Perspectives of FTIR as Promising Tool for Pathogen Diagnosis, Sanitary and Welfare Monitoring in Animal Experimentation Models: A Review Based on Pertinent Literature
Abstract
:1. Introduction
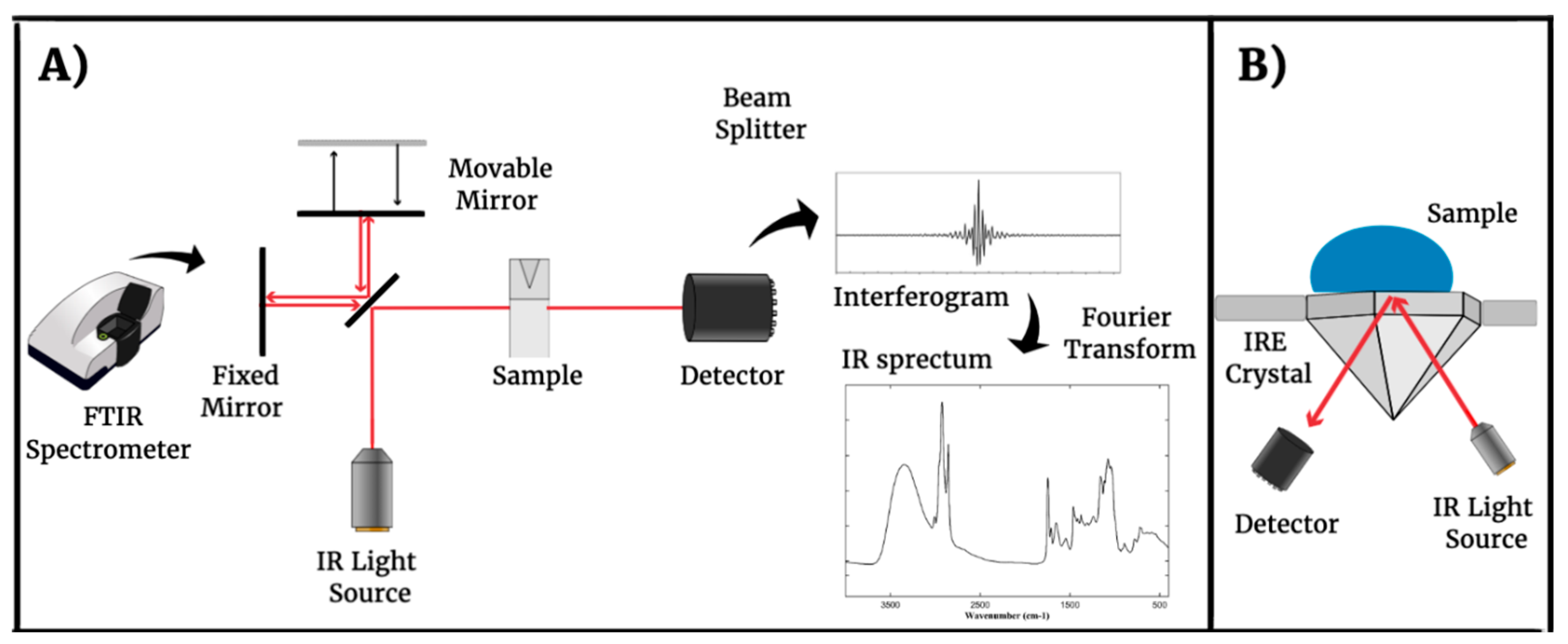
2. Fourier-Transform Infrared Spectroscopy (FTIR) Technique
3. Infections by Different Classes of Pathogens Can Be Diagnosed Using FTIR on Biological Samples
4. Using the FTIR Technique, It Is Possible to Detect Different Types of Molecules in Body Fluids
5. Would FTIR Applied to Sanitary Monitoring in Laboratory Animals Be an Innovation?
6. In Addition to Diagnosing Infectious Diseases, FTIR also Has the Potential to Be Used in Monitoring the Welfare of Laboratory Animals
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rani, M.; Keshu; Shanker, U. Chapter 3—Green nanomaterials: An overview. In Green Functionalized Nanomaterials for Environmental Applications; Shanker, U., Hussain, C.M., Rani, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2022; pp. 43–80. [Google Scholar] [CrossRef]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Kourkoumelis, N.; Zhang, X.; Lin, Z.; Wang, J. Fourier Transform Infrared Spectroscopy of Bone Tissue: Bone Quality Assessment in Preclinical and Clinical Applications of Osteoporosis and Fragility Fracture. Clin. Rev. Bone Miner. Metab. 2019, 17, 24–39. [Google Scholar] [CrossRef]
- Wang, R.; Wang, Y. Fourier Transform Infrared Spectroscopy in Oral Cancer Diagnosis. Int. J. Mol. Sci. 2021, 22, 1206. [Google Scholar] [CrossRef] [PubMed]
- Fabian, H.; Naumann, D. Methods to study protein folding by stopped-flow FT-IR. Methods 2004, 34, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Petibois, C.; Déléris, G. Evidence that erythrocytes are highly susceptible to exercise oxidative stress: FT-IR spectrometric studies at the molecular level. Cell Biol. Int. 2005, 29, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Buchheister, S.; Bleich, A. Health Monitoring of Laboratory Rodent Colonies-Talking about (R)evolution. Animals 2021, 11, 1410. [Google Scholar] [CrossRef] [PubMed]
- Lupini, L.; Bassi, C.; Guerriero, P.; Raspa, M.; Scavizzi, F.; Sabbioni, S. Microbiota and environmental health monitoring of mouse colonies by metagenomic shotgun sequencing. World J. Microbiol. Biotechnol. 2022, 39, 37. [Google Scholar] [CrossRef]
- Choudhary, A.; Ibdah, J.A. Animal models in today’s translational medicine world. Mo. Med. 2013, 110, 220–222. [Google Scholar] [PubMed]
- Robinson, N.B.; Krieger, K.; Khan, F.M.; Huffman, W.; Chang, M.; Naik, A.; Yongle, R.; Hameed, I.; Krieger, K.; Girardi, L.N.; et al. The current state of animal models in research: A review. Int. J. Surg. 2019, 72, 9–13. [Google Scholar] [CrossRef]
- Barré-Sinoussi, F.; Montagutelli, X. Animal models are essential to biological research: Issues and perspectives. Future Sci. OA 2015, 1, Fso63. [Google Scholar] [CrossRef]
- Lewis, D.I. Animal experimentation: Implementation and application of the 3Rs. Emerg. Top. Life Sci. 2019, 3, 675–679. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.L.; Winter, L.M.F. Animal models in biological and biomedical research—Experimental and ethical concerns. An. Acad. Bras. Ciênc. 2019, 91, e20170238. [Google Scholar] [CrossRef] [PubMed]
- Strech, D.; Dirnagl, U. 3Rs missing: Animal research without scientific value is unethical. BMJ Open Sci. 2019, 3, bmjos-2018-000048. [Google Scholar] [CrossRef] [PubMed]
- Nicklas, W. International Harmonization of Health Monitoring. ILAR J. 2008, 49, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Ahrens Kress, A.P.; Zhang, Y.; Kaiser-Vry, A.R.; Sauer, M.B. A Comparison of Blood Collection Techniques in Mice and their Effects on Welfare. J. Am. Assoc. Lab. Anim. Sci. 2022, 61, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, D.B.; Metzdorff, S.B.; Jensen, L.K.; Andersen, K.H.; Teilmann, A.C.; Jensen, H.E.; Frøkiaer, H. Time-dependent Pathologic and Inflammatory Consequences of Various Blood Sampling Techniques in Mice. J. Am. Assoc. Lab. Anim. Sci. 2019, 58, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Baier, J.; Rix, A.; Drude, N.I.; Darguzyte, M.; Baues, M.; May, J.N.; Schipper, S.; Möckel, D.; Palme, R.; Tolba, R.; et al. Influence of MRI Examinations on Animal Welfare and Study Results. Investig. Radiol. 2020, 55, 507–514. [Google Scholar] [CrossRef] [PubMed]
- Shek, W.R.; Smith, A.L.; Pritchett-Corning, K.R. Chapter 11—Microbiological Quality Control for Laboratory Rodents and Lagomorphs. In Laboratory Animal Medicine, 3rd ed.; Fox, J.G., Anderson, L.C., Otto, G.M., Pritchett-Corning, K.R., Whary, M.T., Eds.; Academic Press: Boston, MA, USA, 2015; pp. 463–510. [Google Scholar] [CrossRef]
- Wenning, M.; Scherer, S. Identification of microorganisms by FTIR spectroscopy: Perspectives and limitations of the method. Appl. Microbiol. Biotechnol. 2013, 97, 7111–7120. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.M.C.; Creaser, C.S. Analytical Applications of Spectroscopy II: Proceedings of the Second International Conference on Held 9–12th July, 1990 in Hatfield, England; Royal Society of Chemistry: London, UK, 1991. [Google Scholar]
- Grasselli, J. On the Relative Motion of the Earth and the Luminiferous Ether. Appl. Spectrosc. 1987, 41, 933–935. [Google Scholar] [CrossRef]
- Velasco, A.V.; Cheben, P.; Florjańczyk, M.; Calvo, M.L. Chapter 3—Spatial Heterodyne Fourier-Transform Waveguide Spectrometers. In Progress in Optics; Wolf, E., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 59, pp. 159–208. [Google Scholar]
- Fadlelmoula, A.; Pinho, D.; Carvalho, V.H.; Catarino, S.O.; Minas, G. Fourier Transform Infrared (FTIR) Spectroscopy to Analyse Human Blood over the Last 20 Years: A Review towards Lab-on-a-Chip Devices. Micromachines 2022, 13, 187. [Google Scholar] [CrossRef]
- Grdadolnik, J. ATR-FTIR spectroscopy: Its advantages and limitations. Acta Chim. Slov. 2002, 49, 631–642. [Google Scholar]
- Lin, S.Y.; Li, M.J.; Wei, Y.S. Ethanol or/and captopril-induced precipitation and secondary conformational changes of human serum albumin. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2004, 60, 3107–3111. [Google Scholar] [CrossRef] [PubMed]
- West, T.S.; Harrick, N.J. Internal Reflection Spectroscopy: Interscience Publishers-J. Wiley and Sons, Inc., New York, 1967, xiv+327 pp., price 132 s. Anal. Chim. Acta 1968, 42, 186. [Google Scholar] [CrossRef]
- Larkin, P. Chapter 3—Instrumentation and Sampling Methods. In Infrared and Raman Spectroscopy; Larkin, P., Ed.; Elsevier: Oxford, UK, 2011; pp. 27–54. [Google Scholar] [CrossRef]
- Schatz, G.C.; Van Duyne, R.P.; Chalmers, J.M.; Griffiths, P.R. Handbook of Vibrational Spectroscopy; Wiley: New York, NY, USA, 2006; Volume 1. [Google Scholar]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef]
- Novais, Â.; Freitas, A.R.; Rodrigues, C.; Peixe, L. Fourier transform infrared spectroscopy: Unlocking fundamentals and prospects for bacterial strain typing. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 427–448. [Google Scholar] [CrossRef]
- Rebuffo, C.A.; Schmitt, J.; Wenning, M.; von Stetten, F.; Scherer, S. Reliable and rapid identification of Listeria monocytogenes and Listeria species by artificial neural network-based Fourier transform infrared spectroscopy. Appl. Environ. Microbiol. 2006, 72, 994–1000. [Google Scholar] [CrossRef] [PubMed]
- Nitrosetein, T.; Wongwattanakul, M.; Chonanant, C.; Leelayuwat, C.; Charoensri, N.; Jearanaikoon, P.; Lulitanond, A.; Wood, B.R.; Tippayawat, P.; Heraud, P. Attenuated Total Reflection Fourier Transform Infrared Spectroscopy combined with chemometric modelling for the classification of clinically relevant Enterococci. J. Appl. Microbiol. 2021, 130, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Guliev, R.R.; Suntsova, A.Y.; Vostrikova, T.Y.; Shchegolikhin, A.N.; Popov, D.A.; Guseva, M.A.; Shevelev, A.B.; Kurochkin, I.N. Discrimination of Staphylococcus aureus Strains from Coagulase-Negative Staphylococci and Other Pathogens by Fourier Transform Infrared Spectroscopy. Anal. Chem. 2020, 92, 4943–4948. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Lozano, I.; Galán-Sánchez, F.; Rodríguez-Iglesias, M. Fourier transform infrared spectroscopy as a new tool for surveillance in local stewardship antimicrobial program: A retrospective study in a nosocomial Acinetobacter baumannii outbreak. Braz. J. Microbiol. 2022, 53, 1349–1353. [Google Scholar] [CrossRef]
- Lombardo, D.; Cordovana, M.; Deidda, F.; Pane, M.; Ambretti, S. Application of Fourier transform infrared spectroscopy for real-time typing of Acinetobacter baumannii outbreak in intensive care unit. Future Microbiol. 2021, 16, 1239–1250. [Google Scholar] [CrossRef]
- AlRabiah, H.; Correa, E.; Upton, M.; Goodacre, R. High-throughput phenotyping of uropathogenic E. coli isolates with Fourier transform infrared spectroscopy. Analyst 2013, 138, 1363–1369. [Google Scholar] [CrossRef]
- Passaris, I.; Mauder, N.; Kostrzewa, M.; Burckhardt, I.; Zimmermann, S.; van Sorge, N.M.; Slotved, H.C.; Desmet, S.; Ceyssens, P.J. Validation of Fourier Transform Infrared Spectroscopy for Serotyping of Streptococcus pneumoniae. J. Clin. Microbiol. 2022, 60, e0032522. [Google Scholar] [CrossRef] [PubMed]
- Oligbu, G.; Fry, N.K.; Ladhani, S.N. The Epidemiology and Biostatistics of Pneumococcus. Methods Mol. Biol. 2019, 1968, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Manzulli, V.; Cordovana, M.; Serrecchia, L.; Rondinone, V.; Pace, L.; Farina, D.; Cipolletta, D.; Caruso, M.; Fraccalvieri, R.; Difato, L.M.; et al. Application of Fourier Transform Infrared Spectroscopy to Discriminate Two Closely Related Bacterial Species: Bacillus anthracis and Bacillus cereus Sensu Stricto. Microorganisms 2024, 12, 183. [Google Scholar] [CrossRef] [PubMed]
- Melin, A.-M.; Allery, A.; Perromat, A.; Bébéar, C.; Déléris, G.; de Barbeyrac, B. Fourier transform infrared spectroscopy as a new tool for characterization of mollicutes. J. Microbiol. Methods 2004, 56, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Santos, C.; Fraga, M.E.; Kozakiewicz, Z.; Lima, N. Fourier transform infrared as a powerful technique for the identification and characterization of filamentous fungi and yeasts. Res. Microbiol. 2010, 161, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Tralamazza, S.M.; Bozza, A.; Destro, J.G.R.; Rodríguez, J.I.; Dalzoto, P.d.R.; Pimentel, I.C. Potential of Fourier Transform Infrared Spectroscopy (FT-IR) to Differentiate Environmental Aspergillus Fungi Species A. niger, A. ochraceus, and A. westerdijkiae Using Two Different Methodologies. Appl. Spectrosc. 2013, 67, 274–278. [Google Scholar] [CrossRef] [PubMed]
- Silva, L.G.; Péres, A.F.S.; Freitas, D.L.D.; Morais, C.L.M.; Martin, F.L.; Crispim, J.C.O.; Lima, K.M.G. ATR-FTIR spectroscopy in blood plasma combined with multivariate analysis to detect HIV infection in pregnant women. Sci. Rep. 2020, 10, 20156. [Google Scholar] [CrossRef] [PubMed]
- de Souza, N.M.P.; Machado, B.H.; Koche, A.; da Silva Furtado, L.B.F.; Becker, D.; Corbellini, V.A.; Rieger, A. Detection of metabolic syndrome with ATR-FTIR spectroscopy and chemometrics in blood plasma. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 288, 122135. [Google Scholar] [CrossRef]
- Shaikh, S.; Yadav, D.K.; Rawal, R. Saliva based non invasive screening of Oral Submucous Fibrosis using ATR-FTIR spectroscopy. J. Pharm. Biomed. Anal. 2021, 203, 114202. [Google Scholar] [CrossRef]
- Nascimento, M.H.C.; Marcarini, W.D.; Folli, G.S.; da Silva Filho, W.G.; Barbosa, L.L.; Paulo, E.H.d.; Vassallo, P.F.; Mill, J.G.; Barauna, V.G.; Martin, F.L.; et al. Noninvasive Diagnostic for COVID-19 from Saliva Biofluid via FTIR Spectroscopy and Multivariate Analysis. Anal. Chem. 2022, 94, 2425–2433. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.L.; Dickinson, A.W.; Saba, T.; Bongers, T.; Singh, M.N.; Bury, D. ATR-FTIR Spectroscopy with Chemometrics for Analysis of Saliva Samples Obtained in a Lung-Cancer-Screening Programme: Application of Swabs as a Paradigm for High Throughput in a Clinical Setting. J. Pers. Med. 2023, 13, 1039. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, I.C.C.; Aguiar, E.M.G.; Silva, A.T.F.; Santos, L.L.D.; Cardoso-Sousa, L.; Araújo, T.G.; Santos, D.W.; Goulart, L.R.; Sabino-Silva, R.; Maia, Y.C.P. Attenuated Total Reflection-Fourier Transform Infrared (ATR-FTIR) Spectroscopy Analysis of Saliva for Breast Cancer Diagnosis. J. Oncol. 2020, 2020, 4343590. [Google Scholar] [CrossRef] [PubMed]
- Kochan, K.; Bedolla, D.E.; Perez-Guaita, D.; Adegoke, J.A.; Veettil, T.C.P.; Martin, M.; Roy, S.; Pebotuwa, S.; Heraud, P.; Wood, B.R. Infrared Spectroscopy of Blood. Appl. Spectrosc. 2021, 75, 611–646. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Wei, G.; Chen, W.; Lei, C.; Xu, C.; Guan, Y.; Ji, T.; Wang, F.; Liu, H. Fast and Deep Diagnosis Using Blood-Based ATR-FTIR Spectroscopy for Digestive Tract Cancers. Biomolecules 2022, 12, 1815. [Google Scholar] [CrossRef] [PubMed]
- Guang, P.; Huang, W.; Guo, L.; Yang, X.; Huang, F.; Yang, M.; Wen, W.; Li, L. Blood-based FTIR-ATR spectroscopy coupled with extreme gradient boosting for the diagnosis of type 2 diabetes: A STARD compliant diagnosis research. Medicine 2020, 99, e19657. [Google Scholar] [CrossRef]
- Naseer, K.; Ali, S.; Qazi, J. ATR-FTIR spectroscopy as the future of diagnostics: A systematic review of the approach using bio-fluids. Appl. Spectrosc. Rev. 2021, 56, 85–97. [Google Scholar] [CrossRef]
- Caixeta, D.C.; Lima, C.; Xu, Y.; Guevara-Vega, M.; Espindola, F.S.; Goodacre, R.; Zezell, D.M.; Sabino-Silva, R. Monitoring glucose levels in urine using FTIR spectroscopy combined with univariate and multivariate statistical methods. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 290, 122259. [Google Scholar] [CrossRef] [PubMed]
- Sarigul, N.; Kurultak, İ.; Uslu Gökceoğlu, A.; Korkmaz, F. Urine analysis using FTIR spectroscopy: A study on healthy adults and children. J. Biophotonics 2021, 14, e202100009. [Google Scholar] [CrossRef]
- Sarigul, N.; Bozatli, L.; Kurultak, I.; Korkmaz, F. Using urine FTIR spectra to screen autism spectrum disorder. Sci. Rep. 2023, 13, 19466. [Google Scholar] [CrossRef]
- Saparbaev, E.; Zviagin, A.; Boyarkin, O.V. Identification of Isomeric Biomolecules by Infrared Spectroscopy of Solvent-Tagged Ions. Anal. Chem. 2022, 94, 9514–9518. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Wang, Z.; Luo, Y.; Lin, Z.; Hong, G.; Deng, K.; Huang, P.; Shen, Y. Non/mini-invasive monitoring of diabetes-induced myocardial damage by Fourier transform infrared spectroscopy: Evidence from biofluids. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, L.; Qi, G.; Zhang, X.; Tian, C. Raman and fourier transform infrared spectroscopy techniques for detection of coronavirus (COVID-19): A mini review. Front. Chem. 2023, 11, 1193030. [Google Scholar] [CrossRef] [PubMed]
- Kazmer, S.T.; Hartel, G.; Robinson, H.; Richards, R.S.; Yan, K.; van Hal, S.J.; Chan, R.; Hind, A.; Bradley, D.; Zieschang, F.; et al. Pathophysiological Response to SARS-CoV-2 Infection Detected by Infrared Spectroscopy Enables Rapid and Robust Saliva Screening for COVID-19. Biomedicines 2022, 10, 351. [Google Scholar] [CrossRef]
- Korb, E.; Bağcıoğlu, M.; Garner-Spitzer, E.; Wiedermann, U.; Ehling-Schulz, M.; Schabussova, I. Machine Learning-Empowered FTIR Spectroscopy Serum Analysis Stratifies Healthy, Allergic, and SIT-Treated Mice and Humans. Biomolecules 2020, 10, 1058. [Google Scholar] [CrossRef] [PubMed]
- Lemes, L.; Júnior, P.C.; Ferreira-Strixino, J.; Aguiar, J.; Raniero, L. Analysis of serum cortisol levels by Fourier Transform Infrared Spectroscopy for diagnosis of stress in athletes. Res. Biomed. Eng. 2016, 32, 293–300. [Google Scholar] [CrossRef]
- Alcicek, F.C.; Blat, A.; Rutkowska, W.; Bulat, K.; Szczesny-Malysiak, E.; Franczyk-Zarow, M.; Kostogrys, R.; Dybas, J.; Marzec, K.M. Secondary structure alterations of RBC assessed by FTIR-ATR in correlation to 2,3-DPG levels in ApoE/LDLR–/– Mice. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 284, 121819. [Google Scholar] [CrossRef] [PubMed]
- FELASA Working Group on Revision of Guidelines for Health Monitoring of Rodents and Rabbits; Mähler, M.; Berard, M.; Feinstein, R.; Gallagher, A.; Illgen-Wilcke, B.; Pritchett-Corning, K.; Raspa, M. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab. Anim. 2014, 48, 178–192. [Google Scholar] [CrossRef]
- Enriquez, J.; Mims, B.M.D.; Trasti, S.; Furr, K.L.; Grisham, M.B. Genomic, microbial and environmental standardization in animal experimentation limiting immunological discovery. BMC Immunol. 2020, 21, 50. [Google Scholar] [CrossRef]
- Whitfield, L. Handbook of Laboratory Animal Science: Essential Principles and Practices, 4th edition Edited by J Hau and SJ Schapiro (2021). Published by CRC Press, Boca Raton, FL 33487, USA. 994 pages Hardback (ISBN: 978-1138341807). Price £141.64. Anim. Welf. 2022, 31, 561–563. [Google Scholar] [CrossRef]
- Boot, R. Development and validation of ELISAs for monitoring bacterial and parasitic infections in laboratory rodents and rabbits. Scand. J. Lab. Anim. Sci. 2001, 28, 44–50. [Google Scholar]
- Tyler, J.W.; Cullor, J.S. Titers, tests, and truisms: Rational interpretation of diagnostic serologic testing. J. Am. Vet. Med. Assoc. 1989, 194, 1550–1558. [Google Scholar] [PubMed]
- Jacobson, R.H.; Romatowski, J. Assessing the validity of serodiagnostic test results. Semin. Vet. Med. Surg. Small Anim. 1996, 11, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Janus, L.M.; Mähler, M.; Köhl, W.; Smoczek, A.; Hedrich, H.J.; Bleich, A. Minute virus of mice: Antibody response, viral shedding, and persistence of viral DNA in multiple strains of mice. Comp. Med. 2008, 58, 360–368. [Google Scholar] [PubMed]
- Besselsen, D.G.; Myers, E.L.; Franklin, C.L.; Korte, S.W.; Wagner, A.M.; Henderson, K.S.; Weigler, B.J. Transmission probabilities of mouse parvovirus 1 to sentinel mice chronically exposed to serial dilutions of contaminated bedding. Comp. Med. 2008, 58, 140–144. [Google Scholar] [PubMed]
- Bauer, B.A.; Besch-Williford, C.L.; Riley, L.K. Comparison of the mouse antibody production (MAP) assay and polymerase chain reaction (PCR) assays for the detection of viral contaminants. Biologicals 2004, 32, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gao, W.; Tan, X.; Han, Y.; Jiao, F.; Feng, B.; Xie, J.; Li, B.; Zhao, H.; Tu, H.; et al. MALDI-TOF MS Is an Effective Technique to Classify Specific Microbiota. Microbiol. Spectr. 2023, 11, e00307–e00323. [Google Scholar] [CrossRef]
- Turner, P.V.; Barbee, R.W. Responsible Science and Research Animal Use. ILAR J. 2019, 60, 1–4. [Google Scholar] [CrossRef]
- Bayne, K.; Turner, P.V. Animal Welfare Standards and International Collaborations. ILAR J. 2019, 60, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Veissier, I.; Aubert, A.; Boissy, A. Animal welfare: A result of animal background and perception of its environment. Anim. Front. 2012, 2, 7–15. [Google Scholar] [CrossRef]
- McCausland, C. The Five Freedoms of Animal Welfare are Rights. J. Agric. Environ. Ethics 2014, 27, 649–662. [Google Scholar] [CrossRef]
- Mellor, D.J. Moving beyond the “Five Freedoms” by Updating the “Five Provisions” and Introducing Aligned “Animal Welfare Aims”. Animals 2016, 6, 59. [Google Scholar] [CrossRef]
- Kim, K.; Jo, W.; Lee, G.-H. Why do we always care about the welfare of laboratory animals? Open Access Gov. 2023, 39, 518–519. [Google Scholar] [CrossRef]
- Díaz, L.; Zambrano, E.; Flores, M.E.; Contreras, M.; Crispín, J.C.; Alemán, G.; Bravo, C.; Armenta, A.; Valdés, V.J.; Tovar, A.; et al. Ethical Considerations in Animal Research: The Principle of 3R’s. Rev. Investig. Clin. 2020, 73, 199–209. [Google Scholar] [CrossRef] [PubMed]
- Langford, D.J.; Bailey, A.L.; Chanda, M.L.; Clarke, S.E.; Drummond, T.E.; Echols, S.; Glick, S.; Ingrao, J.; Klassen-Ross, T.; LaCroix-Fralish, M.L.; et al. Coding of facial expressions of pain in the laboratory mouse. Nat. Methods 2010, 7, 447–449. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, A.L.; Liu, Y.; Barker, T.H. Methods Used and Application of the Mouse Grimace Scale in Biomedical Research 10 Years on: A Scoping Review. Animals 2021, 11, 673. [Google Scholar] [CrossRef] [PubMed]
- Foley, P.L.; Kendall, L.V.; Turner, P.V. Clinical Management of Pain in Rodents. Comp. Med. 2019, 69, 468–489. [Google Scholar] [CrossRef]
- Pereira, C.; Kunczik, J.; Bleich, A.; Haeger, C.; Kiessling, F.; Thum, T.; Tolba, R.; Lindauer, U.; Treue, S.; Czaplik, M. Perspective review of optical imaging in welfare assessment in animal-based research. J. Biomed. Opt. 2019, 24, 070601-11. [Google Scholar] [CrossRef]
- Lim, M.A.; Louie, B.; Ford, D.; Heath, K.; Cha, P.; Betts-Lacroix, J.; Lum, P.Y.; Robertson, T.L.; Schaevitz, L. Development of the Digital Arthritis Index, a Novel Metric to Measure Disease Parameters in a Rat Model of Rheumatoid Arthritis. Front. Pharmacol. 2017, 8, 818. [Google Scholar] [CrossRef]
- Do, J.P.; Defensor, E.B.; Ichim, C.V.; Lim, M.A.; Mechanic, J.A.; Rabe, M.D.; Schaevitz, L.R. Automated and Continuous Monitoring of Animal Welfare through Digital Alerting. Comp. Med. 2020, 70, 313–327. [Google Scholar] [CrossRef]
- Ross, S.R.; Lake, B.R.; Fultz, A.; Hopper, L.M. An evaluation of thermal imaging as a welfare monitoring tool for captive chimpanzees. Primates 2021, 62, 919–927. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.B.; Kunczik, J.; Zieglowski, L.; Tolba, R.; Abdelrahman, A.; Zechner, D.; Vollmar, B.; Janssen, H.; Thum, T.; Czaplik, M. Remote Welfare Monitoring of Rodents Using Thermal Imaging. Sensors 2018, 18, 3653. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.F.; Oparaeke, A.; Gallagher, R.; Karimi, A.; Tariq, F.; Smith, M.L. Towards Machine Vision for Insect Welfare Monitoring and Behavioural Insights. Front. Vet. Sci. 2022, 9, 835529. [Google Scholar] [CrossRef] [PubMed]
- Andresen, N.; Wöllhaf, M.; Hohlbaum, K.; Lewejohann, L.; Hellwich, O.; Thöne-Reineke, C.; Belik, V. Towards a fully automated surveillance of well-being status in laboratory mice using deep learning: Starting with facial expression analysis. PLoS ONE 2020, 15, e0228059. [Google Scholar] [CrossRef] [PubMed]
- Van der Mierden, S.; Leenaars, C.H.C.; Boyle, E.C.; Ripoli, F.L.; Gass, P.; Durst, M.; Goerlich-Jansson, V.C.; Jirkof, P.; Keubler, L.M.; Talbot, S.R.; et al. Measuring endogenous corticosterone in laboratory mice—A mapping review, meta-analysis, and open source database. Altex 2021, 38, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Feige-Diller, J.; Palme, R.; Kaiser, S.; Sachser, N.; Richter, S.H. The impact of varying food availability on health and welfare in mice: Testing the Match-Mismatch hypothesis. Physiol. Behav. 2021, 228, 113193. [Google Scholar] [CrossRef] [PubMed]
- Harikrishnan, V.S.; Hansen, A.K.; Abelson, K.S.; Sørensen, D.B. A comparison of various methods of blood sampling in mice and rats: Effects on animal welfare. Lab. Anim. 2018, 52, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Palme, R. Monitoring stress hormone metabolites as a useful, non-invasive tool for welfare assessment in farm animals. Anim. Welf. 2012, 21, 331–337. [Google Scholar] [CrossRef]
- Bahadi, M.; Ismail, A.A.; Vasseur, E. Fourier Transform Infrared Spectroscopy as a Tool to Study Milk Composition Changes in Dairy Cows Attributed to Housing Modifications to Improve Animal Welfare. Foods 2021, 10, 450. [Google Scholar] [CrossRef]
- de Magalhães, C.R.; Carrilho, R.; Schrama, D.; Cerqueira, M.; Rosa da Costa, A.M.; Rodrigues, P.M. Mid-infrared spectroscopic screening of metabolic alterations in stress-exposed gilthead seabream (Sparus aurata). Sci. Rep. 2020, 10, 16343. [Google Scholar] [CrossRef]
- Milekhin, I.A.; Cherkasova, O.P.; Milekhin, A.G.; Kuznetsov, S.A.; Rodyakina, E.E.; Minaeva, V.A.; Latyshev, A.V. Surface-enhanced infrared spectroscopy for cortisol analysis. In Proceedings of the 2018 International Conference Laser Optics (ICLO) 2018, Saint Petersburg, Russia, 4–8 June 2018; p. 551. [Google Scholar]
- Cherkasova, O.; Milekhin, A.; Milekhin, I.; Kuznetsov, S.; Rodyakina, E.; Latyshev, A. Application of surface-enhanced infrared spectroscopy for steroids analysis. In Proceedings of the 2016 International Conference Laser Optics (LO), Saint Petersburg, Russia, 27 June–1 July 2016; pp. S2–S29. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Neves, M.M.; Guerra, R.F.; de Lima, I.L.; Arrais, T.S.; Guevara-Vega, M.; Ferreira, F.B.; Rosa, R.B.; Vieira, M.S.; Fonseca, B.B.; Sabino da Silva, R.; et al. Perspectives of FTIR as Promising Tool for Pathogen Diagnosis, Sanitary and Welfare Monitoring in Animal Experimentation Models: A Review Based on Pertinent Literature. Microorganisms 2024, 12, 833. https://doi.org/10.3390/microorganisms12040833
Neves MM, Guerra RF, de Lima IL, Arrais TS, Guevara-Vega M, Ferreira FB, Rosa RB, Vieira MS, Fonseca BB, Sabino da Silva R, et al. Perspectives of FTIR as Promising Tool for Pathogen Diagnosis, Sanitary and Welfare Monitoring in Animal Experimentation Models: A Review Based on Pertinent Literature. Microorganisms. 2024; 12(4):833. https://doi.org/10.3390/microorganisms12040833
Chicago/Turabian StyleNeves, Matheus Morais, Renan Faria Guerra, Isabela Lemos de Lima, Thomas Santos Arrais, Marco Guevara-Vega, Flávia Batista Ferreira, Rafael Borges Rosa, Mylla Spirandelli Vieira, Belchiolina Beatriz Fonseca, Robinson Sabino da Silva, and et al. 2024. "Perspectives of FTIR as Promising Tool for Pathogen Diagnosis, Sanitary and Welfare Monitoring in Animal Experimentation Models: A Review Based on Pertinent Literature" Microorganisms 12, no. 4: 833. https://doi.org/10.3390/microorganisms12040833
APA StyleNeves, M. M., Guerra, R. F., de Lima, I. L., Arrais, T. S., Guevara-Vega, M., Ferreira, F. B., Rosa, R. B., Vieira, M. S., Fonseca, B. B., Sabino da Silva, R., & Silva, M. V. d. (2024). Perspectives of FTIR as Promising Tool for Pathogen Diagnosis, Sanitary and Welfare Monitoring in Animal Experimentation Models: A Review Based on Pertinent Literature. Microorganisms, 12(4), 833. https://doi.org/10.3390/microorganisms12040833






