.; Oliveira, A.R.; Feitosa, A.P.S. In vitro evaluation of cytotoxicity and leishmanicidal activity of phthalimido-thiazole derivatives. Eur. J. Pharm. Sci. 2017, 105, 1–10. [Google Scholar] [CrossRef]
- El-Sayed, E.R.; Ahmed, A.S.; Al-Hagar, O.E.A. Agro-industrial wastes for production of paclitaxel by irradiated Aspegillus fumigatus under solid-state fermentation. J. Appl. Microbiol. 2020, 128, 1427–1439. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Liu, K.S.; Sung, K.C.; Tsai, C.Y.; Fang, J.Y. Lipid nanoparticles with different oil/ fatty ester ratios as carriers of buprenorphine and its prodrugs for injection. Eur. J. Pharm. Sci. 2010, 38, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Tunes, L.G. Investigação da Atividade e Mecanismos de Ação Leishmanicida e/ou Tripanocida de Produtos Naturais e um Derivado. Master’s Thesis, Fundação Oswaldo Cruz, Rio de Janeiro, Brazil, 2015. [Google Scholar]
- Cruz, E.F. Modelos Markovianos para Canais Iônicos em Membranas Celulares. Master’s Thesis, Universidade de São Paulo, São Paulo, Brazil, 2006. [Google Scholar]
- Saraswathi, K.; Mahalakshmi, S.; Khusro, A.; Arumugam, P.; Mohammed, A.K.; Alkufeidy, R.M. In vitro biological properties of Streptomyces cangkringensis isolated from the floral rhizosphere regions. Saudi J. Biol. Sci. 2020, 27, 3249–3257. [Google Scholar] [CrossRef] [PubMed]
- Ensign, J.C. Formation, properties, and germinations of Actinomycete spores. Ann. Rev. Microb. 1978, 32, 185–219. [Google Scholar] [CrossRef] [PubMed]
- Sholkamy, E.N.; Muthukrishnan, P.; Abdel-Raouf, N.; Nandhini, X.; Ibraheem, I.B.; Mostafa, A.A. Antimicrobial and antinematicidal metabolites from Streptomyces cuspidosporus strain SA4 against selected pathogenic bacteria, fungi and nematode. Saudi J. Biol. Sci. 2020, 27, 3208–3220. [Google Scholar] [CrossRef]
- Abba, C.; Nwachukwu, C.; Okoye, F.; Eboka, C.; Eze, P.; Abonyi, D.; Proksch, P. Phenolic compounds from endophytic Pseudofusicoccum sp. isolated from Annona muricata. Trop. J. Nat. Prod. Res. 2018, 3, 332. [Google Scholar] [CrossRef]
- Huang, X.; Ren, J.; Li, P.; Feng, S.; Dong, P.; Ren, M. Potential of microbial endophytes to enhance the resistance to postharvest diseases of fruit and vegetables. J. Sci. Food Agric. 2021, 101, 1744–1757. [Google Scholar] [CrossRef]
- Liu, C.; Ye, L.; Li, Y.; Jiang, S.; Liu, H.; Yan, K.; Xiang, W.; Wang, X. Streptomyces kronopolitis sp. nov., an actinomycete that produces phoslactomycins isolated from a millipede (Kronopolites svenhedind Verhoeff). Int. J. Syst. Evol. Microb. 2016, 66, 5352–5357. [Google Scholar] [CrossRef]
- Sreedharan, V.; Rao, K.V.B. Efficacy of protease inhibitor from marine Streptomyces sp. VITBVK2 against Leishmania donovani—An in vitro study. Exp. Parasitol. 2017, 174, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Aliança, A.S.D.S.; dos Anjos, K.F.L.; Reis, T.N.D.V.; Higino, T.M.M.; Brelaz-de-Castro, M.C.A.; Bianco, É.M.; De Figueiredo, R.C.B.Q. The in vitro biological activity of the brazilian brown seaweed Dictyota mertensii against Leishmania amazonensis. Molecules 2014, 19, 14052–14065. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.; Acharaya, K. Mushrooms: An emerging resource for therapeutic terpenoids. 3 Biotech 2019, 9, 1–14. [Google Scholar] [CrossRef]
- Szabo, G. Gut–liver axis in alcoholic liver disease. Rev. Arq. Gastroenterol. 2015, 148, 30–36. [Google Scholar] [CrossRef]
- Rosa, F.C.; Mota, A.G.; de Almeida, B.L.; de Souza, G.D.S.; Araújo, M.C.; Melo, M.H.F.; dos Santos Aliança, A.S.; de Mendonça, R.d.C.M. Importance of secondary metabolites produced by actinobacteria. Rev. Ciências Saúde CEUMA 2023, 1, 72–87. [Google Scholar]
- Jose, P.A.; Maharshi, A.; Jha, B. Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 2021, 246, 126708. [Google Scholar] [CrossRef] [PubMed]
| Description | Max Score | Percent Identity | Accession |
|---|---|---|---|
| Streptomyces kronopolitis strain NEAU-ML8.T | 2750 | 99.77% | NR_153682.1 |
| Streptomyces nigrescens strain DSM 40276 | 2734 | 99.20% | NR_117748.1 |
| Streptomyces lydicus strain ATCC 25470 | 2715 | 99.60% | NR_026444.1 |
| Streptomyces chattanoogensis strain DSM 40002 | 2712 | 99.28% | NR_114918.1 |
| Streptomyces chrestomyceticus JCM 4735 strain DSM 40545 | 2706 | 99.14% | NR_025621.1 |
| Clinical Pathogens | Inhibition Halos (mm) |
|---|---|
| Staphylococcus aureus (ATCC 6538) | 25.3 b ± 6.8 |
| Corynebacterium diphtheriae (27012) | 29 b ± 1 |
| Corynebacterium diphteriae (27010) | 36 a ± 5.6 |
| Mycobacterium abscessus | 33 a ± 3 |
| Clinical Pathogens | Inhibition Halos (mm) |
|---|---|
| Staphylococcus aureus (ATCC 6538) | 20.3 b ± 1.5 |
| Corynebacterium diphtheriae (27012) | 20.6 b ± 1.1 |
| Corynebacterium diphteriae (27010) | 28.6 a ± 0.57 |
| Mycobacterium abscessus | 30.6 a ± 3 |
| Metabolites | Clinical Pathogens | |||
|---|---|---|---|---|
| M. abscessos (IC) | S. aureus ATCC 6538 | C. diphterium ATCC 27010 | C. diphterium ATCC 27012 | |
| Streptomyces sp. ARH (A3) | 12.5 µg/mL | 0.048 µg/mL | 12.5 µg/mL | 12.5 µg/mL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Azevedo, V.L.S.; Rosa, F.C.; Dias, L.R.L.; Batista, L.A.; Melo, M.C.; Sales, L.A.T.; Branco, A.d.J.M.; Araújo, T.R.R.; de Miranda, R.d.C.M.; Aliança, A.S.d.S. An Evaluation of the Antibacterial, Antileishmanial, and Cytotoxic Potential of the Secondary Metabolites of Streptomyces sp. ARH (A3). Microorganisms 2024, 12, 476. https://doi.org/10.3390/microorganisms12030476
de Azevedo VLS, Rosa FC, Dias LRL, Batista LA, Melo MC, Sales LAT, Branco AdJM, Araújo TRR, de Miranda RdCM, Aliança ASdS. An Evaluation of the Antibacterial, Antileishmanial, and Cytotoxic Potential of the Secondary Metabolites of Streptomyces sp. ARH (A3). Microorganisms. 2024; 12(3):476. https://doi.org/10.3390/microorganisms12030476
Chicago/Turabian Stylede Azevedo, Virlanna Larissa Santos, Fernanda Costa Rosa, Leo Ruben Lopes Dias, Lucas Abrantes Batista, Mariana Costa Melo, Luis Alfredo Torres Sales, Abia de Jesus Martins Branco, Thalison Rômulo Rocha Araújo, Rita de Cássia Mendonça de Miranda, and Amanda Silva dos Santos Aliança. 2024. "An Evaluation of the Antibacterial, Antileishmanial, and Cytotoxic Potential of the Secondary Metabolites of Streptomyces sp. ARH (A3)" Microorganisms 12, no. 3: 476. https://doi.org/10.3390/microorganisms12030476
APA Stylede Azevedo, V. L. S., Rosa, F. C., Dias, L. R. L., Batista, L. A., Melo, M. C., Sales, L. A. T., Branco, A. d. J. M., Araújo, T. R. R., de Miranda, R. d. C. M., & Aliança, A. S. d. S. (2024). An Evaluation of the Antibacterial, Antileishmanial, and Cytotoxic Potential of the Secondary Metabolites of Streptomyces sp. ARH (A3). Microorganisms, 12(3), 476. https://doi.org/10.3390/microorganisms12030476







