EnvC Homolog Encoded by Xanthomonas citri subsp. citri Is Necessary for Cell Division and Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids and Culture Condition
2.2. Mutant Construction
2.2.1. Partial Deletion of XAC0024 Nucleotide Sequence
2.2.2. Deletion Vector Construction
2.2.3. Mutant Generation
2.3. Mutant Complementation
2.4. Sub-Cellular Localization
2.4.1. Vector Construction
2.4.2. Fluorescence Microscopy
2.5. Pathogenicity and Bacterial Viability Analyses
2.6. Growth Curves
2.7. Phylogenetic Analyses and Protein Modeling
3. Results
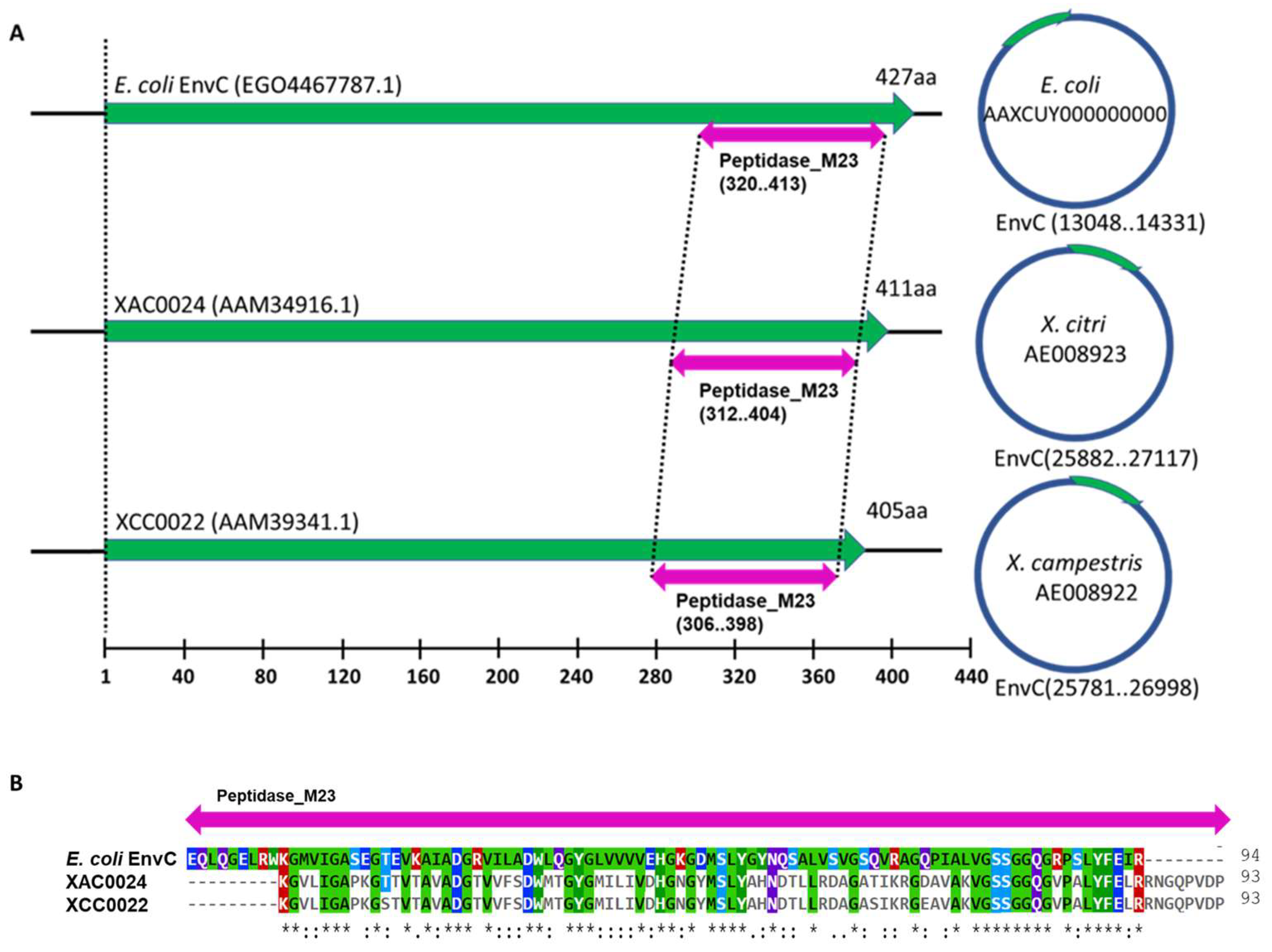
3.1. X. citri Encodes an EnvC Homolog, Which Is Conserved in MANY Bacteria
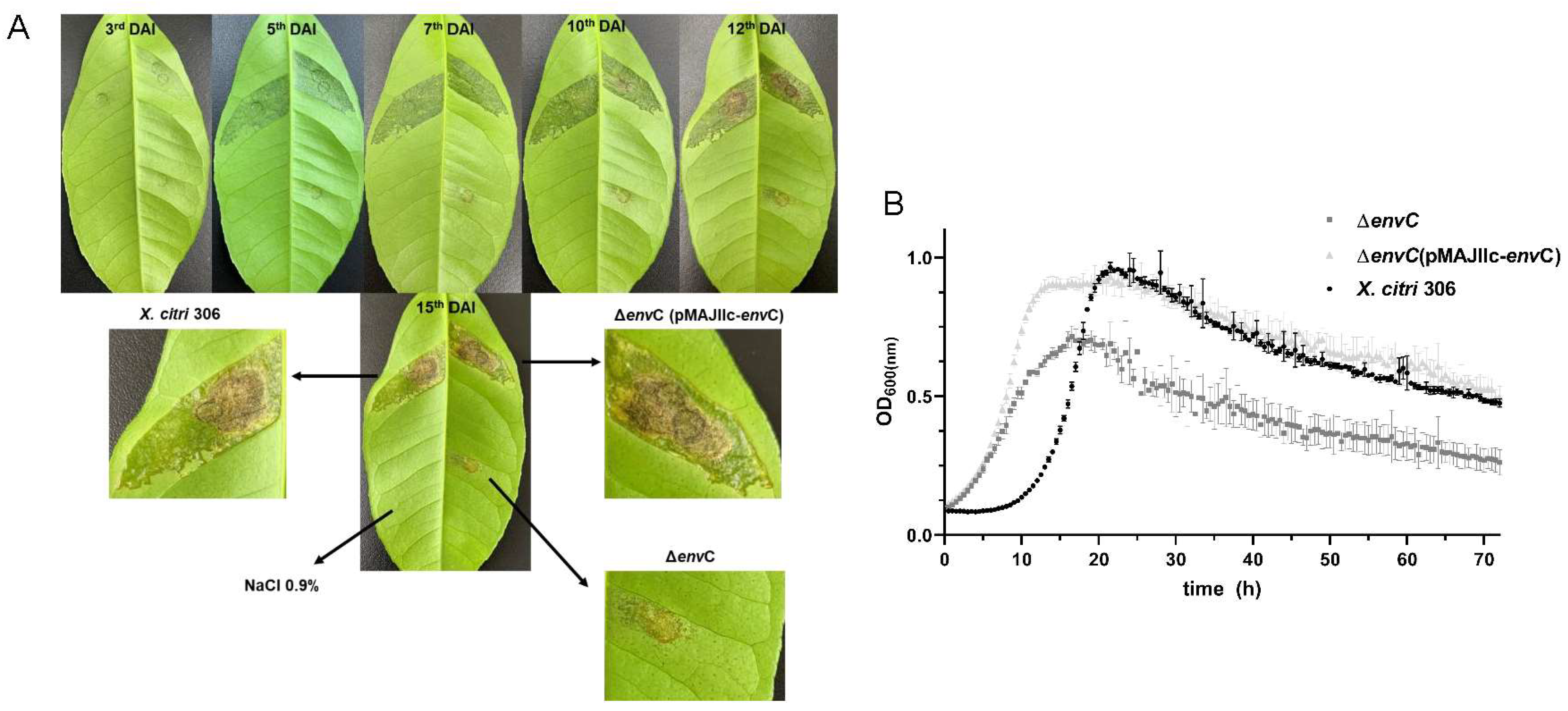
3.2. Disruption of envC Affects X. citri Virulence
3.3. EnvC Is Required for X. citri Daughter-Cell Separation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Das, A.K. Citrus Canker—A review. J. Appl. Hortic. 2003, 5, 52–60. [Google Scholar] [CrossRef]
- Vollmer, W.; Blanot, D.; de Pedro, M.A. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008, 32, 149–167. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Frirdich, E.; Gaynor, E.C. Peptidoglycan hydrolases, bacterial shape, and pathogenesis. Curr. Opin. Microbiol. 2013, 16, 767–778. [Google Scholar] [CrossRef]
- Firczuk, M.; Bochtler, M. Folds and activities of peptidoglycan amidases. FEMS Microbiol. Rev. 2007, 31, 676–691. [Google Scholar] [CrossRef]
- Bernhardt, T.G.; de Boer, P.A. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 2003, 5, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Dinh, T.; Bernhardt, T.G. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J. Bacteriol. 2009, 191, 5094–5107. [Google Scholar] [CrossRef] [PubMed]
- Uehara, T.; Parzych, K.R.; Dinh, T.; Bernhardt, T.G. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010, 29, 1412–1422. [Google Scholar] [CrossRef]
- Wu, C.; Al Mamun, A.A.M.; Luong, T.T.; Hu, B.; Gu, J.; Lee, J.H.; D’Amore, M.; Das, A.; Ton-That, H. Forward Genetic Dissection of Biofilm Development by Fusobacterium nucleatum: Novel Functions of Cell Division Proteins FtsX and EnvC. mBio 2018, 9, e00360-18. [Google Scholar] [CrossRef]
- Möll, A.; Dörr, T.; Alvarez, L.; Chao, M.C.; Davis, B.M.; Cava, F.; Waldor, M.K. Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. J. Bacteriol. 2014, 196, 3937–3948. [Google Scholar] [CrossRef]
- Warr, A.R.; Hubbard, T.P.; Munera, D.; Blondel, C.; Wiesch, P.A.Z.; Abel, S.; Wang, X.; Davis, B.M.; Waldor, M.K. Transposon-insertion sequencing screens unveil requirements for EHEC growth and intestinal colonization. PLoS Pathog. 2019, 15, e1007652. [Google Scholar] [CrossRef] [PubMed]
- Yakhnina, A.A.; Mcmanus, H.R.; Bernhardt, T.G. The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability. Mol. Microbiol. 2015, 97, 957–973. [Google Scholar] [CrossRef]
- Yang, L.C.; Gan, Y.L.; Yang, L.Y.; Jiang, B.L.; Tang, J.L. Peptidoglycan hydrolysis mediated by the amidase AmiC and its LytM activator NlpD is critical for cell separation and virulence in the phytopathogen Xanthomonas campestris. Mol. Plant Pathol. 2018, 19, 1705–1718. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.C.; Ferro, J.A.; Reinach, F.C.; Farah, C.S.; Furlan, L.R.; Quaggio, R.B.; Monteiro-Vitorello, C.B.; Van Sluys, M.A.; Almeida, N.F.; Alves, L.M.; et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 2002, 417, 459–463. [Google Scholar] [CrossRef]
- Huguet, E.; Hahn, K.; Wengelnik, K.; Bonas, U. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiol. 1998, 29, 1379–1390. [Google Scholar] [CrossRef]
- Pena, M.M.; Teper, D.; Ferreira, H.; Wang, N.; Sato, K.U.; Ferro, M.I.T.; Ferro, J.A. mCherry fusions enable the subcellular localization of periplasmic and cytoplasmic proteins in Xanthomonas sp. PLoS ONE 2020, 15, e0236185. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; Volume 1. [Google Scholar]
- Lee, J.; Lee, H.-J.; Shin, M.-K.; Ryu, W.-S. Versatile PCR-mediated insertion or deletion mutagenesis. BioTechniques 2004, 36, 398–400. [Google Scholar] [CrossRef]
- do Amaral, A.M.; Toledo, C.P.; Baptista, J.C.; Machado, M.A. Transformation of Xanthomonas axonopodis pv. citri by electroporation. Fitopatol. Bras. 2005, 30, 292–294. [Google Scholar] [CrossRef]
- Kaniga, K.; Delor, I.; Cornelis, G.R. A wide-host-range suicide vector for improving reverse genetics in Gram-negative bacteria: Inactivation of the blaA gene of Yersinia enterocolitica. Gene 1991, 109, 137–141. [Google Scholar] [CrossRef]
- Martins, P.M.M.; Lau, I.F.; Bacci, M., Jr.; Belasque, J.; Do Amaral, A.M.; Taboga, S.R.; Ferreira, H. Subcellular localization of proteins labeled with GFP in Xanthomonas citri ssp. citri: Targeting the division septum. FEMS Microbiol. Lett. 2010, 310, 76–83. [Google Scholar] [CrossRef]
- Morão, L.G.; Polaquini, C.R.; Kopacz, M.; Torrezan, G.S.; Ayusso, G.M.; Dilarri, G.; Cavalca, L.B.; Zielinska, A.; Scheffers, D.-J.; Regasini, L.O.; et al. A simplified curcumin targets the membrane of Bacillus subtilis. MicrobiologyOpen 2019, 8, e683. [Google Scholar] [CrossRef] [PubMed]
- Lacerda, L.A.; Cavalca, L.B.; Martins, P.M.M.; Govone, J.S.; Bacci, M., Jr.; Ferreira, H. Protein depletion using the arabinose promoter in Xanthomonas citri subsp. citri. Plasmid 2017, 90, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the Gateway Computing Environments Workshop, New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Meng, E.C.; Goddard, T.D.; Pettersen, E.F.; Couch, G.S.; Pearson, Z.J.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Tools for structure building and analysis. Protein Sci. 2023, 32, e4792. [Google Scholar] [CrossRef]
- The Xanthomonas sp. Portal—INRA. Available online: https://iant.toulouse.inra.fr/bacteria/annotation/cgi/xansp.cgi (accessed on 24 July 2023).
- Timilsina, S.; Potnis, N.; Newberry, E.A.; Liyanapathiranage, P.; Iruegas-Bocardo, F.; White, F.F.; Goss, E.M.; Jones, J.B. Xanthomonas diversity, virulence and plant–pathogen interactions. Nat. Rev. Microbiol. 2020, 18, 415–427. [Google Scholar] [CrossRef]
- Rodriguez-R, L.M.; Grajales, A.; Arrieta-Ortiz, M.L.; Salazar, C.; Restrepo, S.; Bernal, A. Genomes-based phylogeny of the genus Xanthomonas. BMC Microbiol. 2012, 12, 43. [Google Scholar] [CrossRef]
- Timilsina, S.; Kara, S.; Jacques, M.A.; Potnis, N.; Minsavage, G.V.; Vallad, G.E.; Jones, J.B.; Fischer-Le Saux, M. Reclassification of Xanthomonas gardneri (ex Šutič 1957) Jones et al. 2006 as a later heterotypic synonym of Xanthomonas cynarae Trébaol et al. 2000 and description of X. cynarae pv. cynarae and X. cynarae pv. gardneri based on whole genome analyses. Int. J. Syst. Evol. Microbiol. 2019, 69, 343–349. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Studer, R.A.; Robinson-Rechavi, M.; Dessimoz, C. Resolving the ortholog conjecture: Orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 2012, 8, e1002514. [Google Scholar] [CrossRef] [PubMed]
- Freiesleben, U.V.; Krekling, M.A.; Hansen, F.G.; Lobner-Olesen, A. The eclipse period of Escherichia coli. EMBO J. 2000, 19, 6240–6248. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lahti, J.M.; Air, G.M.; Burrows, P.D.; Cooper, M.D. Molecular cloning of the murine BP-1/6C3 antigen: A member of the zinc-dependent metallopeptidase family. Proc. Natl. Acad. Sci. USA 1990, 87, 993–997. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, C.; Templin, M.F.; Ursinus, A.; Merdanovic, M.; Berger, J.; SCHWARZ, H.; de Pedro, M.A.; Holtje, J.V. Involvement of N-acetylmuramyl-L-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli. Mol. Microbiol. 2001, 41, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Peterson, T.N.; Winther, O.; Brunak, S.; Heijne, G.V.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Brunings, A.M.; Gabriel, D.W. Xanthomonas citri: Breaking the surface. Mol. Plant Pathol. 2003, 4, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Jha, G.; Rajeshwari, R.; Sonti, R.V. Bacterial Type Two Secretion System Secreted Proteins: Double-Edged Swords for Plant Pathogens. Mol. Plant Microbe Interact. 2005, 18, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Büttner, D.; Bonas, U. Regulation and secretion of Xanthomonas virulence factors. FEMS Microbiol. Rev. 2010, 34, 107–133. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.R.; Facincani, A.P.; Ferreira, R.M.; Moreira, L.M.; De Oliveira, J.C.F.; Ferro, J.A.; Ferro, M.I.T.; Meneghini, R.; Gozzo, F.C. Proteome of the phytopathogen Xanthomonas citri subsp. citri: A global expression profile. Proteome Sci. 2010, 8, 55. [Google Scholar] [CrossRef]
- Ryan, R.P.; Vorhölter, F.J.; Potnis, N.; Jones, J.B.; Sluys, M.-A.V.; Bogdanove, A.J.; Dow, J.M. Pathogenomics of Xanthomonas: Understanding bacterium–plant interactions. Nat. Rev. Microbiol. 2011, 9, 344–355. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Wang, N. The ColR/ColS two-component system plays multiple roles in the pathogenicity of the citrus canker pathogen Xanthomonas citri subsp. citri. J. Bacteriol. 2011, 193, 1590–1599. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hu, X.; Li, J.; Wang, N. A Novel Periplasmic Protein, VrpA, Contributes to Efficient Protein Secretion by the Type III Secretion System in Xanthomonas spp. Mol. Plant Microbe Interact. 2015, 28, 143–153. [Google Scholar] [CrossRef] [PubMed]
- Zimaro, T.; Thomas, L.; Marondedze, C.; Sgro, G.G.; Garofalo, C.G.; Ficarra, F.A.; Gehring, C.; Ottado, J.; Gottig, N. The type III protein secretion system contributes to Xanthomonas citri subsp. citri biofilm formation. BMC Microbiol. 2014, 14, 96. [Google Scholar] [CrossRef] [PubMed]



| Cell Length µm | Filaments % | Minicells % | |
|---|---|---|---|
| X. citri 306 | 1.18 ± 0.22 a | 0 | 0 |
| ΔenvC | 1.89 ± 0.38 b | 55.5 | 5.25 |
| ΔenvC pMAJIIc-envC | 1.22 ± 0.29 a | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pena, M.M.; Martins, T.Z.; Teper, D.; Zamuner, C.; Alves, H.A.; Ferreira, H.; Wang, N.; Ferro, M.I.T.; Ferro, J.A. EnvC Homolog Encoded by Xanthomonas citri subsp. citri Is Necessary for Cell Division and Virulence. Microorganisms 2024, 12, 691. https://doi.org/10.3390/microorganisms12040691
Pena MM, Martins TZ, Teper D, Zamuner C, Alves HA, Ferreira H, Wang N, Ferro MIT, Ferro JA. EnvC Homolog Encoded by Xanthomonas citri subsp. citri Is Necessary for Cell Division and Virulence. Microorganisms. 2024; 12(4):691. https://doi.org/10.3390/microorganisms12040691
Chicago/Turabian StylePena, Michelle M., Thaisa Z. Martins, Doron Teper, Caio Zamuner, Helen A. Alves, Henrique Ferreira, Nian Wang, Maria Inês T. Ferro, and Jesus A. Ferro. 2024. "EnvC Homolog Encoded by Xanthomonas citri subsp. citri Is Necessary for Cell Division and Virulence" Microorganisms 12, no. 4: 691. https://doi.org/10.3390/microorganisms12040691
APA StylePena, M. M., Martins, T. Z., Teper, D., Zamuner, C., Alves, H. A., Ferreira, H., Wang, N., Ferro, M. I. T., & Ferro, J. A. (2024). EnvC Homolog Encoded by Xanthomonas citri subsp. citri Is Necessary for Cell Division and Virulence. Microorganisms, 12(4), 691. https://doi.org/10.3390/microorganisms12040691





