Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling of Biofilms
2.2. Preparation of Culture Medium
2.3. Ex Situ Biofilm Model
2.4. Surfactants
2.5. Extraction of DNA
2.6. qPCR and High-Resolution Melt Analysis (HRMA)
2.7. Determination of Relative Class I Integron Prevalence
2.8. Calculation of Euclidian Distance from HRMA as a Measure of Bacterial Community Dissimilarity
2.9. 16S rRNA Gene Sequencing and Determination of the Dissimilarity of Bacterial Communities
2.10. Statistics
3. Results
3.1. Influence of Surfactants on intI1 Prevalence in a Developing Biofilm
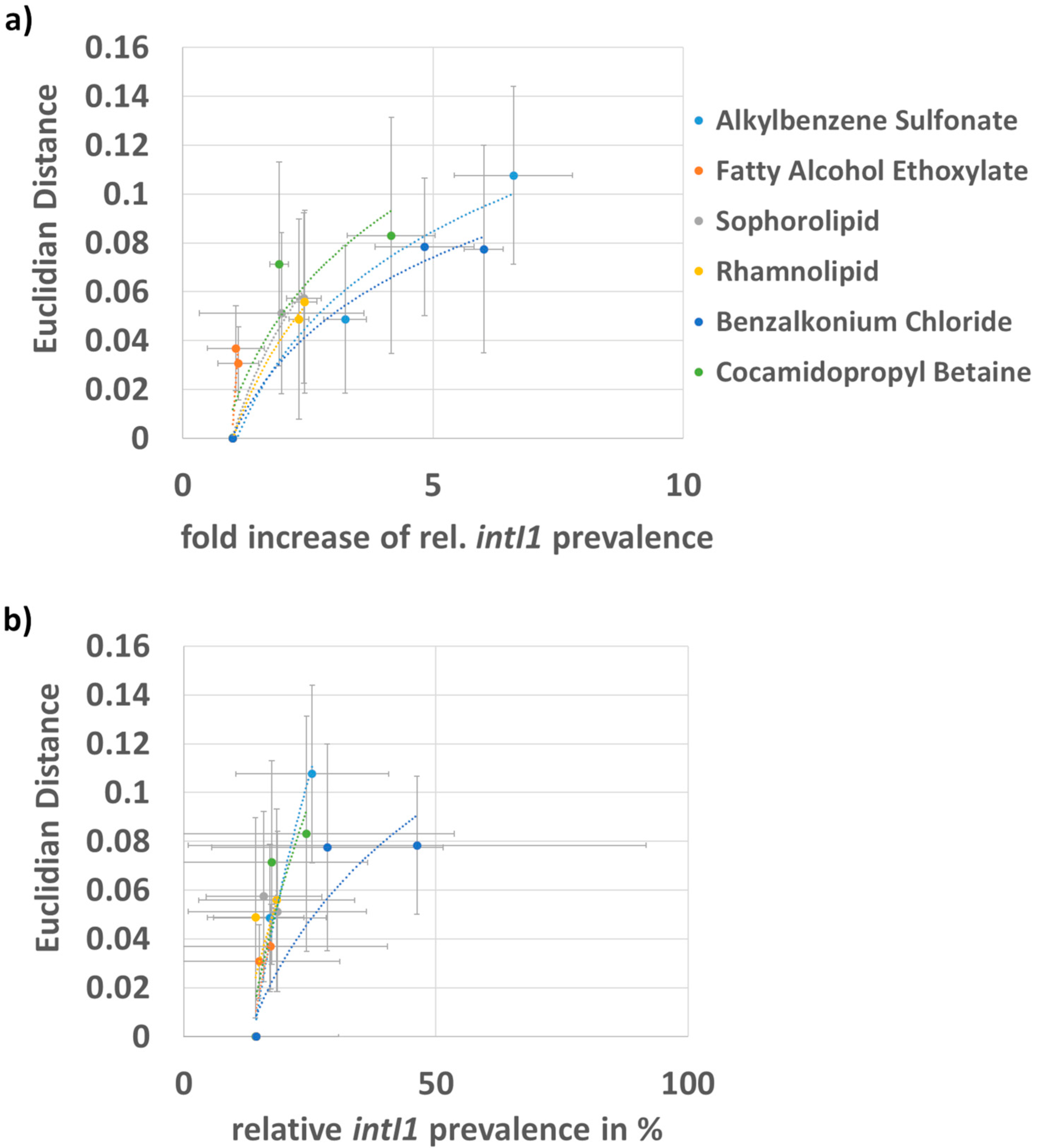
3.2. Correlation of the Fold Increase in intI1 Prevalence and Shift in Bacterial Community
3.3. 16S rRNA Gene Sequencing Results
3.4. Correlation of CMC and intI1 Prevalence
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance Collaborators. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 339, 629–655. [Google Scholar] [CrossRef]
- Liu, C.; Goh, S.G.; You, L.; Yuan, Q.; Mohapatra, S.; Gin, K.Y.; Chen, B. Low concentration quaternary ammonium compounds promoted antibiotic resistance gene transfer via plasmid conjugation. Sci. Total Environ. 2023, 887, 163781. [Google Scholar] [CrossRef] [PubMed]
- Lucassen, R.; Rehberg, L.; Heyden, M.; Bockmühl, D. Strong correlation of total phenotypic resistance of samples from household environments and the prevalence of class 1 integrons suggests for the use of the relative prevalence of intI1 as a screening tool for multi-resistance. PLoS ONE 2019, 14, e0218277. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Schages, L.; Lucassen, R.; Wichern, F.; Kalscheuer, R.; Bockmühl, D. The Household Resistome: Frequency of β-Lactamases, Class 1 Integrons, and Antibiotic-Resistant Bacteria in the Domestic Environment and Their Reduction during Automated Dishwashing and Laundering. Appl. Environ. Microbiol. 2020, 86, e02062-20. [Google Scholar] [CrossRef] [PubMed]
- Kalia, V.C.; Patel, S.K.S.; Lee, J.K. Bacterial biofilm inhibitors: An overview. Ecotoxicol. Environ. Saf. 2023, 264, 115389. [Google Scholar] [CrossRef]
- Hamzah, M.A.A.M.; Aruldass, C.A.; Ahmad, W.A.; Setu, S.A. Effects of surfactants on antibacterial drugs–A brief review. Malays. J. Fundam. Appl. Sci. 2017, 13, 118–123. [Google Scholar] [CrossRef]
- Falk, N.A. Surfactants as Antimicrobials: A Brief Overview of Microbial Interfacial Chemistry and Surfactant Antimicrobial Activity. J. Surfactants Deterg. 2019, 22, 1119–1127. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.P.; Snelling, W.J.; Dooley, J.S.; Ternan, N.G.; Banat, I.M.; Arnscheidt, J.; Hunter, W.R. Biological and synthetic surfactant exposure increases antimicrobial gene occurrence in a freshwater mixed microbial biofilm environment. Microbiologyopen 2023, 12, e1351. [Google Scholar] [CrossRef]
- Zheng, C.W.; Luo, Y.H.; Long, X.; Gu, H.; Cheng, J.; Zhang, L.; Lai, Y.J.S.; Rittmann, B.E. The structure of biodegradable surfactants shaped the microbial community, antimicrobial resistance, and potential for horizontal gene transfer. Water Res. 2023, 236, 119944. [Google Scholar] [CrossRef]
- Rojas-Herrera, R.A.; Ramos-Castillo, A.S.; Estrada-Medina, H.; De los Santos-Briones, C.; Keb-Llanes, M.A.; Barrientos-Medina, R.C.; Peña-Ramírez, Y.J.; O’Connor-Sánchez, A. Living with detergents: Pyrosequencing-based assessment of bacterial community structures in soils subjected for decades to contamination by detergents. Ann. Microbiol. 2015, 65, 1313–1322. [Google Scholar] [CrossRef]
- Marathe, N.P.; Shetty, S.A.; Shouche, Y.S.; Larsson, D.G.J. Limited Bacterial Diversity within a Treatment Plant Receiving Antibiotic-Containing Waste from Bulk Drug Production. PLoS ONE 2016, 11, e0165914. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.W.; Alkatheeri, A.H.S.; Ali, N.; Tay, Z.H.; Lee, Y.L.; Paramasivam, S.J.; Jeevaratnam, K.; Low, W.Y.; Lim, S.H.E. Association of antimicrobial resistance and gut microbiota composition in human and non-human primates at an urban ecotourism site. Gut Pathog. 2020, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Du, G.; Qiao, Z.; Yang, Y.; Shi, H.; Zhang, D.; Pan, X. Environmental concentrations of surfactants as a trigger for climax of horizonal gene transfer of antibiotic resistance. Heliyon 2023, 9, e17034. [Google Scholar] [CrossRef] [PubMed]
- Hocquet, D.; Llanes, C.; Thouverez, M.; Kulasekara, H.D.; Bertrand, X.; Plésiat, P.; Mazel, D.; Miller, S.I. Evidence for induction of integron-based antibiotic resistance by the SOS response in a clinical setting. PLoS Pathog. 2012, 8, e1002778. [Google Scholar] [CrossRef] [PubMed]
- Baltazar, M.; Bourgeois-Nicolaos, N.; Larroudé, M.; Couet, W.; Uwajeneza, S.; Doucet-Populaire, F.; Ploy, M.C.; Da Re, S. Activation of class 1 integron integrase is promoted in the intestinal environment. PLOS Genet. 2022, 18, e1010177. [Google Scholar] [CrossRef] [PubMed]
- Ledwoch, K.; Robertson, A.; Lauran, J.; Norville, P.; Maillard, J.-Y. It’s a trap! The development of a versatile drain biofilm model and its susceptibility to disinfection. J. Hosp. Infect. 2020, 106, 757–764. [Google Scholar] [CrossRef]
- van Leuven, N.; Zinn, M.K.; Lucassen, R.; Lipski, A.; Flemming, H.C.; Bockmühl, D. High resolution ITS amplicon melting analysis as a tool to analyse microbial communities of household biofilms in ex-situ models. J. Microbiol. Methods 2023, 212, 106806. [Google Scholar] [CrossRef]
- Nagtode, V.S.; Cardoza, C.; Yasin, H.K.A.; Mali, S.N.; Tambe, S.M.; Roy, P.; Singh, K.; Goel, A.; Amin, P.D.; Thorat, B.R.; et al. Green Surfactants (Biosurfactants): A Petroleum-Free Substitute for Sustainability—Comparison, Applications, Market, and Future Prospects. ACS Omega 2023, 8, 11674–11699. [Google Scholar] [CrossRef]
- Georgiev, G.A.; Yokoi, N.; Koev, K.; Kutsarova, E.; Ivanova, S.; Kyumurkov, A.; Jordanova, A.; Krastev, R.; Lalchev, Z. Surface chemistry study of the interactions of benzalkonium chloride with films of meibum, corneal cells lipids, and whole tears. Invest. Ophthalmol. Vis. Sci. 2011, 52, 4645–4654. [Google Scholar] [CrossRef]
- Mathioudakis, G.N.; Soto Beobide, A.; Bokias, G.; Koutsoukos, P.G.; Voyiatzis, G.A. Surface-enhanced Raman scattering as a tool to study cationic surfactants exhibiting low critical micelle concentration. J. Raman Spectrosc. 2019, 51, 452–460. [Google Scholar] [CrossRef]
- Zheng, W.; Huyan, J.; Tian, Z.; Zhang, Y.; Wen, X. Clinical class 1 integron-integrase gene-A promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant. Environ. Int. 2020, 135, 105372. [Google Scholar] [CrossRef] [PubMed]
- Lebuhn, M.; Hanreich, A.; Klocke, M.; Schlüter, A.; Bauer, C.; Pérez, C.M. Towards molecular biomarkers for biogas production from lignocellulose-rich substrates. Anaerobe 2014, 29, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Andini, N.; Wang, B.; Athamanolap, P.; Hardick, J.; Masek, B.J.; Thair, S.; Hu, A.; Avornu, G.; Peterson, S.; Cogill, S.; et al. Microbial Typing by Machine Learned DNA Melt Signatures. Sci. Rep. 2017, 7, 42097. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Zhang, L.; Abdouslam, N.A.; Hawkey, P.M.; Calvo-Bado, L.; Royle, J.; Brown, H.; Davis, S.; Kay, P.; Boxall, A.B.; et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011, 5, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Stalder, T.; Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.C. Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 2012, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.J.; Wearsch, P.A.; Veloo, A.C.M.; Rodriguez-Palacios, A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020, 11, 906. [Google Scholar] [CrossRef] [PubMed]
- Jabeen, I.; Islam, S.; Hassan, A.I.; Tasnim, Z.; Shuvo, S.R. A Brief Insight into Citrobacter Species-a Growing Threat to Public Health. Frontiers 2023, 2, 1276982. Available online: https://www.frontiersin.org/articles/10.3389/frabi.2023.1276982/full (accessed on 1 February 2024). [CrossRef]
- Marathe, N.P.; Salvà-Serra, F.; Karlsson, R.; Larsson, D.G.J.; Moore, E.R.B.; Svensson-Stadler, L.; Jakobsson, H.E. Scandinavium goeteborgense gen. nov., sp. nov., a New Member of the Family Enterobacteriaceae Isolated From a Wound Infection, Carries a Novel Quinolone Resistance Gene Variant. Front. Microbiol. 2019, 10, 2511. [Google Scholar] [CrossRef]
- Gao, X.; Wei, J.; Hao, T.; Yang, T.; Han, X.; Li, M.; Li, X.; Xiong, D.; Zhang, X. Dysgonomonas mossii Strain Shenzhen WH 0221, a New Member of the Genus Dysgonomonas Isolated from the Blood of a Patient with Diabetic Nephropathy, Exhibits Multiple Antibiotic Resistance. Microbiol Spectr. 2022, 10, e0238121. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Vaiwala, R.; Parthasarathi, S.; Patil, N.; Verma, A.; Waskar, M.; Raut, J.S.; Basu, J.K.; Ayappa, K.G. Interactions of surfactants with the bacterial cell wall and inner membrane: Revealing the link between aggregation and antimicrobial activity. bioRxiv 2021. [CrossRef] [PubMed]



| Surfactant | CMC [g/L] | CMC [%] | Source |
|---|---|---|---|
| Fatty alcohol ethoxylate (Marlipal 24/70) | 0.012 | 0.0012 | TDS |
| Rhamnolipid (Rewoferm RL 100) | 0.02 | 0.002 | [19] |
| Sophorolipid (Rewoferm SL ONE) | 0.08 | 0.008 | [19] |
| Cocamidopropyl betaine (Dehyton PK 45) | 0.1 | 0.01 | TDS |
| Alkylbenzene sulfonate (Disponil LDBS 55) | 1 | 0.1 | TDS |
| Benzalkonium chloride | 0.05–0.1 | 0.005–0.1 | [20,21] |
| UniFrac Distance (Unweighted) | Mean |
|---|---|
| Medium + alkylbenzene sulfonate 0.1% | 0.65 |
| Medium + benzalkonium chloride 0.01% | 0.51 |
| Medium + cocamidopropyl betaine 0.1% | 0.37 |
| Medium + sophorolipid 0.1% | 0.34 |
| Medium + rhamnolipid 0.1% | 0.33 |
| Medium + fatty alcohol ethoxylate 0.1% | 0.26 |
| Medium | 0.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lucassen, R.; van Leuven, N.; Bockmühl, D. Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms. Microorganisms 2024, 12, 712. https://doi.org/10.3390/microorganisms12040712
Lucassen R, van Leuven N, Bockmühl D. Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms. Microorganisms. 2024; 12(4):712. https://doi.org/10.3390/microorganisms12040712
Chicago/Turabian StyleLucassen, Ralf, Nicole van Leuven, and Dirk Bockmühl. 2024. "Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms" Microorganisms 12, no. 4: 712. https://doi.org/10.3390/microorganisms12040712
APA StyleLucassen, R., van Leuven, N., & Bockmühl, D. (2024). Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms. Microorganisms, 12(4), 712. https://doi.org/10.3390/microorganisms12040712





