Understanding Salinity-Driven Modulation of Microbial Interactions: Rhizosphere versus Edaphic Microbiome Dynamics
Abstract
1. Introduction
2. Materials and Methods
2.1. Geological Setting and Soil Sampling
2.2. Soil Physicochemical Analysis
2.3. Soil DNA Extraction and Bioinformatics Analysis
2.4. Statistical Analysis
3. Results
3.1. Contrasting Physicochemical Profiles of Non-Rhizosphere and Rhizosphere Soils
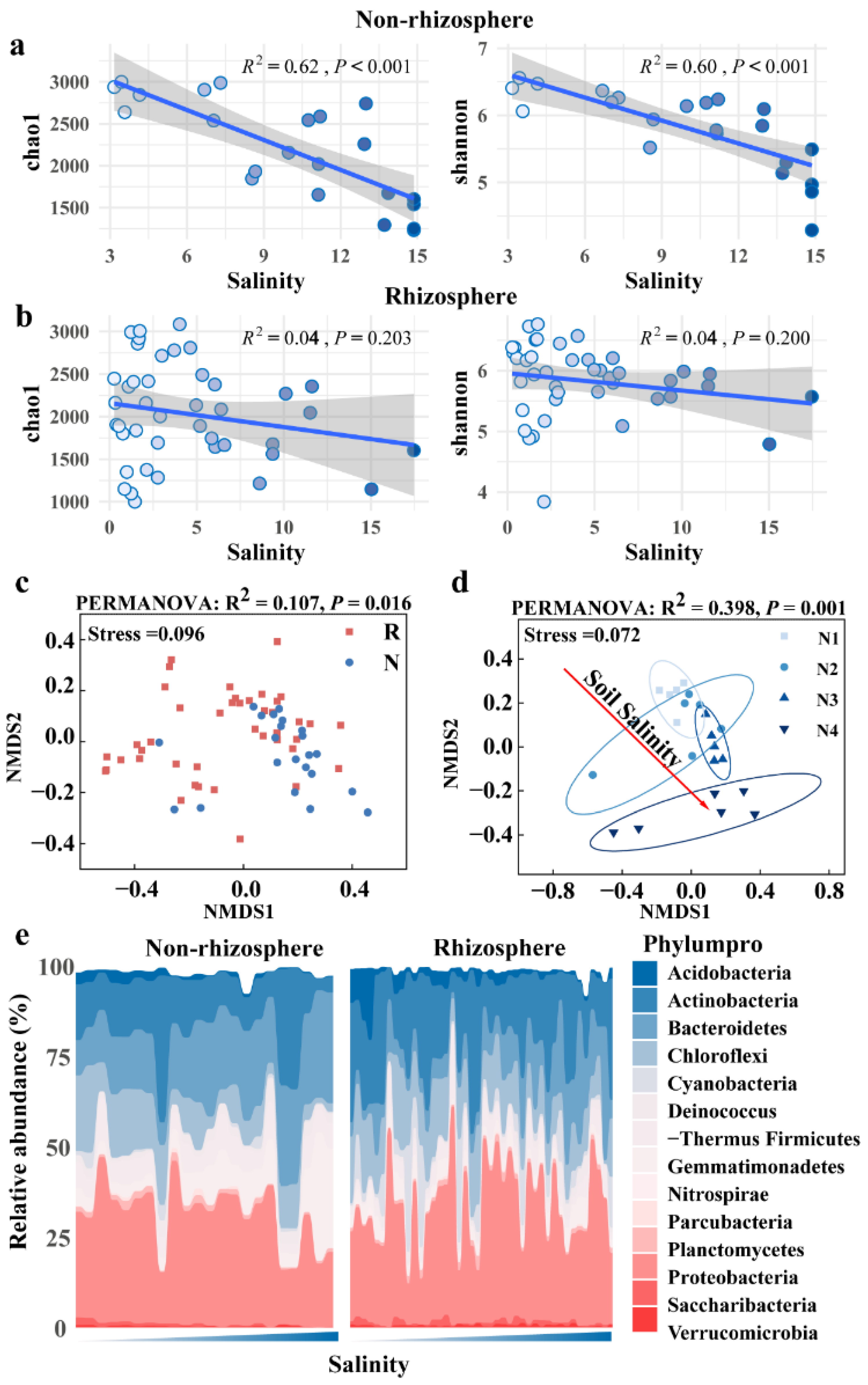
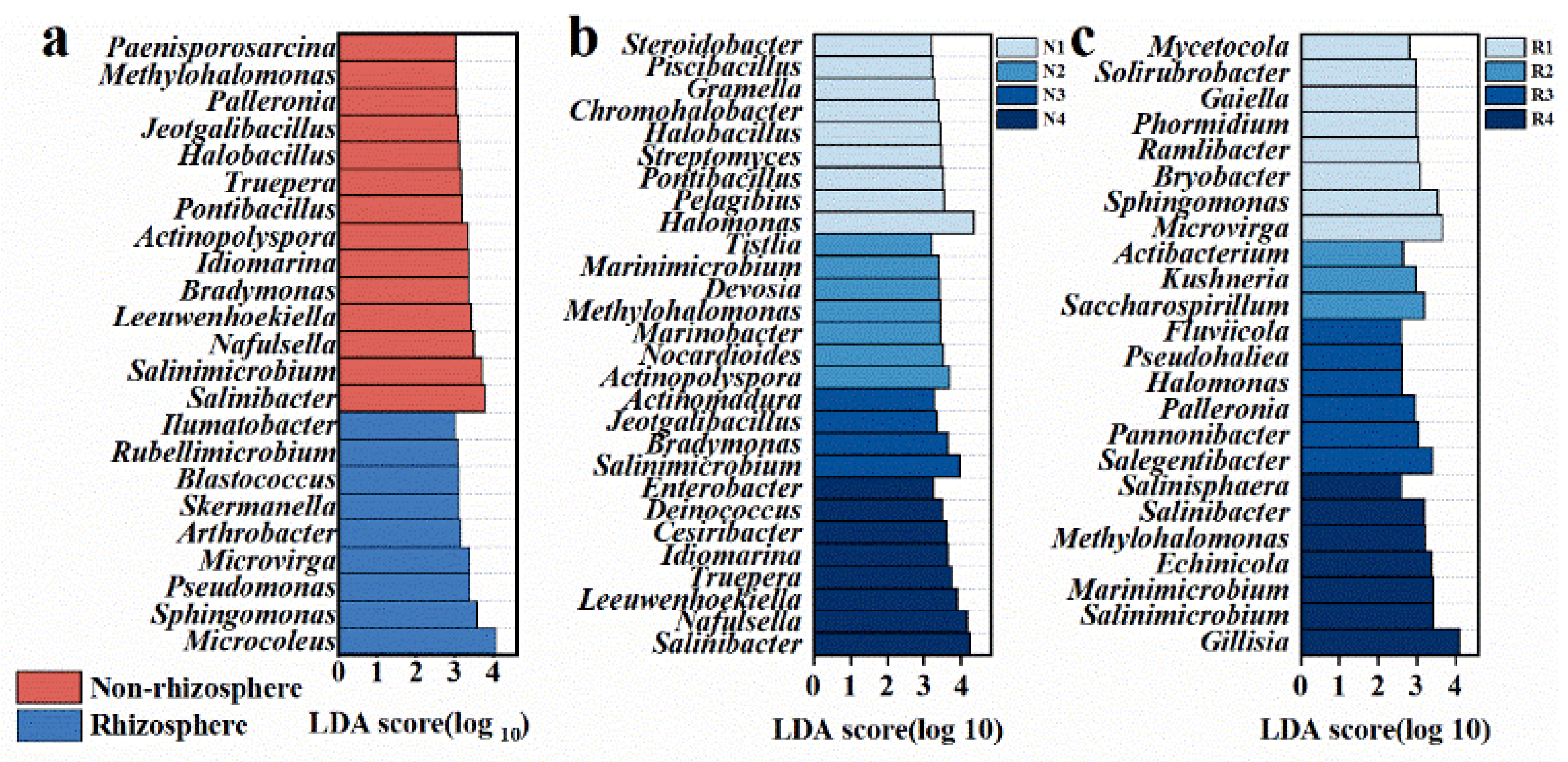
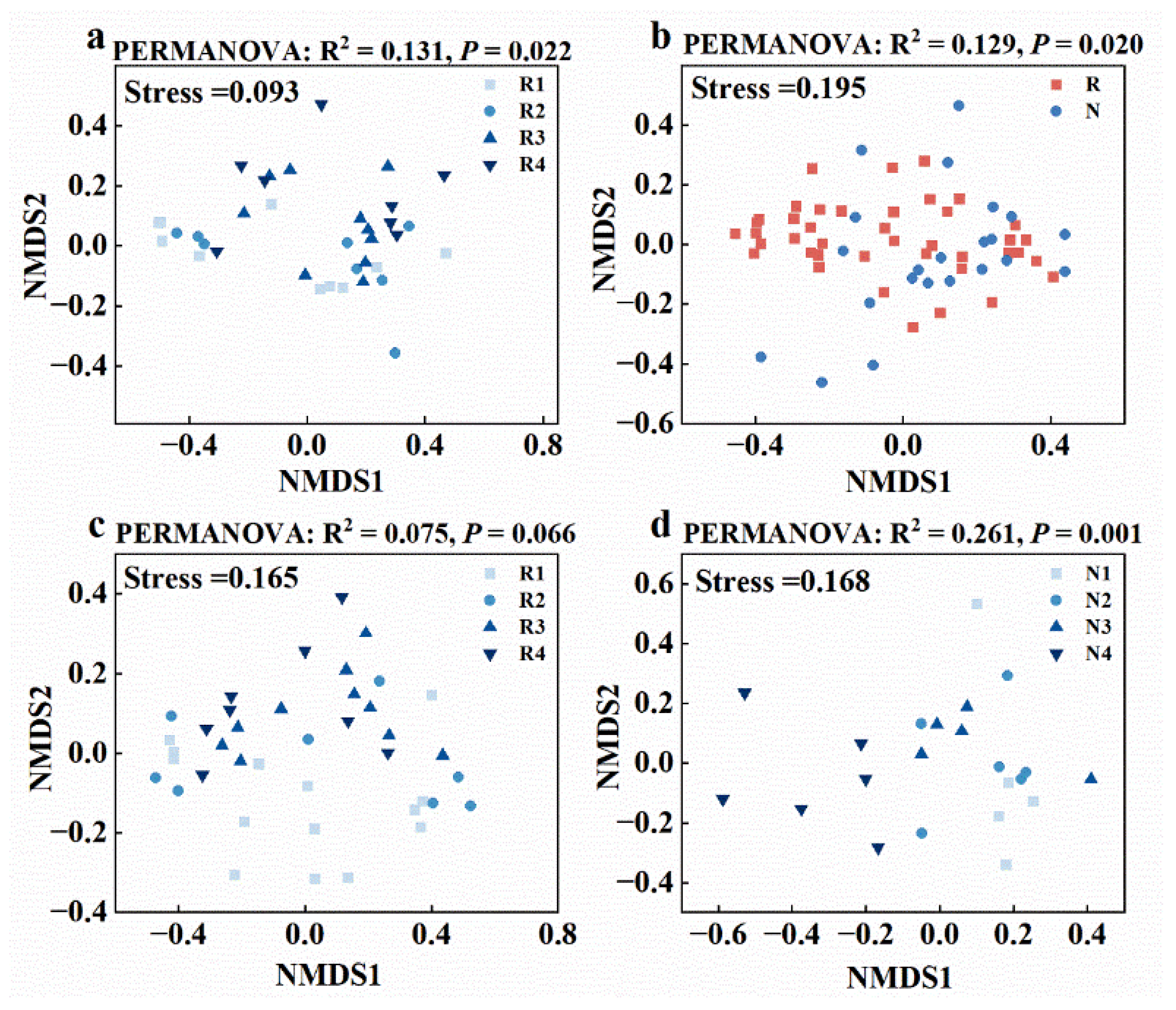
3.2. Distinct Microbiomes in Non-Rhizosphere and Rhizosphere Soils Respond to Salt Stress
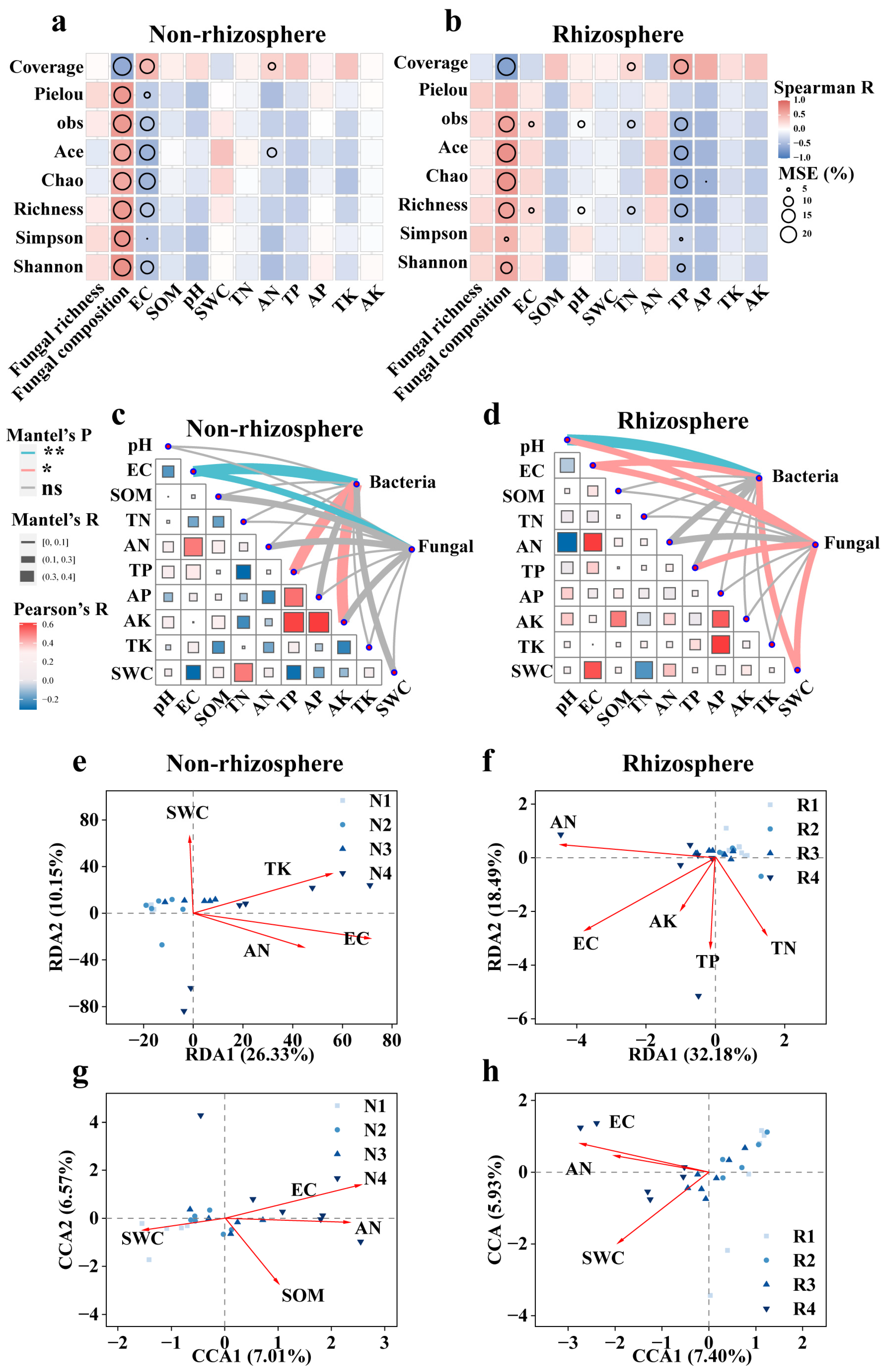
3.3. Various Factors Potentially Drive Microbiome Diversity in Saline Soils
3.4. Salt Stress Coupled to Biotic Factors Shapes Microbiome Assembly
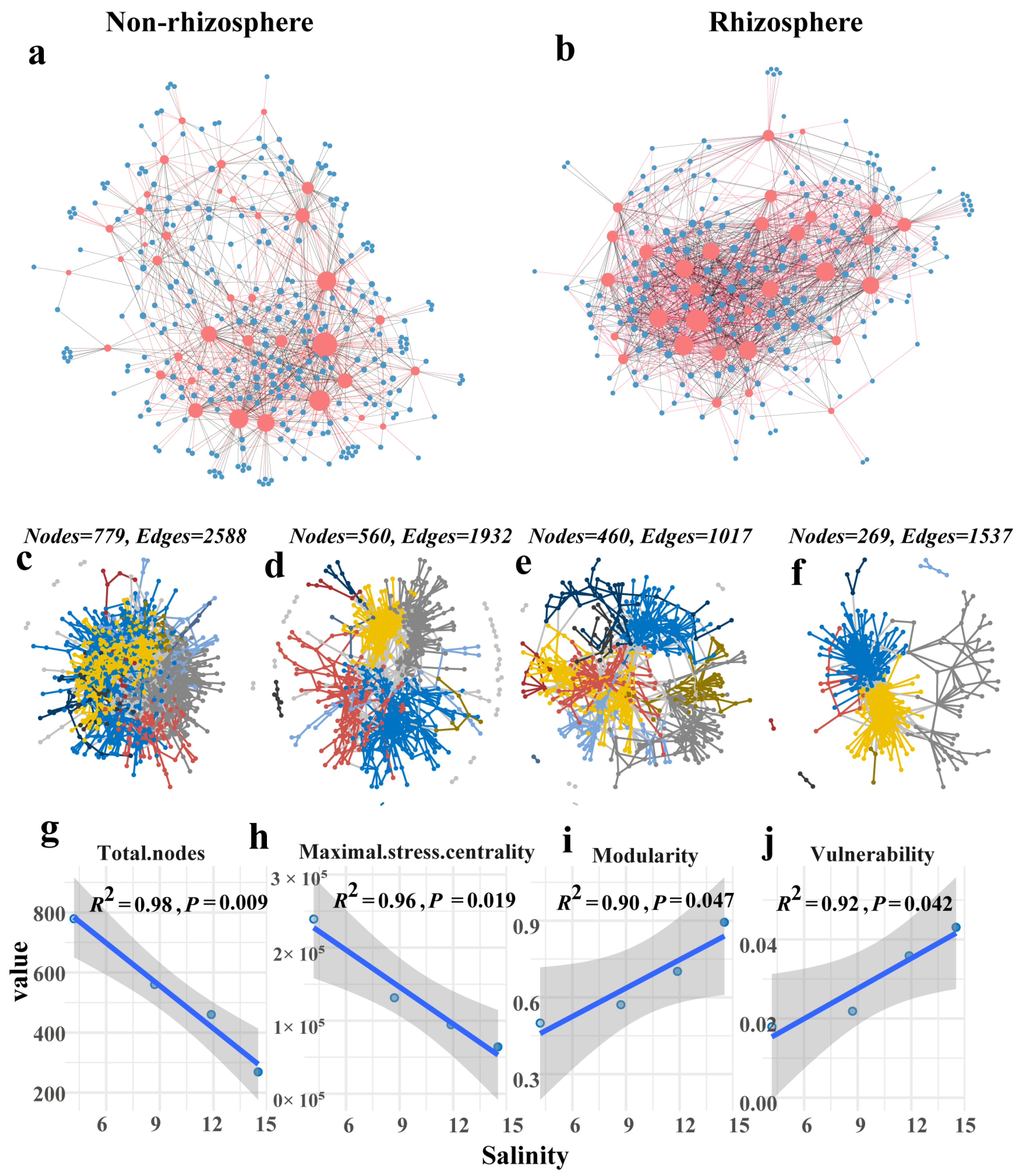
3.5. Salt Stress Destabilizes Microbiome Networks
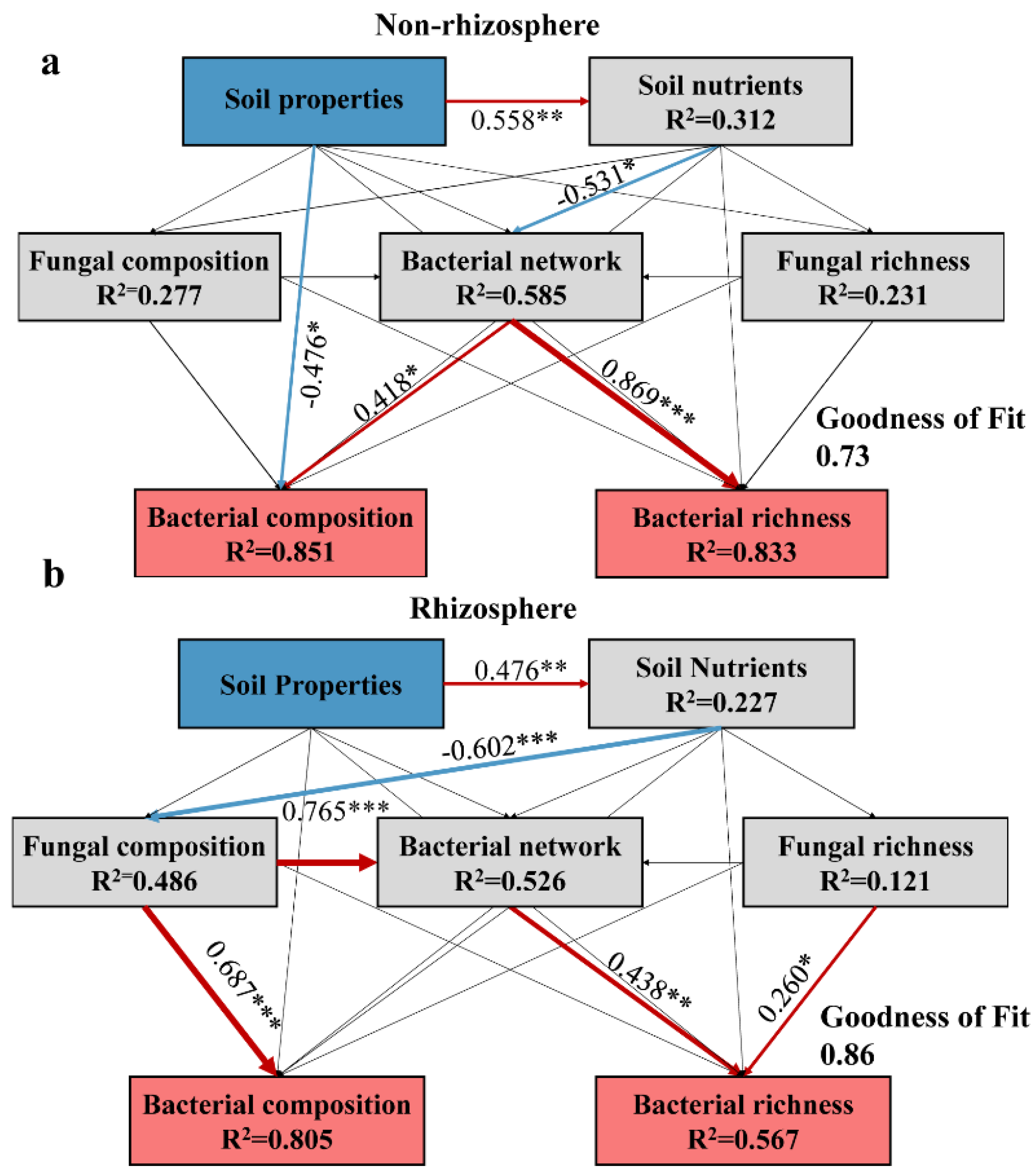
3.6. Biotic and Abiotic Factors Influence Microbiomes under Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hassani, A.; Azapagic, A.; Shokri, N. Global Predictions of Primary Soil Salinization under Changing Climate in the 21st Century. Nat. Commun. 2021, 12, 6663. [Google Scholar] [CrossRef] [PubMed]
- Saberi Riseh, R.; Ebrahimi-Zarandi, M.; Tamanadar, E.; Moradi Pour, M.; Thakur, V.K. Salinity Stress: Toward Sustainable Plant Strategies and Using Plant Growth-Promoting Rhizobacteria Encapsulation for Reducing It. Sustainability 2021, 13, 12758. [Google Scholar] [CrossRef]
- Haj-Amor, Z.; Araya, T.; Kim, D.-G.; Bouri, S.; Lee, J.; Ghiloufi, W.; Yang, Y.; Kang, H.; Jhariya, M.K.; Banerjee, A.; et al. Soil Salinity and Its Associated Effects on Soil Microorganisms, Greenhouse Gas Emissions, Crop Yield, Biodiversity and Desertification: A Review. Sci. Total Environ. 2022, 843, 156946. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, G.; Xia, Y.; Chen, L.; Jiang, Z.; Zheng, H.; Wang, Z. Use of Biochar-Compost to Improve Properties and Productivity of the Degraded Coastal Soil in the Yellow River Delta, China. J. Soils Sediments 2017, 17, 780–789. [Google Scholar] [CrossRef]
- Wichern, J.; Wichern, F.; Joergensen, R.G. Impact of Salinity on Soil Microbial Communities and the Decomposition of Maize in Acidic Soils. Geoderma 2006, 137, 100–108. [Google Scholar] [CrossRef]
- Wang, X.; Xing, X.; Zhang, F.; Xin, K. Biological Improvement of Saline Alkali Soil Reference System: A Review. Sci. Cold Arid. Reg. 2018, 10, 516–521. [Google Scholar] [CrossRef]
- Qadir, M.; Ghafoor, A.; Murtaza, G. Amelioration Strategies for Saline Soils: A Review. Land. Degrad. Dev. 2000, 11, 501–521. [Google Scholar] [CrossRef]
- FAO Global Map of Salt Affected Soils. 2021. Available online: https://www.fao.org/global-soil-partnership/gsasmap/en (accessed on 1 July 2022).
- FAO Economical Losses Due to Soil Salinization. 2021. Available online: https://www.fao.org/global-soil-partnership/areas-of-work/soil-salinity/en/ (accessed on 1 July 2022).
- Lakhdar, A.; Rabhi, M.; Ghnaya, T.; Montemurro, F.; Jedidi, N.; Abdelly, C. Effectiveness of Compost Use in Salt-Affected Soil. J. Hazard. Mater. 2009, 171, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Negacz, K.; Malek, Ž.; de Vos, A.; Vellinga, P. Saline Soils Worldwide: Identifying the Most Promising Areas for Saline Agriculture. J. Arid. Environ. 2022, 203, 104775. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, N.; Tang, H.; Adams, J.M.; Sun, B.; Liang, Y. Straw Biochar Strengthens the Life Strategies and Network of Rhizosphere Fungi in Manure Fertilized Soils. Soil. Ecol. Lett. 2019, 1, 72–84. [Google Scholar] [CrossRef]
- Morrissey, E.M.; Gillespie, J.L.; Morina, J.C.; Franklin, R.B. Salinity Affects Microbial Activity and Soil Organic Matter Content in Tidal Wetlands. Glob. Chang. Biol. 2014, 20, 1351–1362. [Google Scholar] [CrossRef]
- Hollister, E.B.; Engledow, A.S.; Hammett, A.J.M.; Provin, T.L.; Wilkinson, H.H.; Gentry, T.J. Shifts in Microbial Community Structure along an Ecological Gradient of Hypersaline Soils and Sediments. ISME J. 2010, 4, 829–838. [Google Scholar] [CrossRef]
- Yang, J.; Ma, L.; Jiang, H.; Wu, G.; Dong, H. Salinity Shapes Microbial Diversity and Community Structure in Surface Sediments of the Qinghai-Tibetan Lakes. Sci. Rep. 2016, 6, 25078. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lv, D.; Jiang, S.; Lin, H.; Sun, J.; Li, K.; Sun, J. Soil Salinity Regulation of Soil Microbial Carbon Metabolic Function in the Yellow River Delta, China. Sci. Total Environ. 2021, 790, 148258. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Jiang, N.; Wu, Y.; Yang, Z.; Bello, A.; Yang, W. Disentangling the Role of Salinity-Sodicity in Shaping Soil Microbiome along a Natural Saline-Sodic Gradient. Sci. Total Environ. 2021, 765, 142738. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Wang, T.; Kuzyakov, Y. Rhizosphere Bacteriome Structure and Functions. Nat. Commun. 2022, 13, 836. [Google Scholar] [CrossRef] [PubMed]
- Shu, W.-S.; Huang, L.-N. Microbial Diversity in Extreme Environments. Nat. Rev. Microbiol. 2022, 20, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Sokol, N.W.; Slessarev, E.; Marschmann, G.L.; Nicolas, A.; Blazewicz, S.J.; Brodie, E.L.; Firestone, M.K.; Foley, M.M.; Hestrin, R.; Hungate, B.A.; et al. Life and Death in the Soil Microbiome: How Ecological Processes Influence Biogeochemistry. Nat. Rev. Microbiol. 2022, 20, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Wagg, C.; Schlaeppi, K.; Banerjee, S.; Kuramae, E.E.; van der Heijden, M.G.A. Fungal-Bacterial Diversity and Microbiome Complexity Predict Ecosystem Functioning. Nat. Commun. 2019, 10, 4841. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, Variation, and Assembly of the Root-Associated Microbiomes of Rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Human Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and Heritability of the Maize Rhizosphere Microbiome under Field Conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, Y.; Li, T.; Zhao, D.; Liao, Y. Tillage Practices with Different Soil Disturbance Shape the Rhizosphere Bacterial Community throughout Crop Growth. Soil. Tillage Res. 2020, 197, 104501. [Google Scholar] [CrossRef]
- Li, Y.; Li, T.; Zhao, D.; Wang, Z.; Liao, Y. Different Tillage Practices Change Assembly, Composition, and Co-Occurrence Patterns of Wheat Rhizosphere Diazotrophs. Sci. Total Environ. 2021, 767, 144252. [Google Scholar] [CrossRef] [PubMed]
- He, R.; Zeng, J.; Zhao, D.; Wang, S.; Wu, Q.L. Decreased Spatial Variation and Deterministic Processes of Bacterial Community Assembly in the Rhizosphere of Phragmites Australis across the Middle–Lower Yangtze Plain. Mol. Ecol. 2022, 31, 1180–1195. [Google Scholar] [CrossRef] [PubMed]
- Fan, M.; Li, J.; Luan, X.; Yang, L.; Chen, W.; Ma, X.; Yang, Z.; Shangguan, Z. Biogeographical Patterns of Rhizosphere Microbial Communities in Robinia Pseudoacacia Forests along a North–South Transect in the Loess Plateau. Geoderma 2023, 435, 116516. [Google Scholar] [CrossRef]
- de Vries, F.T.; Wallenstein, M.D. Below-Ground Connections Underlying above-Ground Food Production: A Framework for Optimising Ecological Connections in the Rhizosphere. J. Ecol. 2017, 105, 913–920. [Google Scholar] [CrossRef]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The Interconnected Rhizosphere: High Network Complexity Dominates Rhizosphere Assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, J.; Liu, Y.; Shi, P.; Wei, G. Co-Occurrence Patterns of Soybean Rhizosphere Microbiome at a Continental Scale. Soil. Biol. Biochem. 2018, 118, 178–186. [Google Scholar] [CrossRef]
- Wang, N.; Zhao, Z.; Zhang, X.; Liu, S.; Zhang, K.; Hu, M. Plant Growth, Salt Removal Capacity, and Forage Nutritive Value of the Annual Euhalophyte Suaeda Salsa Irrigated with Saline Water. Front. Plant Sci. 2023, 13, 1040520. [Google Scholar] [CrossRef]
- Wang, S.; Zhao, Z.; Ge, S.; Zhang, K.; Tian, C.; Mai, W. The Effects of Suaeda Salsa/Zea Mays L. Intercropping on Plant Growth and Soil Chemical Characteristics in Saline Soil. Agriculture 2022, 12, 107. [Google Scholar] [CrossRef]
- Cheng, Z.; Han, Q.; He, Y.; Shi, J.; Wu, L.; Xu, J. Contrasting Response of Rice Rhizosphere Microbiomes to in Situ Cadmium-Contaminated Soil Remediation. Soil. Ecol. Lett. 2023, 6, 230203. [Google Scholar] [CrossRef]
- Guo, K.; Liu, X. Effect of Initial Soil Water Content and Bulk Density on the Infiltration and Desalination of Melting Saline Ice Water in Coastal Saline Soil. Eur. J. Soil. Sci. 2019, 70, 1249–1266. [Google Scholar] [CrossRef]
- Scales, F.; Harrison, A.P. Boric Acid Modification of the Kjeldahl Method for Crop and Soil Analysis. Ind. Eng. Chem. 1920, 12, 350–352. [Google Scholar] [CrossRef][Green Version]
- Xue, L.; Sun, B.; Yang, Y.; Jin, B.; Zhuang, G.; Bai, Z.; Zhuang, X. Efficiency and Mechanism of Reducing Ammonia Volatilization in Alkaline Farmland Soil Using Bacillus amyloliquefaciens Biofertilizer. Environ. Res. 2021, 202, 111672. [Google Scholar] [CrossRef]
- Wan, W.; Hao, X.; Xing, Y.; Liu, S.; Zhang, X.; Li, X.; Chen, W.; Huang, Q. Spatial Differences in Soil Microbial Diversity Caused by pH-Driven Organic Phosphorus Mineralization. Land. Degrad. Dev. 2021, 32, 766–776. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Y.; Song, L.; Yuan, J.; Li, W.; Zhu, Y.; Chang, S.X.; Luo, Y.; Ciais, P.; Peñuelas, J.; et al. Reduced Phosphorus Availability in Paddy Soils under Atmospheric CO2 Enrichment. Nat. Geosci. 2023, 16, 162–168. [Google Scholar] [CrossRef]
- Gosling, P.; Shepherd, M. Long-Term Changes in Soil Fertility in Organic Arable Farming Systems in England, with Particular Reference to Phosphorus and Potassium. Agric. Ecosyst. Environ. 2005, 105, 425–432. [Google Scholar] [CrossRef]
- Palanivell, P.; Ahmed, O.H.; Omar, L.; Abdul Majid, N.M. Nitrogen, Phosphorus, and Potassium Adsorption and Desorption Improvement and Soil Buffering Capacity Using Clinoptilolite Zeolite. Agronomy 2021, 11, 379. [Google Scholar] [CrossRef]
- Degnan, P.H.; Ochman, H. Illumina-Based Analysis of Microbial Community Diversity. ISME J. 2012, 6, 183–194. [Google Scholar] [CrossRef]
- Xue, F.; Liu, T. DNA Sequence and Community Structure Diversity of Multi-Year Soil Fungi in Grape of Xinjiang. Sci. Rep. 2021, 11, 16367. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Accuracy of Taxonomy Prediction for 16S rRNA and Fungal ITS Sequences. PeerJ 2018, 6, e4652. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023. [Google Scholar]
- Cao, Y.; Dong, Q.; Wang, D.; Zhang, P.; Liu, Y.; Niu, C. microbiomeMarker: An R/Bioconductor Package for Microbiome Marker Identification and Visualization. Bioinformatics 2022, 38, 4027–4029. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Shen, X.; Zhang, N.; Sun, C.; Shao, Y. Smartphones as an Ecological Niche of Microorganisms: Microbial Activities, Assembly, and Opportunistic Pathogens. Microbiol. Spectr. 2022, 10, e0150822. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Chen, L.; Ma, T.; Li, X.; Zheng, M.; Zhou, X.; Chen, L.; Qian, X.; Xi, J.; Lu, H.; et al. EasyAmplicon: An Easy-to-Use, Open-Source, Reproducible, and Community-Based Pipeline for Amplicon Data Analysis in Microbiome Research. iMeta 2023, 2, e83. [Google Scholar] [CrossRef]
- Guo, Y.; Gu, S.; Wu, K.; Tanentzap, A.J.; Yu, J.; Liu, X.; Li, Q.; He, P.; Qiu, D.; Deng, Y.; et al. Temperature-mediated Microbial Carbon Utilization in China’s Lakes. Glob. Chang. Biol. 2023, 29, 5044–5061. [Google Scholar] [CrossRef]
- Sun, B.; Bai, Z.; Li, Y.; Li, R.; Song, M.; Xu, S.; Zhang, H.; Zhuang, X. Emission Mitigation of CH4 and N2O during Semi-Permeable Membrane Covered Hyperthermophilic Aerobic Composting of Livestock Manure. J. Clean. Prod. 2022, 379, 134850. [Google Scholar] [CrossRef]
- Hartmann, A.C.; Petras, D.; Quinn, R.A.; Protsyuk, I.; Archer, F.I.; Ransome, E.; Williams, G.J.; Bailey, B.A.; Vermeij, M.J.A.; Alexandrov, T.; et al. Meta-Mass Shift Chemical Profiling of Metabolomes from Coral Reefs. Proc. Natl. Acad. Sci. USA 2017, 114, 11685–11690. [Google Scholar] [CrossRef]
- Jiao, S.; Chu, H.; Zhang, B.; Wei, X.; Chen, W.; Wei, G. Linking Soil Fungi to Bacterial Community Assembly in Arid Ecosystems. iMeta 2022, 1, e2. [Google Scholar] [CrossRef]
- Huang, H. LinkET: Everything Is Linkable. R Package Version 0.0.3 2021, 2. Available online: https://github.com/Hy4m/linkET (accessed on 1 November 2022).
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2023, 14, 927–930. [Google Scholar] [CrossRef]
- Ning, D.; Yuan, M.; Wu, L.; Zhang, Y.; Guo, X.; Zhou, X.; Yang, Y.; Arkin, A.P.; Firestone, M.K.; Zhou, J. A Quantitative Framework Reveals Ecological Drivers of Grassland Microbial Community Assembly in Response to Warming. Nat. Commun. 2020, 11, 4717. [Google Scholar] [CrossRef]
- He, J.; Zhang, N.; Muhammad, A.; Shen, X.; Sun, C.; Li, Q.; Hu, Y.; Shao, Y. From Surviving to Thriving, the Assembly Processes of Microbial Communities in Stone Biodeterioration: A Case Study of the West Lake UNESCO World Heritage Area in China. Sci. Total Environ. 2022, 805, 150395. [Google Scholar] [CrossRef]
- He, J.; Zhang, N.; Shen, X.; Muhammad, A.; Shao, Y. Deciphering Environmental Resistome and Mobilome Risks on the Stone Monument: A Reservoir of Antimicrobial Resistance Genes. Sci. Total Environ. 2022, 838, 156443. [Google Scholar] [CrossRef] [PubMed]
- Watts, S.C.; Ritchie, S.C.; Inouye, M.; Holt, K.E. FastSpar: Rapid and Scalable Correlation Estimation for Compositional Data. Bioinformatics 2019, 35, 1064–1066. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Peng, X.; Zhang, Z.; Gu, S.; He, Q.; Shen, W.; Wang, Z.; Wang, D.; Hu, Q.; Li, Y.; et al. iNAP: An Integrated Network Analysis Pipeline for Microbiome Studies. iMeta 2022, 1, e13. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T.; Traag, V.; Horvát, S.; Zanini, F.; Noom, D.; Müller, K. Igraph: Network Analysis and Visualization in R. 2023. Available online: https://zenodo.org/records/10681749 (accessed on 1 October 2022).
- Liao, H.; Zhao, Q.; Cui, P.; Chen, Z.; Yu, Z.; Geisen, S.; Friman, V.-P.; Zhou, S. Efficient Reduction of Antibiotic Residues and Associated Resistance Genes in Tylosin Antibiotic Fermentation Waste Using Hyperthermophilic Composting. Environ. Int. 2019, 133, 105203. [Google Scholar] [CrossRef]
- Liao, H.; Lu, X.; Rensing, C.; Friman, V.P.; Geisen, S.; Chen, Z.; Yu, Z.; Wei, Z.; Zhou, S.; Zhu, Y. Hyperthermophilic Composting Accelerates the Removal of Antibiotic Resistance Genes and Mobile Genetic Elements in Sewage Sludge. Environ. Sci. Technol. 2018, 52, 266–276. [Google Scholar] [CrossRef]
- Mo, Y.; Peng, F.; Gao, X.; Xiao, P.; Logares, R.; Jeppesen, E.; Ren, K.; Xue, Y.; Yang, J. Low Shifts in Salinity Determined Assembly Processes and Network Stability of Microeukaryotic Plankton Communities in a Subtropical Urban Reservoir. Microbiome 2021, 9, 128. [Google Scholar] [CrossRef]
- Neill, M. A Method to Determine Which Nutrient Is Limiting for Plant Growth in Estuarine Waters—At Any Salinity. Mar. Pollut. Bull. 2005, 50, 945–955. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, G.; Smolders, A.J.P.; Loeb, R.; Bout, A.; Roelofs, J.G.M.; Lamers, L.P.M. Salinization of Coastal Freshwater Wetlands; Effects of Constant versus Fluctuating Salinity on Sediment Biogeochemistry. Biogeochemistry 2015, 126, 71–84. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.; Banerjee, S.; Zhou, N.; Zhao, Z.; Zhang, K.; Hu, M.; Tian, C. Biogeographical Distribution of Bacterial Communities in Saline Agricultural Soil. Geoderma 2020, 361, 114095. [Google Scholar] [CrossRef]
- Yang, T.; Tedersoo, L.; Liu, X.; Gao, G.-F.; Dong, K.; Adams, J.M.; Chu, H. Fungi Stabilize Multi-Kingdom Community in a High Elevation Timberline Ecosystem. iMeta 2022, 1, e49. [Google Scholar] [CrossRef]
- Boer, W.d.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a Fungal World: Impact of Fungi on Soil Bacterial Niche Development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Xu, L.; Montoya, L.; Madera, M.; Hollingsworth, J.; Chen, L.; Purdom, E.; Singan, V.; Vogel, J.; Hutmacher, R.B.; et al. Co-Occurrence Networks Reveal More Complexity than Community Composition in Resistance and Resilience of Microbial Communities. Nat. Commun. 2022, 13, 3867. [Google Scholar] [CrossRef] [PubMed]
- Ehrenfeld, J.G.; Ravit, B.; Elgersma, K. Feedback in the Plant-Soil System. Annu. Rev. Environ. Resour. 2005, 30, 75–115. [Google Scholar] [CrossRef]
- Dong, C.-J.; Wang, L.-L.; Li, Q.; Shang, Q.-M. Bacterial Communities in the Rhizosphere, Phyllosphere and Endosphere of Tomato Plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef]
- Fu, W.; Chen, B.; Rillig, M.C.; Jansa, J.; Ma, W.; Xu, C.; Luo, W.; Wu, H.; Hao, Z.; Wu, H.; et al. Community Response of Arbuscular Mycorrhizal Fungi to Extreme Drought in a Cold-Temperate Grassland. New Phytol. 2022, 234, 2003–2017. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.-G.; Wang, J.-T.; Singh, B.; Han, L.-L.; Shen, J.-P.; Li, P.-P.; Wang, G.-B.; Wu, C.-F.; Ge, A.-H.; et al. Host Selection Shapes Crop Microbiome Assembly and Network Complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhang, L.; Zhou, J.; George, T.S.; Feng, G. Arbuscular Mycorrhizal Fungi Enhance Mineralisation of Organic Phosphorus by Carrying Bacteria along Their Extraradical Hyphae. New Phytol. 2021, 230, 304–315. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Jiao, H.; Sun, B.; Song, M.; Yan, G.; Bai, Z.; Wang, J.; Zhuang, X.; Hu, Q. Understanding Salinity-Driven Modulation of Microbial Interactions: Rhizosphere versus Edaphic Microbiome Dynamics. Microorganisms 2024, 12, 683. https://doi.org/10.3390/microorganisms12040683
Li R, Jiao H, Sun B, Song M, Yan G, Bai Z, Wang J, Zhuang X, Hu Q. Understanding Salinity-Driven Modulation of Microbial Interactions: Rhizosphere versus Edaphic Microbiome Dynamics. Microorganisms. 2024; 12(4):683. https://doi.org/10.3390/microorganisms12040683
Chicago/Turabian StyleLi, Rui, Haihua Jiao, Bo Sun, Manjiao Song, Gaojun Yan, Zhihui Bai, Jiancheng Wang, Xuliang Zhuang, and Qing Hu. 2024. "Understanding Salinity-Driven Modulation of Microbial Interactions: Rhizosphere versus Edaphic Microbiome Dynamics" Microorganisms 12, no. 4: 683. https://doi.org/10.3390/microorganisms12040683
APA StyleLi, R., Jiao, H., Sun, B., Song, M., Yan, G., Bai, Z., Wang, J., Zhuang, X., & Hu, Q. (2024). Understanding Salinity-Driven Modulation of Microbial Interactions: Rhizosphere versus Edaphic Microbiome Dynamics. Microorganisms, 12(4), 683. https://doi.org/10.3390/microorganisms12040683








