Cell-Free Supernatant from a Strain of Bacillus siamensis Isolated from the Skin Showed a Broad Spectrum of Antimicrobial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation of Microorganisms from Different Skin Sites
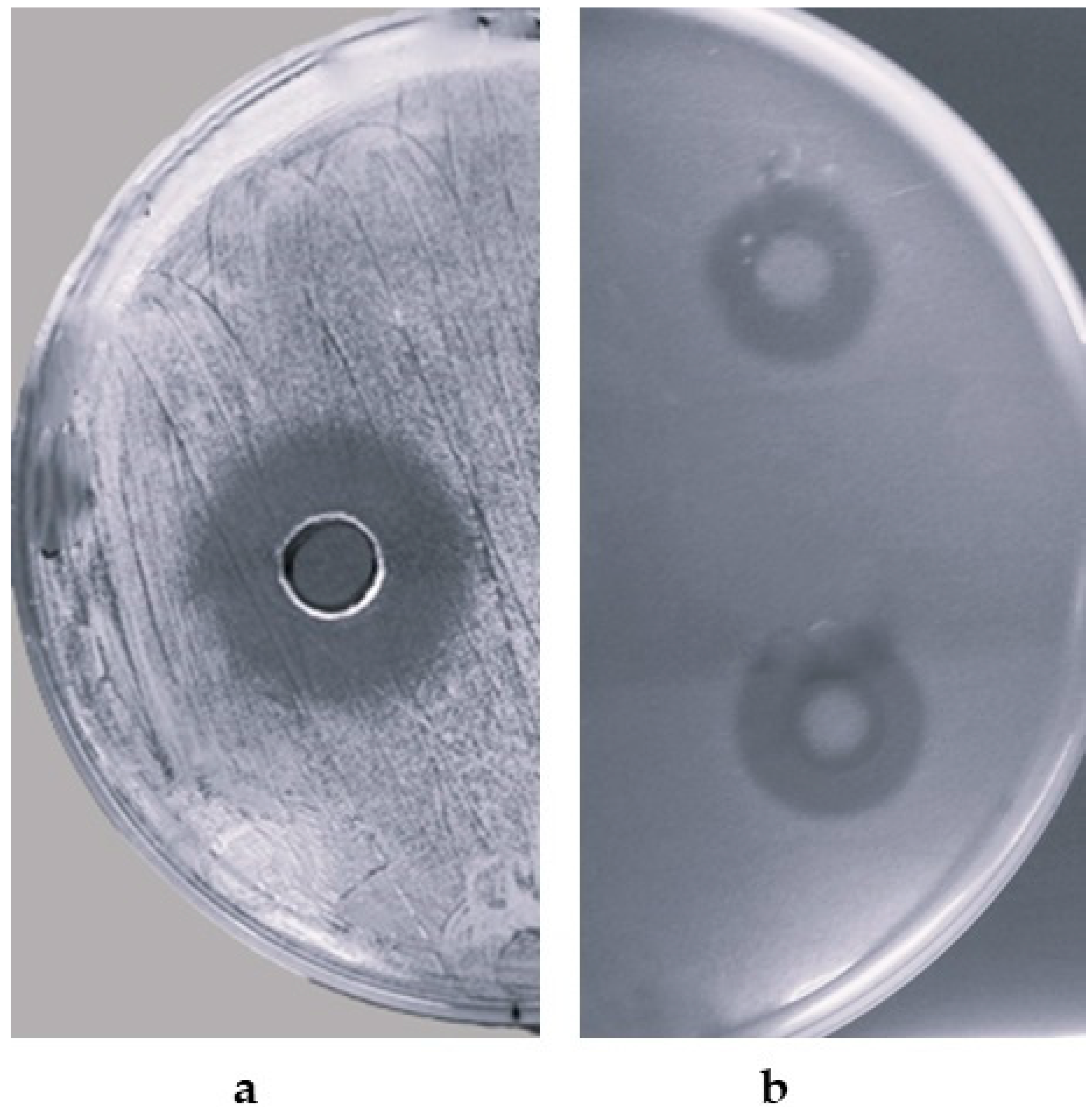
2.2. Detection of Antimicrobial Activity of B. siamensis CPAY1
2.3. Kinetics of Growth and CPAY1 CFS Biosynthesis
2.4. CPAY1 CFS Heat Stability, and Protease and pH Sensitivity
2.5. Time–Kill Studies
2.6. Anti-Biofilm Activity
2.7. The 16S rRNA Sequencing
2.8. Antibiotic Resistance
2.9. Plasmid Isolation
2.10. Statistical Analysis
3. Results
3.1. Isolation of Microorganisms from Different Skin Sites
3.2. Detection of Antimicrobial Activity
3.3. Kinetics of Growth and Biosynthesis
3.4. Effect of Heat Stability, and Protease and pH Sensitivity
3.5. Time–Kill Studies
3.6. Anti-Biofilm Activity
3.7. The 16S rRNA Sequencing
3.8. Antibiotic Resistance
3.9. Plasmid Isolation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yang, Y.; Qu, L.; Mijakovic, I.; Wei, Y. Advances in the human skin microbiota and its roles in cutaneous diseases. Microb. Cell Fact. 2022, 21, 176. [Google Scholar] [CrossRef] [PubMed]
- Mousa, W.K.; Athar, B.; Merwin, N.J.; Magarvey, N.A. Antibiotics and specialized metabolites from the human microbiota. Nat. Prod. Rep. 2017, 34, 1302–1331. [Google Scholar] [CrossRef]
- WHO. Guidelines on Hand Hygiene in Health Care: First Global Patient Safety Challenge Clean Care Is Safer Care; WHO: Geneva, Switzerland, 2009. [Google Scholar]
- Schmid-Wendtner, M.H.; Korting, H.C. The pH of the skin surface and its impact on the barrier function. Skin Pharmacol. Physiol. 2006, 19, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Newstead, L.L.; Varjonen, K.; Nuttall, T.; Paterson, G.K. Staphylococcal-produced bacteriocins and antimicrobial peptides: Their potential as alternative treatments for Staphylococcus aureus infections. Antibiotics 2020, 9, 40. [Google Scholar] [CrossRef]
- Anthony, W.E.; Burnham, C.D.; Dantas, G.; Kwon, J.H. The Gut Microbiome as a Reservoir for Antimicrobial Resistance. J. Infect. Dis. 2021, 223, S209–S213. [Google Scholar] [CrossRef] [PubMed]
- Ellis, J.C.; Ross, R.P.; Hill, C. Nisin Z and lacticin 3147 improve efficacy of antibiotics against clinically significant bacteria. Future Microbiol. 2019, 14, 1573–1587. [Google Scholar] [CrossRef]
- Twomey, E.; Hill, C.; Field, D.; Begley, M. Recipe for Success: Suggestions and Recommendations for the Isolation and Characterisation of Bacteriocins. Int. J. Microbiol. 2021, 2021, 9990635. [Google Scholar] [CrossRef]
- Rogers, A.M.; Montville, T.J. Improved agar diffusion assay for nisin quantification. Food Biotechnol. 1991, 5, 161–168. [Google Scholar] [CrossRef]
- Hernández, D.; Cardell, E.; Zárate, V. Antimicrobial activity of lactic acid bacteria isolated from Tenerife cheese: Initial characterization of plantaricin TF711, a bacteriocin-like substance produced by Lactobacillus plantarum TF711. J. Appl. Microbiol. 2005, 99, 77–84. [Google Scholar] [CrossRef]
- Sabia, C.; Anacarso, I.; Bergonzini, A.; Gargiulo, R.; Sarti, M.; Condò, C.; Messi, P.; de Niederhausern, S.; Iseppi, R.; Bondi, M. Detection and partial characterization of a bacteriocin-like substance produced by Lactobacillus fermentum CS57 isolated from human vaginal secretions. Anaerobe 2014, 26, 41–45. [Google Scholar] [CrossRef]
- Stepanovic, S.; Vukovic, D.; Hola, V.; Di Bonaventura, G.; Djukic, S.; Cirkovic, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Humphries, R.M.; Ambler, J.; Mitchell, S.L.; Castanheira, M.; Dingle, T.; Hindler, J.A.; Koeth, L.; Sei, K.; Development, C.M.; Standardization Working Group of the Subcommittee on Antimicrobial Susceptibility Testing. CLSI Methods Development and Standardization Working Group Best Practices for Evaluation of Antimicrobial Susceptibility Tests. J. Clin. Microbiol. 2018, 56, e01934-17, Erratum in J. Clin. Microbiol. 2023, 61, e0073923. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority (EFSA). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- O’Sullivan, D.J.; Klaenhammer, T.R. Rapid mini-prep isolation of high-quality pPlasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 1993, 59, 2730–2733. [Google Scholar] [CrossRef] [PubMed]
- Macrina, F.L.; Kopecko, D.J.; Jones, K.R.; Ayers, D.J.; McCowen, S.M. A multiple plasmid-containing Escherichia coli strain: Convenient source of size reference plasmid molecules. Plasmid 1978, 1, 417–420. [Google Scholar] [CrossRef] [PubMed]
- Motulsky, H. The GraphPad Guide to Comparing Dose-Response or Kinetic Curves; GraphPad Software: San Diego, CA, USA, 1998. [Google Scholar]
- Christensen, G.J.; Brüggemann, H. Bacterial skin commensals and their role as host guardians. Benef. Microbes. 2014, 5, 201–215. [Google Scholar] [CrossRef]
- Hong, H.; Linlin, S.; Yuanyuan, R.; Xi, T.; Wei, L.; Zhi, L. Applications of Human Skin Microbiota in the Cutaneous Disorders for Ecology-Based Therapy. Front. Cell Infect. Microbiol. 2020, 10, 570261. [Google Scholar] [CrossRef]
- Ferček, I.; Lugović-Mihić, L.; Tambić-Andrašević, A.; Ćesić, D.; Grginić, A.G.; Bešlić, I.; Mravak-Stipetić, M.; Mihatov-Štefanović, I.; Buntić, A.M.; Čivljak, R. Features of the skin microbiota in common inflammatory skin diseases. Life 2021, 11, 962. [Google Scholar] [CrossRef]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecińska-Piróg, J.; Wałecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human skin microbiome: Impact of intrinsic and extrinsic factors on skin microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef] [PubMed]
- Rungsirivanich, P.; Parlindungan, E.; O’Connor, P.M.; Field, D.; Mahony, J.; Thongwai, N.; van Sinderen, D. Simultaneous production of multiple antimicrobial compounds by Bacillus velezensis ML122-2 isolated from Assam tea leaf [Camellia sinensis var. assamica (J.W.Mast.) Kitam.]. Front. Microbiol. 2021, 12, 789362. [Google Scholar] [CrossRef]
- Caulier, S.; Nannan, C.; Gillis, A.; Licciardi, F.; Bragard, C.; Mahillon, J. Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front. Microbiol. 2019, 10, 302. [Google Scholar] [CrossRef] [PubMed]
- Andersson, D.I.; Hughes, D.; Kubicek-Sutherland, J.Z. Mechanisms and consequences of bacterial resistance to antimicrobial peptides. Drug Resist. Updates 2016, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Li, S.; Li, S.Y.; Zhang, H.; Jiang, M. The siderophore-producing bacterium, Bacillus siamensis Gxun-6, has an antifungal activity against Fusarium oxysporum and promotes the growth of banana. Egypt. J. Biol. Pest Control 2022, 32, 34. [Google Scholar] [CrossRef]
- Sumpavapol, P.; Tongyonk, L.; Tanasupawat, S.; Chokesajjawatee, N.; Luxananil, P.; Visessanguan, W. Bacillus siamensis sp. nov., isolated from salted crab (poo-khem) in Thailand. Int. J. Syst. Evol. Microbiol. 2010, 60, 2364–2370. [Google Scholar] [CrossRef] [PubMed]
- Nandhini, P.; Kumar, P.; Mickymaray, S.; Alothaim, A.S.; Somasundaram, J.; Rajan, M. Recent developments in methicillin-resistant Staphylococcus aureus (MRSA) treatment: A review. Antibiotics 2022, 11, 606. [Google Scholar] [CrossRef]
- Piper, C.; Draper, L.A.; Paul Cotter, P.D.; Ross, R.P.; Hill, C. A comparison of the activities of lacticin 3147 and nisin against drug-resistant Staphylococcus aureus and Enterococcus species. J. Antimicrob. Chemother. 2009, 64, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.V.; Baranwal, G.; Chatterjee, M.; Sachu, A.; Vasudevan, A.K.; Bose, C.; Banerji, A.; Biswas, R. Antimicrobial activity of plumbagin, a naturally occurring naphthoquinone from Plumbago rosea, against Staphylococcus aureus and Candida albicans. Int. J. Med. Microbiol. 2016, 306, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Moll, G.N.; Konings, W.N.; Driessen, A.J. Bacteriocins: Mechanism of membrane insertion and pore formation. Antonie Van Leeuwenhoek 1999, 76, 185–198. [Google Scholar] [CrossRef]
- Sablon, E.; Contreras, B.; Vandamme, E. Antimicrobial peptides of lactic acid bacteria: Mode of action, genetics and biosynthesis. Adv. Biochem. Eng. Biotechnol. 2000, 68, 21–60. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Wang, Y.; Shi, J.; Zhang, L.; Ding, Z.; Qu, X.; Cui, H. Class IIa bacteriocins: Diversity and new developments. Int. J. Mol. Sci. 2012, 6, 16668–16707. [Google Scholar] [CrossRef]
- Rani, A.; Saini, K.C.; Bast, F.; Varjani, S.; Mehariya, S.; Bhatia, S.K.; Sharma, N.; Funk, C. A Review on Microbial Products and Their Perspective Application as Antimicrobial Agents. Biomolecules 2021, 11, 1860. [Google Scholar] [CrossRef] [PubMed]
- Percival, S.L.; McCarty, S.M.; Lipsky, B. Biofilms and wounds: An overview of the evidence. Adv. Wound Care 2015, 4, 373–381. [Google Scholar] [CrossRef]
- Di Domenico, E.G.; Farulla, I.; Prignano, G.; Gallo, M.T.; Vespaziani, M.; Cavallo, I.; Sperduti, I.; Pontone, M.; Bordignon, V.; Cilli, L.; et al. Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. Int. J. Mol. Sci. 2017, 18, 1077. [Google Scholar] [CrossRef] [PubMed]
- Heo, S.; Kim, J.H.; Kwak, M.S.; Jeong, D.W.; Sung, M.H. Functional genomic insights into probiotic Bacillus siamensis strain B28 from traditional korean fermented Kimchi. Foods 2021, 10, 1906. [Google Scholar] [CrossRef] [PubMed]
- Tarek, H.; Nam, K.B.; Kim, Y.K.; Suchi, S.A.; Yoo, J.C. Biochemical characterization and application of a detergent stable, antimicrobial and antibiofilm potential protease from Bacillus siamensis. Int. J. Mol. Sci. 2023, 24, 5774. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Ye, Z.W.; Zheng, Q.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Isolation and characterization of cyclic lipopeptides with broad-spectrum antimicrobial activity from Bacillus siamensis JFL15. 3 Biotech 2018, 8, 444. [Google Scholar] [CrossRef]
- Sanders, M.E.; Akkermans, L.M.; Haller, D.; Hammerman, C.; Heimbach, J.; Hörmannsperger, G.; Huys, G.; Levy, D.D.; Lutgendorff, F.; Mack, D.; et al. Safety assessment of probiotics for human use. Gut Microbes 2010, 1, 164–185. [Google Scholar] [CrossRef]






| Indicator Strains | IZ (mm) | Indicator Strains | IZ (mm) |
|---|---|---|---|
| Staphylococcus aureus ATCC 6538 | +++ | Escherichia coli ATCC 25922 | + |
| Staphylococcus aureus MRSA M1 | +++ | Pseudomonas aeruginosa ATCC 9027 | − |
| Staphylococcus aureus MRSA H1 | +++ | Klebsiella pneumoniae ATCC 33495 | − |
| Staphylococcus epidermidis IM10 | + | Candida albicans ATCC 10231 | + |
| Streptococcus pneumoniae ATCC 46916 | − | Candida albicans IM56 | ++ |
| Streptococcus agalactiae V1 | + | Candida albicans IM 57 | ++ |
| Enterococcus faecalis ATCC 29212 | + | Candida parapsilosis ATCC 22019 | + |
| Enterococcus faecalis VRE A | + | Candida parapsilosis IM63 | ++ |
| Enterococcus faecalis VRE B | + | Candida parapsilosis IM64 | ++ |
| Enterococcus faecalis VRE C | ++ | Candida rugosa IM67 | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedretti, N.; Iseppi, R.; Condò, C.; Spaggiari, L.; Messi, P.; Pericolini, E.; Di Cerbo, A.; Ardizzoni, A.; Sabia, C. Cell-Free Supernatant from a Strain of Bacillus siamensis Isolated from the Skin Showed a Broad Spectrum of Antimicrobial Activity. Microorganisms 2024, 12, 718. https://doi.org/10.3390/microorganisms12040718
Pedretti N, Iseppi R, Condò C, Spaggiari L, Messi P, Pericolini E, Di Cerbo A, Ardizzoni A, Sabia C. Cell-Free Supernatant from a Strain of Bacillus siamensis Isolated from the Skin Showed a Broad Spectrum of Antimicrobial Activity. Microorganisms. 2024; 12(4):718. https://doi.org/10.3390/microorganisms12040718
Chicago/Turabian StylePedretti, Natalia, Ramona Iseppi, Carla Condò, Luca Spaggiari, Patrizia Messi, Eva Pericolini, Alessandro Di Cerbo, Andrea Ardizzoni, and Carla Sabia. 2024. "Cell-Free Supernatant from a Strain of Bacillus siamensis Isolated from the Skin Showed a Broad Spectrum of Antimicrobial Activity" Microorganisms 12, no. 4: 718. https://doi.org/10.3390/microorganisms12040718
APA StylePedretti, N., Iseppi, R., Condò, C., Spaggiari, L., Messi, P., Pericolini, E., Di Cerbo, A., Ardizzoni, A., & Sabia, C. (2024). Cell-Free Supernatant from a Strain of Bacillus siamensis Isolated from the Skin Showed a Broad Spectrum of Antimicrobial Activity. Microorganisms, 12(4), 718. https://doi.org/10.3390/microorganisms12040718












