A Journey into the Evolution of Human Host-Oral Microbiome Relationship through Ancient Dental Calculus: A Scoping Review
Abstract
:1. Introduction
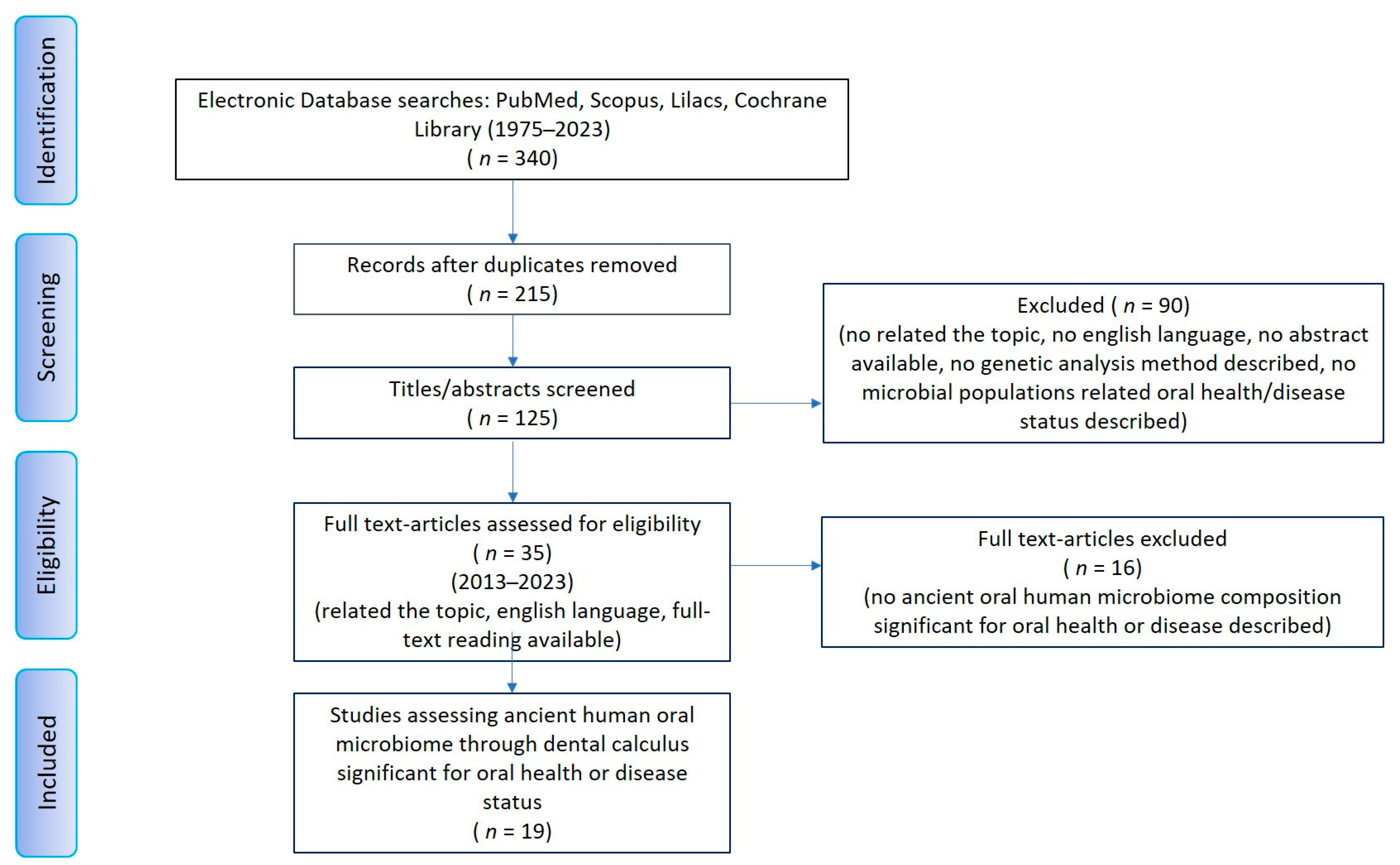
2. Materials and Methods
- Human Microbiome
- Dental Calculus
- Tooth
- Oral health
- Oral disease
- 1 OR 2 OR 3 OR 4 OR 5
- DNA, Ancient
- Metagenomics
- High-Throughput Nucleotide Sequencing
- Biological Evolution
- 7 OR 8 Or 9 OR 10 OR 11
- 6 and 11
3. Results
3.1. Geographical Origin and Historical Dating of Dental Calculus Samples
3.2. Sample Size and Site of Dental Calculus Collection
3.3. DNA Sequencing Technology
3.4. Bacterial Composition Dominance in Ancient Dental Calculus
| Authors—Year | Geographical Origin | Time Period | Sample Size and Site | DNA Sequencing Technology | Bacterial Composition Dominant in Collected Ancient Dental Calculus Samples |
|---|---|---|---|---|---|
| Adler et al., 2013 [37] | Poland | From 10th–6th BCE to 500–1500 CE | Supra and subgingival dental calculus from 34 human skeletons | Extraction of bacterial DNA and generation of PCR amplicon libraries of the 16S rRNA gene; Random Forest analysis | Proteobacteria, Firmicutes, Actinobacteria. Veillonellaceae (tooth decay), P. gingivalis, Tannerella, Treponema (p. disease). S. mutans is not dominant. |
| Warinner et al., 2014 [49] | Germany | From 500 to 1500 CE | Dental tissues of 4 four adult human skeletons | Meta-genomic-shotgun sequencing and 16s rRNA amplicon | Tannerella forsythia, Porphyromonas gingivalis, and Treponema denticola are particularly abundant. Additional pathogens include those implicated in acute dental infections (e.g., Actinomyces odontolyticus) and caries (S. mutans). Filifactor alocis and Olsenella uli have recently been associated with periodontitis and endodontic infections, respectively. |
| Weyrich et al., 2017 [38] | Belgium, Italy, Spain | From 12th to 10th millennium BCE | Dental calculus (24 specimens) from 5 skeletons | Metagenomic-shotgun sequencing and 16s rRNA amplicon (v4 region) | S. mutans presence is irrelevant. P. gingivalis, T. forsythia, T. denticola are abundant. Abundance of Methanobrevibacter oralis. |
| Jersie-Christensen et al., 2018 [39] | Denmark | 1100–1450 CE | Dental calculus (22 specimens) from 21 human remains | Metaproteomic and metagenomic analysis | 3671 protein groups, covering 220 bacterial species and 81 genera across all medieval samples. After Actinomyces spp., the genera Olsenella and Fretibacterium, both of which have been implicated in periodontitis, are the most abundant. Significant contributions from Fretibacterium spp., Porphyromonas spp., Treponema spp., Tannerella spp., and Desulfobulbus sp. oral taxon 041; all of which have been suggested to be involved in clinical periodontitis. |
| Mann et al., 2018 [22] | Miscellaneous worldwide | 500–1500 CE | 48 specimens of dental calculus and dentin | Metagenomic- shotgun sequencing | Methanobrevibacter; Tannerella; Porphyromonas; Actinomyces; Streptomyces |
| Willman et al., 2018 [26] | France | 18th century | Specimens from 9 subjects | High-Throughput DNA sequencing (HTS) | The presence of Streptococcus mutans and also Rothia dentocariosa, Actinomyces viscosus, Porphyromonas gingivalis, Tannerella forsythia, Pseudoramibacter alactolyticus, Olsenella uli and Parvimonas micra was confirmed like specific bacterial signature associated to carious or periodontal pathologies. |
| Velsko et al., 2019 [40] | United Kingdom | 1778–1785 | 48 samples of historic dental calculus | Metagenomic-shotgun sequencing | Many of the taxa with higher abundance in calculus are “late colonizers” (i.e., Desulfobulbus, Methanobrevibacter, Tannerella); P. gingivalis and T. forsythia characterised historic periodontal disease site from historic healthy site calculus as they do in modern plaque. |
| Achtman et al., 2020 [21] | Miscellaneous worldwide | Not specified | 110 samples of ancient dental calculus | Meta-genomic-shotgun sequencing data sets analysed with new bioinformatic tools (SPARSE, EToKi, GrapeTree) | Streptococcus sanguinis and Tannerella forsythia were most abundant in historical dental calculus (periodontal disease). Treponema denticola was most frequently found in historical dental calculus but Porphyromonas gingivalis is most frequent in modern plaque and is generally much less abundant. |
| McLean et al., 2020 [41] | Not Specified | From 6th–10th BCE to 500–1500 CE and 19th century CE | Not Specified | Metagenomic-shotgun sequencing | Saccharibacteria/TM7 phylum is present (G1 group genome) in dental calculus of ancient adult skeletons with evidence of mild to severe periodontal diseases. |
| Neukamm et al., 2020 [42] | Egypt | From 2196 BCE to 395 CE | 5 ancient dental calculus samples | Metagenomic-shotgun sequencing | Red Complex bacteria (Tannerella forsythia, Porphyromonas gingivalis, and Treponema denticola); Two other bacteria (Filifactor alocis and Olsenella uli) associated with periodontitis and endodontic infections were also identified with damage profiles. In general, the calculus samples are dominated by Firmicutes, Actinobacteria, Proteobacteria, Bacteroidetes, Chloroflexi, Fusobacteria, and Spirochetes. |
| Farrer et al., 2021 [43] | United Kingdom | 1170–1290 CE | Dental calculus samples extracted by 26 individuals buried | Metagenomic-shotgun sequencing and decontamination protocols | Three oral species—Actinomyces sp., Olsenella sp. and Streptococcus sanguinis were more likely to be present in the EDTA and UV + NaClO groups than the others. |
| Granehäll et al., 2021 [44] | Italy | 6000–3500 BCE to 400–1000 CE | Dental calculus from 20 ancient human skeletal remains | Metagenomic-shotgun sequencing | Red complex and besides the presence of M. oralis |
| Kazarina et al., 2021 [7] | Latvia | 16–17th century AD | 15 historic dental calculus samples | Metagenomic-shotgun sequencing | Historic dental calculus samples represented a slightly different pattern of the most abundant species, the first 10 of which were Olsenella sp. oral taxon 807, Actinomyces sp. oral taxon 414, Anaerolineaceae bacterium oral taxon 439, Pseudopropionibacterium propionicum, Streptococcus sanguinis, Eubacterium minutum, Desulfobulbus oralis, Lautropia mirabilis, Streptococcus cristatus, and Ottowia sp. oral taxon 894. |
| Ottoni et al., 2021 [45] | Balkans and Italy | From 3rd BCE to 500–1500 CE | Dental Calculus from 44 prehistoric foragers and farmers | Metagenomic-shotgun sequencing | Prevalence of Anaerolineaceae bacterium oral taxon 439, M. oralis, Desulfomicrobium orale, and Desulfobulbus oralis. Neolithic farmers possessed a higher frequency Olsenella sp. oral taxon 807. The analysis of differential species abundances showed that Olsenella sp. oral taxon 807 and Anaerolineaceae bacterium oral taxon 439 were more abundant in the Neolithic farmers, whereas Streptococcus sanguinis was higher in the Mesolithic foragers. |
| Fagernäs et al., 2022 [46] | Spain | 4500–5000 BP | Dental calculus from 4 adults’ skeletons with Brothwell score from 1 (slight) to 4 (gross); | Metagenomic-shotgun sequencing; A total of two separate analyses were conducted, one without occlusal samples and one including occlusal samples. | Methanobrevibacter and Olsenella |
| Garralda et al., 2022 [47] | France, Israel | 75–70/60 ky BP | Dental calculus from the left corpus of a juvenile mandible (France) compared with dental calculus of 2 adult Neanderthals from different locations (Israel) | SEM analysis | Cocci and filamentous types of bacteria in the French sample; more filamentous bacteria are present in the Israeli samples. |
| Scorrano et al., 2022 [48] | Italy | 50–12 ky BP | Dental Calculus from the remains of 2 hunter-gatherers (San Teodoro 3 and San Teodoro 5) | Metagenomic-shotgun sequencing | Abundance in Actinomyces, Streptococcus and Propionibacterium, genera typically associated with the oral microbiome. In one sample abundant Olsenella (known to cause endodontic infections) while in the other was abundant Aggregatibacter and Neisseria, (normal oral microbiome). Most of the ancient calculus samples were also highly abundant in the periodontal component: Prevotella conceptionensis, Porphyromonas gingivalis and Prevotella nigrescens. Found species including Methanobrevibacter oralis and Olsenella sp. Oral taxon 807 among those with significantly higher abundance in the ancient samples. Porphyromonas gingivalis and Treponema denticola were more abundant in samples with periodontal disease and ancient calculus. |
| Velsko et al., 2022 [24] | Netherlands | 19th Century | Dental calculus from 75 skeletal collections | Metagenomic-shotgun sequencing | The top species with strongest negative loadings are largely anaerobic taxa that are dominant in mature oral biofilms, including those in the genera Methanobrevibacter, Eubacterium, Desulfobulbus, Fretibacterium, and Tannerella. |
| Gancz et al., 2023 [50] | United Kingdom | 2200 BCE to 1853 CE | 235 ancient dental calculus samples | Meta-genomic-shotgun sequencing | Methanobrevibacter was not common in the oral microbiome of modern industrialised societies. Its disappearance suggests pre-industrialised microbiomes were more diverse than previously recognised, enhancing our understanding of chronic, non-communicable disease origins in industrialised populations. |
4. Discussion
4.1. Dental Calculus and Oral Health and Disease
4.2. Modern and Ancient Oral Microbiome
4.3. Genomic Analyses on Modern and Ancient Dental Calculus
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dhami, N.K.; Greenwood, P.F.; Poropat, S.F.; Tripp, M.; Elson, A.; Vijay, H.; Brosnan, L.; Holman, A.I.; Campbell, M.; Hopper, P.; et al. Microbially mediated fossil concretions and their characterization by the latest methodologies: A review. Front. Microbiol. 2023, 14, 1225411. [Google Scholar] [CrossRef] [PubMed]
- Panaiotov, S.; Madzharov, D.; Hodzhev, Y. Biodiversity of Mycobacterium tuberculosis in Bulgaria Related to Human Migrations or Ecological Adaptation. Microorganisms 2022, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Reynoso-García, J.; Narganes-Storde, Y.; Santiago-Rodriguez, T.M.; Toranzos, G.A. Mycobiome-Host Coevolution? The Mycobiome of Ancestral Human Populations Seems to Be Different and Less Diverse Than Those of Extant Native and Urban-Industrialized Populations. Microorganisms 2022, 10, 459. [Google Scholar] [CrossRef] [PubMed]
- Logan, A.C.; Katzman, M.A.; Balanzá-Martínez, V. Natural environments, ancestral diets, and microbial ecology: Is there a modern “paleo-deficit disorder”? Part II. J. Physiol. Anthropol. 2015, 34, 9. [Google Scholar] [CrossRef] [PubMed]
- Naud, S.; Ibrahim, A.; Valles, C.; Maatouk, M.; Bittar, F.; Tidjani Alou, M.; Raoult, D. Candidate Phyla Radiation, an Underappreciated Division of the Human Microbiome, and Its Impact on Health and Disease. Clin. Microbiol. Rev. 2022, 35, e0014021. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Luo, K.; Su, Z.; Huang, F.; Wu, Y.; Zhou, F.; Li, Y.; Peng, X.; Li, J.; Ren, B. Dental calculus: A repository of bioinformation indicating diseases and human evolution. Front. Cell. Infect. Microbiol. 2022, 12, 1035324. [Google Scholar] [CrossRef] [PubMed]
- Kazarina, A.; Petersone-Gordina, E.; Kimsis, J.; Kuzmicka, J.; Zayakin, P.; Griškjans, Ž.; Gerhards, G.; Ranka, R. The Postmedieval Latvian Oral Microbiome in the Context of Modern Dental Calculus and Modern Dental Plaque Microbial Profiles. Genes 2021, 12, 309. [Google Scholar] [CrossRef] [PubMed]
- Wasterlain, S.N.; Cunha, E.; Hillson, S. Periodontal disease in a Portuguese identified skeletal sample from the late nineteenth and early twentieth centuries. Am. J. Phys. Anthropol. 2011, 145, 30–42. [Google Scholar] [CrossRef]
- Kambouris, M.E.; Patrinos, G.P.; Velegraki, A.; Manoussopoulos, Y. Historical microbiology: Researching past bioevents by integrating scholarship (re)sources with paleomicrobiology assets. Future Microbiol. 2023, 18, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Arning, N.; Wilson, D.J. The past, present and future of ancient bacterial DNA. Microb. Genom. 2020, 6, mgen000384. [Google Scholar] [CrossRef]
- Bos, K.I.; Kühnert, D.; Herbig, A.; Esquivel-Gomez, L.R.; Andrades Valtueña, A.; Barquera, R.; Giffin, K.; Kumar Lankapalli, A.; Nelson, E.A.; Sabin, S.; et al. Paleomicrobiology: Diagnosis and Evolution of Ancient Pathogens. Annu. Rev. Microbiol. 2019, 73, 639–666. [Google Scholar] [CrossRef] [PubMed]
- Vassallo, A.; Modi, A.; Quagliariello, A.; Bacci, G.; Faddetta, T.; Gallo, M.; Provenzano, A.; La Barbera, A.; Lombardo, G.; Maggini, V.; et al. Novel Sources of Biodiversity and Biomolecules from Bacteria Isolated from a High Middle Ages Soil Sample in Palermo (Sicily, Italy). Microbiol. Spectr. 2023, 11, e0437422. [Google Scholar] [CrossRef] [PubMed]
- Pérez, V.; Liu, Y.; Hengst, M.B.; Weyrich, L.S. A Case Study for the Recovery of Authentic Microbial Ancient DNA from Soil Samples. Microorganisms 2022, 10, 1623. [Google Scholar] [CrossRef] [PubMed]
- Capo, E.; Monchamp, M.E.; Coolen, M.J.L.; Domaizon, I.; Armbrecht, L.; Bertilsson, S. Environmental paleomicrobiology: Using DNA preserved in aquatic sediments to its full potential. Environ. Microbiol. 2022, 24, 2201–2209. [Google Scholar] [CrossRef] [PubMed]
- Colautti, A.; Comi, G.; Peterlunger, E.; Iacumin, L. Ancient Roman bacterium against current issues: Strain Aquil_B6, Paenisporosarcina quisquiliarum, or Psychrobacillus psychrodurans? Microbiol. Spectr. 2023, 11, e0068623. [Google Scholar] [CrossRef] [PubMed]
- Eisenhofer, R.; Weyrich, L.S. Proper Authentication of Ancient DNA Is Still Essential. Genes 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Key, F.M.; Posth, C.; Krause, J.; Herbig, A.; Bos, K.I. Mining Metagenomic Data Sets for Ancient DNA: Recommended Protocols for Authentication. Trends Genet. 2017, 33, 508–520. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M. Paleomicrobiology Data: Authentification and Interpretation. Microbiol. Spectr. 2016, 4, 1110–1128. [Google Scholar] [CrossRef] [PubMed]
- Afouda, P.; Dubourg, G.; Raoult, D. Archeomicrobiology applied to environmental samples. Microb. Pathog. 2020, 143, 104140. [Google Scholar] [CrossRef] [PubMed]
- Alt, K.W.; Al-Ahmad, A.; Woelber, J.P. Nutrition and Health in Human Evolution-Past to Present. Nutrients 2022, 14, 3594. [Google Scholar] [CrossRef]
- Achtman, M.; Zhou, Z. Metagenomics of the modern and historical human oral microbiome with phylogenetic studies on Streptococcus mutans and Streptococcus sobrinus. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190573. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.E.; Sabin, S.; Ziesemer, K.; Vågene, Å.J.; Schroeder, H.; Ozga, A.T.; Sankaranarayanan, K.; Hofman, C.A.; Fellows Yates, J.A.; Salazar-García, D.C.; et al. Differential preservation of endogenous human and microbial DNA in dental calculus and dentin. Sci. Rep. 2018, 8, 9822. [Google Scholar] [CrossRef] [PubMed]
- Warinner, C.; Speller, C.; Collins, M.J. A new era in palaeomicrobiology: Prospects for ancient dental calculus as a long-term record of the human oral microbiome. Philos. Trans. R Soc. Lond. B Biol. Sci. 2015, 370, 20130376. [Google Scholar] [CrossRef] [PubMed]
- Velsko, I.M.; Semerau, L.; Inskip, S.A.; García-Collado, M.I.; Ziesemer, K.; Ruber, M.S.; Benítez de Lugo Enrich, L.; Molero García, J.M.; Valle, D.G.; Peña Ruiz, A.C.; et al. Ancient dental calculus preserves signatures of biofilm succession and interindividual variation independent of dental pathology. PNAS Nexus 2022, 1, pgac148. [Google Scholar] [CrossRef] [PubMed]
- Moore, N.E.; Weyrich, L.S. A Standardized Approach for Shotgun Metagenomic Analysis of Ancient Dental Calculus. Methods Mol. Biol. 2021, 2327, 93–118. [Google Scholar] [CrossRef]
- Willmann, C.; Mata, X.; Hanghoej, K.; Tonasso, L.; Tisseyre, L.; Jeziorski, C.; Cabot, E.; Chevet, P.; Crubézy, E.; Orlando, L.; et al. Oral health status in historic population: Macroscopic and metagenomic evidence. PLoS ONE 2018, 13, e0196482. [Google Scholar] [CrossRef]
- Cai, P.; Sun, X.; Wu, Y.; Gao, C.; Mortimer, M.; Holden, P.A.; Redmile-Gordon, M.; Qiaoyun, H. Soil biofilms: Microbial interactions, challenges, and advanced techniques for ex-situ characterization. Soil Ecol. Lett. 2019, 1, 85–93. [Google Scholar] [CrossRef]
- Pitts, N.B.; Twetman, S.; Fisher, J.; Marsh, P.D. Understanding dental caries as a non-communicable disease. Br. Dent. J. 2021, 231, 749–753. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.F., Jr.; Rôças, I.N. Present status and future directions: Microbiology of endodontic infections. Int. Endod. J. 2022, 55, 512–530. [Google Scholar] [CrossRef]
- Mombelli, A. Microbial colonization of the periodontal pocket and its significance for periodontal therapy. Periodontol. 2000 2018, 76, 85–96. [Google Scholar] [CrossRef]
- Willis, J.R.; Gabaldón, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Solbiati, J.; Frias-Lopez, J. Metatranscriptome of the Oral Microbiome in Health and Disease. J. Dent. Res. 2018, 97, 492–500. [Google Scholar] [CrossRef]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the Oral Microbiota in Health: Mechanisms That Prevent Dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef]
- Gancz, A.S.; Weyrich, L.S. Studying ancient human oral microbiomes could yield insights into the evolutionary history of noncommunicable diseases. F1000Res 2023, 12, 109. [Google Scholar] [CrossRef]
- Tricco, A.C.; Lillie, E.; Zarin, W.; O’Brien, K.K.; Colquhoun, H.; Levac, D.; Moher, D.; Peters, M.D.J.; Horsley, T.; Weeks, L.; et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): Checklist and Explanation. Ann. Intern. Med. 2018, 169, 467–473. [Google Scholar] [CrossRef]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. BMJ 2009, 62, e1–e34. [Google Scholar] [CrossRef] [PubMed]
- Adler, C.J.; Dobney, K.; Weyrich, L.S.; Kaidonis, J.; Walker, A.W.; Haak, W.; Bradshaw, C.J.; Townsend, G.; Sołtysiak, A.; Alt, K.W.; et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat. Genet. 2013, 45, 450–455e1. [Google Scholar] [CrossRef]
- Weyrich, L.S.; Duchene, S.; Soubrier, J.; Arriola, L.; Llamas, B.; Breen, J.; Morris, A.G.; Alt, K.W.; Caramelli, D.; Dresely, V.; et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature 2017, 544, 357–361. [Google Scholar] [CrossRef]
- Jersie-Christensen, R.R.; Lanigan, L.T.; Lyon, D.; Mackie, M.; Belstrøm, D.; Kelstrup, C.D.; Fotakis, A.K.; Willerslev, E.; Lynnerup, N.; Jensen, L.J.; et al. Quantitative metaproteomics of medieval dental calculus reveals individual oral health status. Nat. Commun. 2018, 9, 4744. [Google Scholar] [CrossRef]
- Velsko, I.M.; Fellows Yates, J.A.; Aron, F.; Hagan, R.W.; Frantz, L.A.F.; Loe, L.; Martinez, J.B.R.; Chaves, E.; Gosden, C.; Larson, G.; et al. Microbial differences between dental plaque and historic dental calculus are related to oral biofilm maturation stage. Microbiome 2019, 7, 102. [Google Scholar] [CrossRef]
- McLean, J.S.; Bor, B.; Kerns, K.A.; Liu, Q.; To, T.T.; Solden, L.; Hendrickson, E.L.; Wrighton, K.; Shi, W.; He, X. Acquisition and Adaptation of Ultra-small Parasitic Reduced Genome Bacteria to Mammalian Hosts. Cell Rep. 2020, 32, 107939. [Google Scholar] [CrossRef]
- Neukamm, J.; Pfrengle, S.; Molak, M.; Seitz, A.; Francken, M.; Eppenberger, P.; Avanzi, C.; Reiter, E.; Urban, C.; Welte, B.; et al. 2000-year-old pathogen genomes reconstructed from metagenomic analysis of Egyptian mummified individuals. BMC Biol. 2020, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- Farrer, A.G.; Wright, S.L.; Skelly, E.; Eisenhofer, R.; Dobney, K.; Weyrich, L.S. Effectiveness of decontamination protocols when analyzing ancient DNA preserved in dental calculus. Sci. Rep. 2021, 11, 7456. [Google Scholar] [CrossRef] [PubMed]
- Granehäll, L.; Huang, K.D.; Tett, A.; Manghi, P.; Paladin, A.; O’Sullivan, N.; Rota-Stabelli, O.; Segata, N.; Zink, A.; Maixner, F. Metagenomic analysis of ancient dental calculus reveals unexplored diversity of oral archaeal Methanobrevibacter. Microbiome 2021, 9, 197. [Google Scholar] [CrossRef] [PubMed]
- Ottoni, C.; Borić, D.; Cheronet, O.; Sparacello, V.; Dori, I.; Coppa, A.; Antonović, D.; Vujević, D.; Price, T.D.; Pinhasi, R.; et al. Tracking the transition to agriculture in Southern Europe through ancient DNA analysis of dental calculus. Proc. Natl. Acad. Sci. USA 2021, 118, e2102116118. [Google Scholar] [CrossRef] [PubMed]
- Fagernäs, Z.; Salazar-García, D.C.; Haber Uriarte, M.; Avilés Fernández, A.; Henry, A.G.; Lomba Maurandi, J.; Ozga, A.T.; Velsko, I.M.; Warinner, C. Understanding the microbial biogeography of ancient human dentitions to guide study design and interpretation. FEMS Microbes 2022, 3, xtac006. [Google Scholar] [CrossRef] [PubMed]
- Garralda, M.D.; Weiner, S.; Arensburg, B.; Maureille, B.; Vandermeersch, B. Dental Paleobiology in a Juvenile Neanderthal (Combe-Grenal, Southwestern France). Biology 2022, 11, 1352. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, G.; Nielsen, S.H.; Vetro, D.L.; Sawafuji, R.; Mackie, M.; Margaryan, A.; Fotakis, A.K.; Martínez-Labarga, C.; Fabbri, P.F.; Allentoft, M.E.; et al. Genomic ancestry, diet and microbiomes of Upper Palaeolithic hunter-gatherers from San Teodoro cave. Commun. Biol. 2022, 5, 1262. [Google Scholar] [CrossRef]
- Warinner, C.; Rodrigues, J.F.; Vyas, R.; Trachsel, C.; Shved, N.; Grossmann, J.; Radini, A.; Hancock, Y.; Tito, R.Y.; Fiddyment, S.; et al. Pathogens and host immunity in the ancient human oral cavity. Nat. Genet. 2014, 46, 336–344. [Google Scholar] [CrossRef]
- Gancz, A.S.; Farrer, A.G.; Nixon, M.P.; Wright, S.; Arriola, L.; Adler, C.; Davenport, E.R.; Gully, N.; Cooper, A.; Britton, K.; et al. Ancient dental calculus reveals oral microbiome shifts associated with lifestyle and disease in Great Britain. Nat. Microbiol. 2023, 8, 2315–2325. [Google Scholar] [CrossRef]
- White, D.J. Dental calculus: Recent insights into occurrence, formation, prevention, removal and oral health effects of supragingival and subgingival deposits. Eur. J. Oral Sci. 1997, 105, 508–522. [Google Scholar] [CrossRef]
- Nazir, M.; Al-Ansari, A.; Al-Khalifa, K.; Alhareky, M.; Gaffar, B.; Almas, K. Global Prevalence of Periodontal Disease and Lack of Its Surveillance. Sci. World J. 2020, 2020, 2146160. [Google Scholar] [CrossRef] [PubMed]
- Harrel, S.K.; Cobb, C.M.; Sheldon, L.N.; Rethman, M.P.; Sottosanti, J.S. Calculus as a Risk Factor for Periodontal Disease: Narrative Review on Treatment Indications When the Response to Scaling and Root Planing Is Inadequate. Dent. J. 2022, 10, 195. [Google Scholar] [CrossRef] [PubMed]
- Akcalı, A.; Lang, N.P. Dental calculus: The calcified biofilm and its role in disease development. Periodontol. 2000 2018, 76, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Forshaw, R. Dental calculus—Oral health, forensic studies and archaeology: A review. Br. Dent. J. 2022, 233, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; He, L.; Yan, S.; Chen, X.; Que, G. The impact of caries status on supragingival plaque and salivary microbiome in children with mixed dentition: A cross-sectional survey. BMC Oral Health 2021, 21, 319. [Google Scholar] [CrossRef] [PubMed]
- Yamabe, K.; Maeda, H.; Kokeguchi, S.; Soga, Y.; Meguro, M.; Naruishi, K.; Asakawa, S.; Takashiba, S. Antigenic group II chaperonin in Methanobrevibacter oralis may cross-react with human chaperonin CCT. Mol. Oral Microbiol. 2010, 25, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrowicz, P.; Brzezińska-Błaszczyk, E.; Dudko, A.; Agier, J. Archaea Occurrence in the Subgingival Biofilm in Patients with Peri-Implantitis and Periodontitis. Int. J. Periodontics Restor. Dent. 2020, 40, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wang, L.; Zhang, L.; Tan, X.; Huang, D.; Song, D. Potential relationship between clinical symptoms and the root canal microbiomes of root filled teeth based on the next-generation sequencing. Int. Endod. J. 2022, 55, 18–29. [Google Scholar] [CrossRef]
- Vieira Colombo, A.P.; Magalhães, C.B.; Hartenbach, F.A.; Martins do Souto, R.; Maciel da Silva-Boghossian, C. Periodontal-disease-associated biofilm: A reservoir for pathogens of medical importance. Microb. Pathog. 2016, 94, 27–34. [Google Scholar] [CrossRef]
- Dabney, J.; Meyer, M.; Paabo, S. Ancient DNA Damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012567. [Google Scholar]
- Klapper, M.; Hübner, A.; Ibrahim, A.; Wasmuth, I.; Borry, M.; Haensch, V.G.; Zhang, S.; Al-Jammal, W.K.; Suma, H.; Fellows Yates, J.A.; et al. Natural products from reconstructed bacterial genomes of the Middle and Upper Paleolithic. Science 2023, 380, 619–624. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Putrino, A.; Marinelli, E.; Galeotti, A.; Ferrazzano, G.F.; Ciribè, M.; Zaami, S. A Journey into the Evolution of Human Host-Oral Microbiome Relationship through Ancient Dental Calculus: A Scoping Review. Microorganisms 2024, 12, 902. https://doi.org/10.3390/microorganisms12050902
Putrino A, Marinelli E, Galeotti A, Ferrazzano GF, Ciribè M, Zaami S. A Journey into the Evolution of Human Host-Oral Microbiome Relationship through Ancient Dental Calculus: A Scoping Review. Microorganisms. 2024; 12(5):902. https://doi.org/10.3390/microorganisms12050902
Chicago/Turabian StylePutrino, Alessandra, Enrico Marinelli, Angela Galeotti, Gianmaria Fabrizio Ferrazzano, Massimiliano Ciribè, and Simona Zaami. 2024. "A Journey into the Evolution of Human Host-Oral Microbiome Relationship through Ancient Dental Calculus: A Scoping Review" Microorganisms 12, no. 5: 902. https://doi.org/10.3390/microorganisms12050902
APA StylePutrino, A., Marinelli, E., Galeotti, A., Ferrazzano, G. F., Ciribè, M., & Zaami, S. (2024). A Journey into the Evolution of Human Host-Oral Microbiome Relationship through Ancient Dental Calculus: A Scoping Review. Microorganisms, 12(5), 902. https://doi.org/10.3390/microorganisms12050902









