Genome-Wide Computational Prediction and Analysis of Noncoding RNAs in Oleidesulfovibrio alaskensis G20
Abstract
1. Introduction
2. Materials and Methods
2.1. Prediction and Identification of the ncRNAs
2.2. Secondary Structures of ncRNAs
3. Results
Secondary Structures of ncRNAs of OA G20
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Repoila, F.; Darfeuille, F. Small regulatory non-coding RNAs in bacteria: Physiology and mechanistic aspects. Biol. Cell 2009, 101, 117–131. [Google Scholar] [CrossRef]
- Wootton, L. Non-coding RNAs make E. coli unpalatable. Nat. Rev. Microbiol. 2012, 10, 733. [Google Scholar] [CrossRef]
- Ahmed, W.; Zheng, K.; Liu, Z.F. Small Non-Coding RNAs: New insights in Modulation of Host immune Response by intracellular Bacterial Pathogens. Front. Immunol. 2016, 7, 431. [Google Scholar] [CrossRef]
- Irnov, I.; Sharma, C.M.; Vogel, J.; Winkler, W.C. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010, 38, 6637–6651. [Google Scholar] [CrossRef]
- Dar, D.; Sorek, R. Bacterial Noncoding RNAs Excised from within Protein-Coding Transcripts. mBio 2018, 9, e01730-18. [Google Scholar] [CrossRef]
- Tomasini, A.; Moreau, K.; Chicher, J.; Geissmann, T.; Vandenesch, F.; Romby, P.; Marzi, S.; Caldelari, I. The RNA targetome of Staphylococcus aureus non-coding RNA RsaA: Impact on cell surface properties and defense mechanisms. Nucleic Acids Res. 2017, 45, 6746–6760. [Google Scholar] [CrossRef]
- Saetrom, P.; Sneve, R.; Kristiansen, K.I.; Snove, O., Jr.; Grunfeld, T.; Rognes, T.; Seeberg, E. Predicting non-coding RNA genes in Escherichia coli with boosted genetic programming. Nucleic Acids Res. 2005, 33, 3263–3270. [Google Scholar] [CrossRef]
- Boysen, A.; Moller-Jensen, J.; Kallipolitis, B.; Valentin-Hansen, P.; Overgaard, M. Translational Regulation of Gene Expression by an Anaerobically Induced Small Non-coding RNA in Escherichia coli. J. Biol. Chem. 2010, 285, 10690–10702. [Google Scholar] [CrossRef]
- Storz, G.; Vogel, J.; Wassarman, K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell 2011, 43, 880–891. [Google Scholar] [CrossRef]
- Nitzan, M.; Rehani, R.; Margalit, H. Integration of Bacterial Small RNAs in Regulatory Networks. Annu. Rev. Biophys. 2017, 46, 131–148. [Google Scholar] [CrossRef]
- Pita, T.; Feliciano, J.R.; Leitao, J.H. Small Noncoding Regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia Complex. Int. J. Mol. Sci. 2018, 19, 3759. [Google Scholar] [CrossRef]
- Bordeau, V.; Felden, B. Curli synthesis and biofilm formation in enteric bacteria are controlled by a dynamic small RNA module made up of a pseudoknot assisted by an RNA chaperone. Nucleic Acids Res. 2014, 42, 4682–4696. [Google Scholar] [CrossRef]
- Chen, L.; Gu, L.P.; Geng, X.F.; Xu, G.X.; Huang, X.X.; Zhu, X.J. A novel cis antisense RNA AsfD promotes Salmonella enterica serovar Typhi motility and biofilm formation. Microb Pathog. 2020, 142, 104044. [Google Scholar] [CrossRef]
- De Lay, N.; Gottesman, S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol. Microbiol. 2012, 86, 524–538. [Google Scholar] [CrossRef]
- Thomason, M.K.; Fontaine, F.; De Lay, N.; Storz, G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol. Microbiol. 2012, 84, 17–35. [Google Scholar] [CrossRef]
- Taylor, P.K.; Van Kessel, A.T.M.; Colavita, A.; Hancock, R.E.W.; Mah, T.F. A novel small RNA is important for biofilm formation and pathogenicity in Pseudomonas aeruginosa. PLoS ONE 2017, 12, e0182582. [Google Scholar] [CrossRef]
- Orell, A.; Tripp, V.; Aliaga-Tobar, V.; Albers, S.V.; Maracaja-Coutinho, V.; Randau, L. A regulatory RNA is involved in RNA duplex formation and biofilm regulation in Sulfolobus acidocaldarius. Nucleic Acids Res. 2018, 46, 4794–4806. [Google Scholar] [CrossRef]
- Silvaggi, J.M.; Perkins, J.B.; Losick, R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J. Bacteriol. 2006, 188, 532–541. [Google Scholar] [CrossRef]
- Amarasinghe, J.J.; Connell, T.D.; Scannapieco, F.A.; Haase, E.M. Novel iron-regulated and Fur-regulated small regulatory RNAs in Aggregatibacter actinomycetemcomitans. Mol. Oral Microbiol. 2012, 27, 327–349. [Google Scholar] [CrossRef]
- Tahrioui, A.; Duchesne, R.; Bouffartigues, E.; Rodrigues, S.; Maillot, O.; Tortuel, D.; Hardouin, J.; Taupin, L.; Groleau, M.C.; Dufour, A.; et al. Extracellular DNA release, quorum sensing, and PrrF1/F2 small RNAs are key players in Pseudomonas aeruginosa tobramycin-enhanced biofilm formation. Npj Biofilms Microbiomes 2019, 5, 15. [Google Scholar] [CrossRef]
- Tripathi, A.K.; Thakur, P.; Saxena, P.; Rauniyar, S.; Gopalakrishnan, V.; Singh, R.N.; Gadhamshetty, V.; Gnimpieba, E.Z.; Jasthi, B.K.; Sani, R.K. Gene Sets and Mechanisms of Sulfate-Reducing Bacteria Biofilm Formation and Quorum Sensing with Impact on Corrosion. Front. Microbiol. 2021, 12, 754140. [Google Scholar] [CrossRef] [PubMed]
- Beech, I.B.; Sunner, J.A. Sulphate-reducing bacteria and their role in corrosion of ferrous materials. In Sulphate-Reducing Bacteria: Environmental and Engineered Systems; Barton, L.L., Hamilton, W.A., Eds.; Cambridge University Press: Cambridge, UK, 2007; pp. 459–482. [Google Scholar]
- Procópio, L. The role of biofilms in the corrosion of steel in marine environments. World J. Microbiol. Biotechnol. 2019, 35, 73. [Google Scholar] [CrossRef]
- Clark, M.E.; Edelmann, R.E.; Duley, M.L.; Wall, J.D.; Fields, M.W. Biofilm formation in Desulfovibrio vulgaris Hildenborough is dependent upon protein filaments. Environ. Microbiol. 2007, 9, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.E.; He, Z.L.; Redding, A.M.; Joachimiak, M.P.; Keasling, J.D.; Zhou, J.Z.Z.; Arkin, A.P.; Mukhopadhyay, A.; Fields, M.W. Transcriptomic and proteomic analyses of Desulfovibrio vulgaris biofilms: Carbon and energy flow contribute to the distinct biofilm growth state. BMC Genom. 2012, 13, 138. [Google Scholar] [CrossRef]
- Zhang, Y.; Pei, G.S.; Chen, L.; Zhang, W.W. Metabolic dynamics of Desulfovibrio vulgaris biofilm grown on a steel surface. Biofouling 2016, 32, 725–736. [Google Scholar] [CrossRef]
- Scarascia, G.; Lehmann, R.; Machuca, L.L.; Morris, C.; Cheng, K.Y.; Kaksonen, A.; Hong, P.Y. Effect of Quorum Sensing on the Ability of Desulfovibrio vulgaris to Form Biofilms and to Biocorrode Carbon Steel in Saline Conditions. Appl. Environ. Microbiol. 2020, 86, e01664-19. [Google Scholar] [CrossRef]
- Waters, C.M.; Bassler, B.L. Quorum sensing: Cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 2005, 21, 319–346. [Google Scholar] [CrossRef]
- Qi, Z.H.; Chen, L.; Zhang, W.W. Comparison of Transcriptional Heterogeneity of Eight Genes between Batch Desulfovibrio vulgaris Biofilm and Planktonic Culture at a Single-Cell Level. Front. Microbiol. 2016, 7, 597. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Sweeney, B.A.; Petrov, A.I.; Ribas, C.E.; Finn, R.D.; Bateman, A.; Szymanski, M.; Karlowski, W.M.; Seemann, S.E.; Gorodkin, J.; Cannone, J.J.; et al. RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021, 49, D212–D220. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Sweeney, B.A.; Hoksza, D.; Nawrocki, E.P.; Ribas, C.E.; Madeira, F.; Cannone, J.J.; Gutell, R.; Maddala, A.; Meade, C.D.; Williams, L.D.; et al. R2DT is a framework for predicting and visualising RNA secondary structure using templates. Nat. Commun. 2021, 12, 3494. [Google Scholar] [CrossRef]
- Elias, R.; Hoksza, D. TRAVeLer: A tool for template-based RNA secondary structure visualization. BMC Bioinform. 2017, 18, 487. [Google Scholar] [CrossRef] [PubMed]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef] [PubMed]
- Sprinzl, M.; Horn, C.; Brown, M.; Ioudovitch, A.; Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998, 26, 148–153. [Google Scholar] [CrossRef]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.S.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar] [CrossRef]
- Bernier, C.R.; Petrov, A.S.; Waterbury, C.C.; Jett, J.; Li, F.B.; Freil, L.E.; Xiong, X.; Wang, L.; Migliozzi, B.L.R.; Hershkovits, E.; et al. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss 2014, 169, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Thornlow, B.P.; Armstrong, J.; Holmes, A.D.; Howard, J.M.; Corbett-Detig, R.B.; Lowe, T.M. Predicting transfer RNA gene activity from sequence and genome context. Genome Res. 2020, 30, 85–94. [Google Scholar] [CrossRef]
- Brown, J.W. The Ribonuclease P database. Nucleic Acids Res. 1996, 24, 236–237. [Google Scholar] [CrossRef][Green Version]
- Saxena, P.; Rauniyar, S.; Thakur, P.; Singh, R.N.; Bomgni, A.; Alaba, M.O.; Tripathi, A.K.; Gnimpieba, E.Z.; Lushbough, C.; Sani, R.K. Integration of text mining and biological network analysis: Identification of essential genes in sulfate-reducing bacteria. Front. Microbiol. 2023, 14, 1086021. [Google Scholar] [CrossRef]
- Hartman, T.W.; Radichev, E.; Ali, H.M.; Alaba, M.O.; Hoffman, M.; Kassa, G.; Sani, R.; Gadhamshetty, V.; Ragi, S.; Messerli, S.M.; et al. BASIN: A Semi-automatic Workflow, with Machine Learning Segmentation, for Objective Statistical Analysis of Biomedical and Biofilm Image Datasets. J. Mol. Biol. 2023, 435, 167895. [Google Scholar] [CrossRef]
- Singh, R.N.; Gnimpieba, E.Z.; Sani, R.K. Genome wide computational prediction and analysis of noncoding genome of corrosive biofilm forming Oleidesulfovibrio alaskensis G20. In Proceedings of the 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Istanbul, Turkey, 5–8 December 2023; pp. 4509–4512. [Google Scholar]
- Chelkowska-Pauszek, A.; Kosinski, J.G.; Marciniak, K.; Wysocka, M.; Bakowska-Zywicka, K.; Zywicki, M. The Role of RNA Secondary Structure in Regulation of Gene Expression in Bacteria. Int. J. Mol. Sci. 2021, 22, 7845. [Google Scholar] [CrossRef]
- Lai, E.C. RNA sensors and riboswitches: Self-regulating messages. Curr. Biol. 2003, 13, R285–R291. [Google Scholar] [CrossRef]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef]
- Reyes-Darias, J.A.; Krell, T. Riboswitches as Potential Targets for the Development of Anti-Biofilm Drugs. Curr. Top. Med. Chem. 2017, 17, 1945–1953. [Google Scholar] [CrossRef]
- Serganov, A.; Polonskaia, A.; Phan, A.T.; Breaker, R.R.; Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 2006, 441, 1167–1171. [Google Scholar] [CrossRef]
- Edwards, T.E.; Ferré-D’Amaré, A.R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 2006, 14, 1459–1468. [Google Scholar] [CrossRef]
- Rentmeister, A.; Mayer, G.; Kuhn, N.; Famulok, M. Conformational changes in the expression domain of the Escherichia coli thiM riboswitch. Nucleic Acids Res. 2007, 35, 3713–3722. [Google Scholar] [CrossRef]
- Pavlova, N.; Traykovska, M.; Penchovsky, R. Targeting FMN, TPP, SAM-I, and glmS Riboswitches with Chimeric Antisense Oligonucleotides for Completely Rational Antibacterial Drug Development. Antibiotics 2023, 12, 1607. [Google Scholar] [CrossRef]
- Stav, S.; Atilho, R.M.; Arachchilage, G.M.; Nguyen, G.; Higgs, G.; Breaker, R.R. Genome-wide discovery of structured noncoding RNAs in bacteria. BMC Microbiol. 2019, 19, 66. [Google Scholar] [CrossRef]
- Rentmeister, A.; Mayer, G.; Kuhn, N.; Famulok, M. Secondary structures and functional requirements for thiM riboswitches from Desulfovibrio vulgaris, Erwinia carotovora, and Rhodobacter spheroides. Biol. Chem. 2008, 389, 127–134. [Google Scholar] [CrossRef]
- Panchal, V.; Brenk, R. Riboswitches as Drug Targets for Antibiotics. Antibiotics 2021, 10, 45. [Google Scholar] [CrossRef]
- Pavlova, N.; Penchovsky, R. Genome-wide bioinformatics analysis of FMN, SAM-I, glmS, TPP, lysine, purine, cobalamin, and SAH riboswitches for their applications as allosteric antibacterial drug targets in human pathogenic bacteria. Expert Opin. Ther. Targets 2019, 23, 631–643. [Google Scholar] [CrossRef]
- Kozlowski, P.M.; Garabato, B.D.; Lodowski, P.; Jaworska, M. Photolytic properties of cobalamins: A theoretical perspective. Dalton Trans. 2016, 45, 4457–4470. [Google Scholar] [CrossRef]
- Lobo, S.A.L.; Videira, M.A.M.; Pacheco, I.; Wass, M.N.; Warren, M.J.; Teixeira, M.; Matias, P.M.; Romao, C.V.; Saraiva, L.M. Desulfovibrio vulgaris CbiKP cobaltochelatase: Evolution of a haem binding protein orchestrated by the incorporation of two histidine residues. Environ. Microbiol. 2017, 19, 106–118. [Google Scholar] [CrossRef]
- Choudhary, P.K.; Duret, A.; Rohrbach-Brandt, E.; Holliger, C.; Sigel, R.K.O.; Maillard, J. Diversity of Cobalamin Riboswitches in the Corrinoid-Producing Organohalide Respirer Desulfitobacterium hafniense. J. Bacteriol. 2013, 195, 5186–5195. [Google Scholar] [CrossRef]
- Serrano-Gutiérrez, M.; Merino, E. Antisense-acting riboswitches: A poorly characterized yet important model of transcriptional regulation in prokaryotic organisms. PLoS ONE 2023, 18, e0281744. [Google Scholar] [CrossRef]
- Kuehne, A.; Emmert, H.; Soehle, J.; Winnefeld, M.; Fischer, F.; Wenck, H.; Gallinat, S.; Terstegen, L.; Lucius, R.; Hildebrand, J.; et al. Acute Activation of Oxidative Pentose Phosphate Pathway as First-Line Response to Oxidative Stress in Human Skin Cells. Mol. Cell 2015, 59, 359–371. [Google Scholar] [CrossRef]
- Keshavarz, H.; Moghadam, R.S.G. Seed priming with cobalamin (vitamin B12) provides significant protection against salinity stress in the common bean. Rhizosphere 2017, 3, 143–149. [Google Scholar] [CrossRef]
- Grundy, F.J.; Henkin, T.M. t-RNA as a Positive Regulator of Transcription Antitermination in B. subtilis. Cell 1993, 74, 475–482. [Google Scholar] [CrossRef]
- Barrick, J.E.; Corbino, K.A.; Winkler, W.C.; Nahvi, A.; Mandal, M.; Collins, J.; Lee, M.; Roth, A.; Sudarsan, N.; Jona, I.; et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA 2004, 101, 6421–6426. [Google Scholar] [CrossRef]
- Khani, A.; Popp, N.; Kreikemeyer, B.; Patenge, N. A Glycine Riboswitch in Streptococcus pyogenes Controls Expression of a Sodium: Alanine Symporter Family Protein Gene. Front. Microbiol. 2018, 9, 200. [Google Scholar] [CrossRef]
- Babina, A.M.; Lea, N.E.; Meyer, M.M. In Vivo Behavior of the Tandem Glycine Riboswitch in Bacillus subtilis. mBio 2017, 8, e01602-17. [Google Scholar] [CrossRef]
- Epshtein, V.; Mironov, A.S.; Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. USA 2003, 100, 5052–5056. [Google Scholar] [CrossRef]
- Lee, Y.J.; Wang, C.L. Links between S-adenosylmethionine and Agr-based quorum sensing for biofilm development in Listeria monocytogenes EGD-e. Microbiologyopen 2020, 9, e1015. [Google Scholar] [CrossRef]
- Zhou, J.J.; Deng, Y.C.; Iyamu, I.D.; Horton, J.R.; Yu, D.; Hajian, T.; Vedadi, M.; Rotili, D.; Mai, A.; Blumenthal, R.M.; et al. Comparative Study of Adenosine Analogs as Inhibitors of Protein Arginine Methyltransferases and a Clostridioides difficile-Specific DNA Adenine Methyltransferase. ACS Chem. Biol. 2023, 18, 734–745. [Google Scholar] [CrossRef]
- Serganov, A.; Huang, L.L.; Patel, D.J. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 2009, 458, 233–237. [Google Scholar] [CrossRef]
- Winkler, W.C.; Cohen-Chalamish, S.; Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 2002, 99, 15908–15913. [Google Scholar] [CrossRef]
- Howe, J.A.; Wang, H.; Fischmann, T.O.; Balibar, C.J.; Xiao, L.; Galgoci, A.M.; Malinverni, J.C.; Mayhood, T.; Villafania, A.; Nahvi, A.; et al. Selective small-molecule inhibition of an RNA structural element. Nature 2015, 526, 672–677. [Google Scholar] [CrossRef]
- Lee, E.R.; Blount, K.F.; Breaker, R.R. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009, 6, 187–194. [Google Scholar] [CrossRef]
- Krantz, G.P.; Lucas, K.; Wunderlich, E.L.; Hoang, L.T.; Avci, R.; Siuzdak, G.; Fields, M.W. Bulk phase resource ratio alters carbon steel corrosion rates and endogenously produced extracellular electron transfer mediators in a sulfate-reducing biofilm. Biofouling 2019, 35, 669–683. [Google Scholar] [CrossRef]
- Edel, M.; Sturm, G.; Sturm-Richter, K.; Wagner, M.; Ducassou, J.N.; Couté, Y.; Horn, H.; Gescher, J. Extracellular riboflavin induces anaerobic biofilm formation in Shewanella oneidensis. Biotechnol. Biofuels 2021, 14, 130. [Google Scholar] [CrossRef]
- Pu, Y.A.; Tian, Y.; Hou, S.; Dou, W.W.; Chen, S.G. Enhancement of exogenous riboflavin on microbiologically influenced corrosion of nickel by electroactive Desulfovibrio vulgaris biofilm. Npj Mater. Degrad. 2023, 7, 7. [Google Scholar] [CrossRef]
- Caly, D.L.; Bellini, D.; Walsh, M.A.; Dow, J.M.; Ryan, R.P. Targeting Cyclic di-GMP Signalling: A Strategy to Control Biofilm Formation? Curr. Pharm. Design 2015, 21, 12–24. [Google Scholar] [CrossRef]
- Whiteley, C.G.; Lee, D.J. Bacterial diguanylate cyclases: Structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol. Adv. 2015, 33, 124–141. [Google Scholar] [CrossRef]
- Kampf, J.; Stülke, J. Cyclic-di-GMP signalling meets extracellular polysaccharide synthesis in Bacillus subtilis. Environ. Microbiol. Rep. 2017, 9, 182–185. [Google Scholar] [CrossRef]
- Tuckerman, J.R.; Gonzalez, G.; Gilles-Gonzalez, M.A. Cyclic di-GMP Activation of Polynucleotide Phosphorylase Signal-Dependent RNA Processing. J. Mol. Biol. 2011, 407, 633–639. [Google Scholar] [CrossRef]
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef]
- Pratt, J.T.; Tamayo, R.; Tischler, A.D.; Camilli, A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 2007, 282, 12860–12870. [Google Scholar] [CrossRef]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef]
- Tang, Q.; Yin, K.; Qian, H.L.; Zhao, Y.W.; Wang, W.; Chou, S.H.; Fu, Y.; He, J. Cyclic di-GMP contributes to adaption and virulence of Bacillus thuringiensis through a riboswitch-regulated collagen adhesion protein. Sci. Rep. 2016, 6, 28807. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wang, J.P.; Yin, W.; Shao, X.H.; Zheng, H.J.; Li, M.S.; Zhao, Y.W.; Sun, M.; Wang, S.Y.; Yu, Z.N. Complete Genome Sequence of Bacillus thuringiensis subsp. chinensis Strain CT-43. J. Bacteriol. 2011, 193, 3407–3408. [Google Scholar] [CrossRef]
- da Silva, S.M.; Amaral, C.; Neves, S.S.; Santos, C.; Pimentel, C.; Rodrigues-Pousada, C. An HcpR paralog of Desulfovibrio gigas provides protection against nitrosative stress. FEBS Open Bio 2015, 5, 594–604. [Google Scholar] [CrossRef]
- Hutchings, M.I.; Mandhana, N.; Spiro, S. The NorR protein of Escherichia coli activates expression of the flavorubredoxin gene norV in response to reactive nitrogen species. J. Bacteriol. 2002, 184, 4640–4643. [Google Scholar] [CrossRef]
- Rajeev, L.; Luning, E.G.; Altenburg, S.; Zane, G.M.; Baidoo, E.E.; Catena, M.; Keasling, J.D.; Wall, J.D.; Fields, M.W.; Mukhopadhyay, A. Identification of a cyclic-di-GMP-modulating response regulator that impacts biofilm formation in a model sulfate reducing bacterium. Front. Microbiol. 2014, 5, 382. [Google Scholar] [CrossRef]
- Cone, J.E.; Martindelrio, R.; Davis, J.N.; Stadtman, T.C. Chemical Characterization of Selenoprotein Component of Clostridial Glycine Reductase—Identification of Selenocysteine as Organoselenium Moiety. Proc. Natl. Acad. Sci. USA 1976, 73, 2659–2663. [Google Scholar] [CrossRef]
- Serrao, V.H.B.; Silva, I.R.; da Silva, M.T.A.; Scortecci, J.F.; Fernandes, A.D.; Thiemann, O.H. The unique tRNASec and its role in selenocysteine biosynthesis. Amino Acids 2018, 50, 1145–1167. [Google Scholar] [CrossRef]
- Zhang, Y.; Romero, H.; Salinas, G.; Gladyshev, V.N. Dynamic evolution of selenocysteine utilization in bacteria: A balance between selenoprotein loss and evolution of selenocysteine from redox active cysteine residues. Genome Biol. 2006, 7, R94. [Google Scholar] [CrossRef]
- Nunes, L.G.A.; Cain, A.; Comyns, C.; Hoffmann, P.R.; Krahn, N. Deciphering the Role of Selenoprotein M. Antioxidants 2023, 12, 1906. [Google Scholar] [CrossRef]
- Zinoni, F.; Birkmann, A.; Stadtman, T.C.; Bock, A. Nucleotide-Sequence and Expression of the Selenocysteine-Containing Polypeptide of Formate Dehydrogenase (Formate-Hydrogen-Lyase-Linked) from Escherichia coli. Proc. Natl. Acad. Sci. USA 1986, 83, 4650–4654. [Google Scholar] [CrossRef]
- Peng, T.; Lin, J.; Xu, Y.Z.; Zhang, Y. Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 2016, 10, 2048–2059. [Google Scholar] [CrossRef] [PubMed]
- Sumner, S.E.; Markley, R.L.; Kirimanjeswara, G.S. Role of Selenoproteins in Bacterial Pathogenesis. Biol. Trace Elem. Res. 2019, 192, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, M.; Mallard, C.; Barke, T.; Hancock, L.E.; Self, W.T. A Selenium-Dependent Xanthine Dehydrogenase Triggers Biofilm Proliferation in Enterococcus faecalis through Oxidant Production. J. Bacteriol. 2011, 193, 1643–1652. [Google Scholar] [CrossRef] [PubMed]
- Keiler, K.C.; Waller, P.R.H.; Sauer, R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 1996, 271, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Nevo-Dinur, K.; Nussbaum-Shochat, A.; Ben-Yehuda, S.; Amster-Choder, O. Translation-Independent Localization of mRNA in E. coli. Science 2011, 331, 1081–1084. [Google Scholar] [CrossRef] [PubMed]
- Kannaiah, S.; Livny, J.; Amster-Choder, O. Spatiotemporal Organization of the E. coli Transcriptome: Translation Independence and Engagement in Regulation. Mol. Cell 2019, 76, 574–589.e7. [Google Scholar] [CrossRef]
- Fei, J.Y.; Sharma, C.M. RNA Localization in Bacteria. Microbiol. Spectr. 2018, 6, RWR-0024-2018. [Google Scholar] [CrossRef] [PubMed]
- Malabirade, A.; Habier, J.; Heintz-Buschart, A.; May, P.; Godet, J.; Halder, R.; Etheridge, A.; Galas, D.; Wilmes, P.; Fritz, J.V. The RNA Complement of Outer Membrane Vesicles from Salmonella enterica Serovar Typhimurium under Distinct Culture Conditions. Front. Microbiol. 2018, 9, 2015. [Google Scholar] [CrossRef]
- Alikina, O.V.; Glazunova, O.A.; Bykov, A.A.; Kiselev, S.S.; Tutukina, M.N.; Shavkunov, K.S.; Ozoline, O.N. A cohabiting bacterium alters the spectrum of short RNAs secreted by Escherichia coli. FEMS Microbiol. Lett. 2018, 365, fny262. [Google Scholar] [CrossRef]
- Himeno, H.; Kurita, D.; Muto, A. tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front. Genet. 2014, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.S.; Cao, Q.Q.; Liu, Z.Z.; Chen, J.P.; Yan, P.G.; Li, B.Y.; Xu, Y. Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis. Microorganisms 2022, 10, 1338. [Google Scholar] [CrossRef] [PubMed]
- Prody, G.A.; Bakos, J.T.; Buzayan, J.M.; Schneider, I.R.; Bruening, G. Autolytic Processing of Dimeric Plant-Virus Satellite RNA. Science 1986, 231, 1577–1580. [Google Scholar] [CrossRef] [PubMed]
- Wassarman, K.M. 6S RNA: A regulator of transcription. Mol. Microbiol. 2007, 65, 1425–1431. [Google Scholar] [CrossRef] [PubMed]
- Burenina, O.Y.; Elkina, D.A.; Ovcharenko, A.; Bannikova, V.A.; Schlüter, M.A.C.; Oretskaya, T.S.; Hartmann, R.K.; Kubareva, E.A. Involvement of E. coli 6S RNA in Oxidative Stress Response. Int. J. Mol. Sci. 2022, 23, 3653. [Google Scholar] [CrossRef]
- Trotochaud, A.E.; Wassarman, K.M. 6S RNA regulation of pspF transcription leads to altered cell survival at high pH. J. Bacteriol. 2006, 188, 3936–3943. [Google Scholar] [CrossRef]
- Hoch, P.G.; Burenina, O.Y.; Weber, M.H.W.; Elkina, D.A.; Nesterchuk, M.V.; Sergiev, P.V.; Hartmann, R.K.; Kubareva, E.A. Phenotypic characterization and complementation analysis of Bacillus subtilis 6S RNA single and double deletion mutants. Biochimie 2015, 117, 87–99. [Google Scholar] [CrossRef]
- Haas, E.S.; Brown, J.W. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 1998, 26, 4093–4099. [Google Scholar] [CrossRef]
- Gössinger, M.; Lechner, M.; Brillante, N.; Weber, C.; Rossmanith, W.; Hartmann, R.K. Protein-only RNase P function in Escherichia coli: Viability, processing defects and differences between PRORP isoenzymes. Nucleic Acids Res. 2017, 45, 7441–7454. [Google Scholar] [CrossRef]
- Perreault, J.; Weinberg, Z.; Roth, A.; Popescu, O.; Chartrand, P.; Ferbeyre, G.; Breaker, R.R. Identification of Hammerhead Ribozymes in All Domains of Life Reveals Novel Structural Variations. PLoS Comput. Biol. 2011, 7, e1002031. [Google Scholar] [CrossRef]
- Jimenez, R.M.; Delwart, E.; Lupták, A. Structure-based Search Reveals Hammerhead Ribozymes in the Human Microbiome. J. Biol. Chem. 2011, 286, 7737–7743. [Google Scholar] [CrossRef] [PubMed]
- De Lay, N.; Schu, D.J.; Gottesman, S. Bacterial Small RNA-based Negative Regulation: Hfq and Its Accomplices. J. Biol. Chem. 2013, 288, 7996–8003. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Sundin, G.W. Genome-wide identification of Hfq-regulated small RNAs in the fire blight pathogen Erwinia amylovora discovered small RNAs with virulence regulatory function. BMC Genom. 2014, 15, 414. [Google Scholar] [CrossRef] [PubMed]
- Soper, T.; Mandin, P.; Majdalani, N.; Gottesman, S.; Woodson, S.A. Positive regulation by small RNAs and the role of Hfq. Proc. Natl. Acad. Sci. USA 2010, 107, 9602–9607. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.P.; Tang, D.J.; Chen, X.L.; He, Y.Q.; Feng, J.X.; Jiang, B.L.; Lu, G.T.; Lin, M.; Tang, J.L. Identification of four novel small non-coding RNAs from Xanthomonas campestris pathovar campestris. BMC Genom. 2010, 11, 316. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, C.; Findeiss, S.; Sharma, C.M.; Kuhfuss, J.; Hoffmann, S.; Vogel, J.; Stadler, P.F.; Bonas, U. Genome-wide transcriptome analysis of the plant pathogen Xanthomonas identifies sRNAs with putative virulence functions. Nucleic Acids Res. 2012, 40, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Zhao, Y.T.; Zhang, J.Q.; Wang, X.J.; Fang, R.X.; Jia, Y.T. Identification and functional characterization of small non-coding RNAs in Xanthomonas oryzae pathovar oryzae. BMC Genom. 2011, 12, 87. [Google Scholar] [CrossRef]
- Livny, J.; Brencic, A.; Lory, S.; Waldor, M.K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006, 34, 3484–3493. [Google Scholar] [CrossRef]
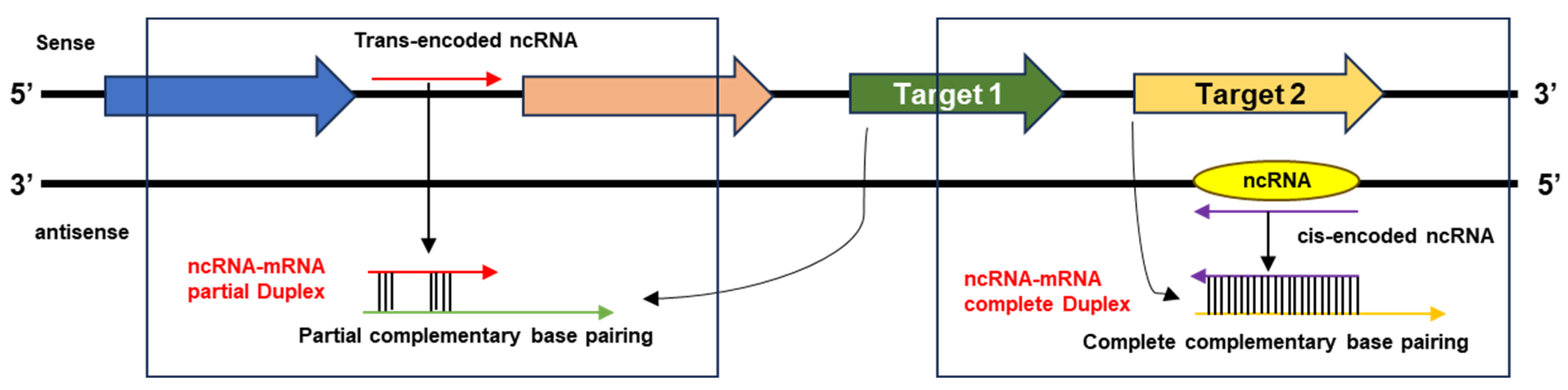

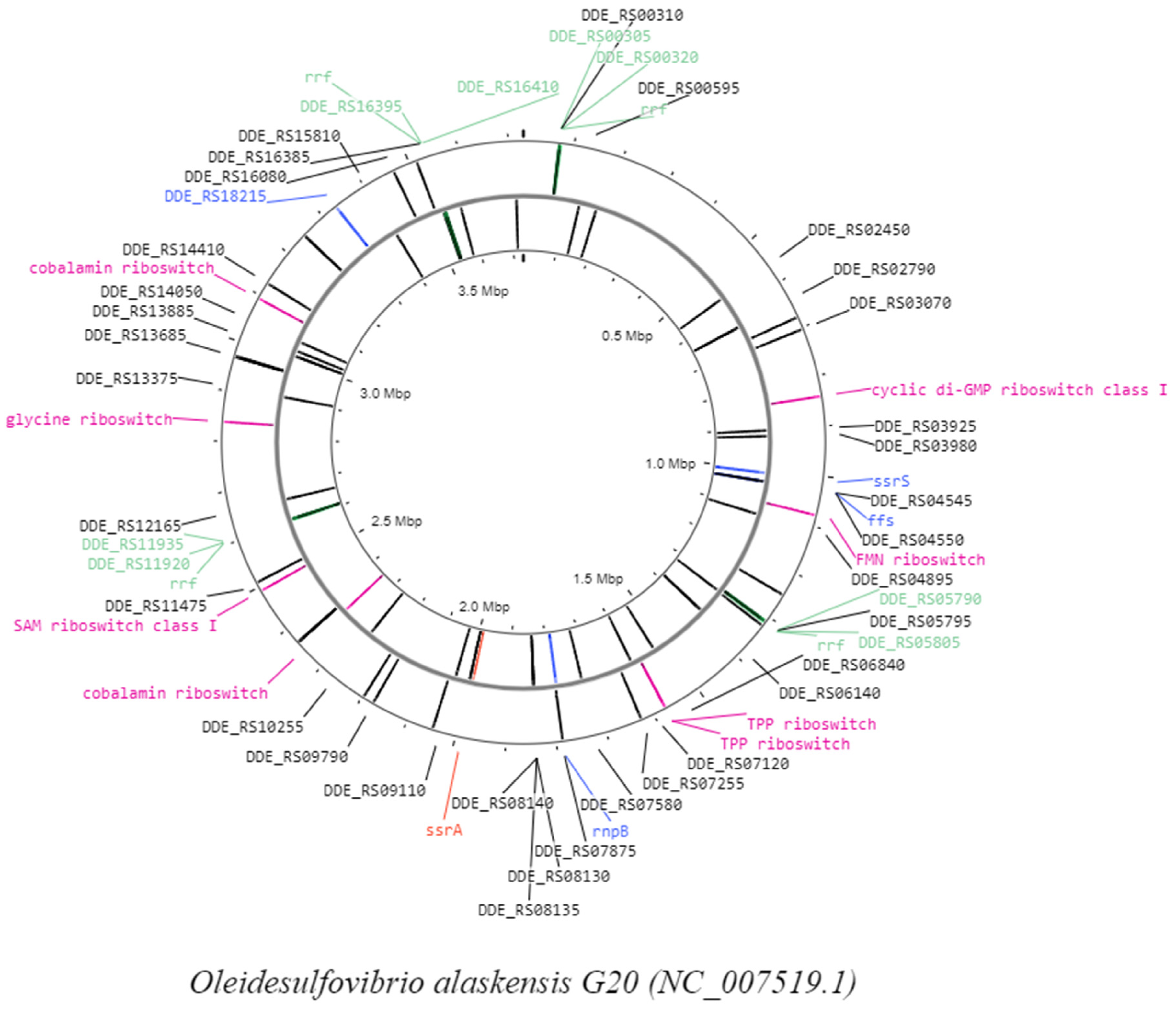
| Approaches | Genome Sequence | Genome Reso. Acc. | Software and Tools |
|---|---|---|---|
| Approach 1 | 3,730,232 bp | NC_007519.1 NCBI | Rfam, cmsscan, infernal, R2DT, RNAcentral |
| Approach 2 | 3,730,232 bp | CP000112.1 ENA/EMBL | Rfam, R2DT, RNAcentral |
| Approach 3 | 3,730,232 bp | NC_007519.1 NCBI | Proksee server, R2DT, RNAcentral |
| Gene Types | |
|---|---|
| Protein coding | 3257 |
| tRNAs | 66 |
| rRNAs | 12 |
| Pseudogenes | 37 |
| Miscellaneous RNAs | 5 |
| ncRNA Family | Rfam ID | Gene Type | Length | Genome Location | Strand | Description |
|---|---|---|---|---|---|---|
| 5S_rRNA | RF00001 | rRNA | 119 | 3,533,731:3,533,617 | − | 5S ribosomal RNA |
| Glycine_riboswitch | RF00504 | Cis-reg; riboswitch | 94 | 2,838,359:2,838,468 | + | Glycine riboswitch |
| TPP_riboswitch | RF00059 | Cis-reg; riboswitch | 105 | 1,573,919:1,574,022 | + | TPP riboswitch (THI element) |
| TPP_riboswitch | RF00059 | Cis-reg; riboswitch | 105 | 1,574,080:1,574,186 | + | TPP riboswitch (THI element) |
| Cobalamin_riboswitch | RF00174 | Cis-reg; riboswitch | 189 | 3,093,545:3,093,727 | + | Cobalamin riboswitch |
| SAM riboswitch | RF00162 | Cis-reg; riboswitch | 108 | 2,490,976:2,491,083 | + | SAM riboswitch (S box leader) |
| SSU_rRNA_bacteria | RF0177 | rRNA | 1533 | 2,609,189:2,607,648 | − | Bacterial small subunit ribosomal RNA |
| SSU_rRNA_archaea | RF01959 | rRNA | 1477 | 1,309,443:1,310,984 | + | Bacterial small subunit ribosomal RNA |
| SSU_rRNA_microsporidia | RF02542 | rRNA | 1311 | 3,538,761:3,537,220 | − | Bacterial small subunit ribosomal RNA |
| SSU_rRNA_eukarya | RF01960 | rRNA | 1831 | 69,852:71,393 | + | Bacterial small subunit ribosomal RNA |
| LSU_rRNA_bacteria | RF02541 | rRNA | 2925 | 2,607,209:2,604,279 | − | Bacterial large subunit ribosomal RNA |
| LSU_rRNA_archaea | RF02540 | rRNA | 2987 | 3,536,781:3,533,851 | − | Bacterial large subunit ribosomal RNA |
| LSU_rRNA_eukarya | RF02543 | rRNA | 3401 | 71,832:74,762 | + | Bacterial large subunit ribosomal RNA |
| tRNA_Sec | RF01852 | tRNA | 91 | 1,531,711:1,531,804 | − | tRNA-Sec |
| tRNA | RF00005 | tRNA | 73 | 2,047,102:2,047,175 | + | tRNA |
| ncRNA Family | Rfam IDs | Gene Type |
|---|---|---|
| Identified in approaches 2 and 3 | ||
| Glycine_riboswitch | RF00504 | cis-reg; riboswitch |
| TPP_riboswitch | RF00059 | cis-reg; riboswitch |
| TPP_riboswitch | RF00059 | cis-reg; riboswitch |
| FMN_riboswitch | RF00050 | cis-reg; riboswitch |
| c-di-GMP-I | RF01051 | cis-reg; riboswitch |
| Cobalamin_riboswitch | RF00174 | cis-reg; riboswitch |
| Cobalamin_riboswitch | RF00174 | cis-reg; riboswitch |
| SAM-Box | RF00162 | cis-reg; riboswitch |
| Bacterial small_SRP | RF00169 | gene |
| 6S | RF000013 | SsrS_RNA |
| 5S_rRNA | RF00001 | rRNA |
| 5S_rRNA | RF00001 | rRNA |
| 5S_rRNA | RF00001 | rRNA |
| 5S_rRNA | RF00001 | rRNA |
| SSU_rRNA_bacteria | RF0177 | rRNA |
| SSU_rRNA_bacteria | RF0177 | rRNA |
| SSU_rRNA_bacteria | RF0177 | rRNA |
| SSU_rRNA_bacteria | RF0177 | rRNA |
| LSU_rRNA_bacteria | RF02541 | rRNA |
| LSU_rRNA_bacteria | RF02541 | rRNA |
| LSU_rRNA_bacteria | RF02541 | rRNA |
| LSU_rRNA_bacteria | RF02541 | rRNA |
| tRNA_Sec | RF01852 | tRNA |
| Only (unique) ncRNAs identified from approach 2 | ||
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| sX4 | RF02223 | sRNA |
| P10 | RF01668 | sRNA |
| P10 | RF01668 | sRNA |
| STnc490 | RF01405 | sRNA |
| tRNA_Sec | RF01852 | tRNA |
| Only (unique) ncRNAs identified from approach 3 | ||
| tmRNA | RF00023 | SsrA |
| rnpB | RF00010 | rnpB |
| DDE_RS18215 | RF02276 | hammerhead |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, R.N.; Sani, R.K. Genome-Wide Computational Prediction and Analysis of Noncoding RNAs in Oleidesulfovibrio alaskensis G20. Microorganisms 2024, 12, 960. https://doi.org/10.3390/microorganisms12050960
Singh RN, Sani RK. Genome-Wide Computational Prediction and Analysis of Noncoding RNAs in Oleidesulfovibrio alaskensis G20. Microorganisms. 2024; 12(5):960. https://doi.org/10.3390/microorganisms12050960
Chicago/Turabian StyleSingh, Ram Nageena, and Rajesh K. Sani. 2024. "Genome-Wide Computational Prediction and Analysis of Noncoding RNAs in Oleidesulfovibrio alaskensis G20" Microorganisms 12, no. 5: 960. https://doi.org/10.3390/microorganisms12050960
APA StyleSingh, R. N., & Sani, R. K. (2024). Genome-Wide Computational Prediction and Analysis of Noncoding RNAs in Oleidesulfovibrio alaskensis G20. Microorganisms, 12(5), 960. https://doi.org/10.3390/microorganisms12050960







