Forensic Microbiology: When, Where and How
Abstract
1. Introduction
2. Patterns of Microbial Dynamics after Death
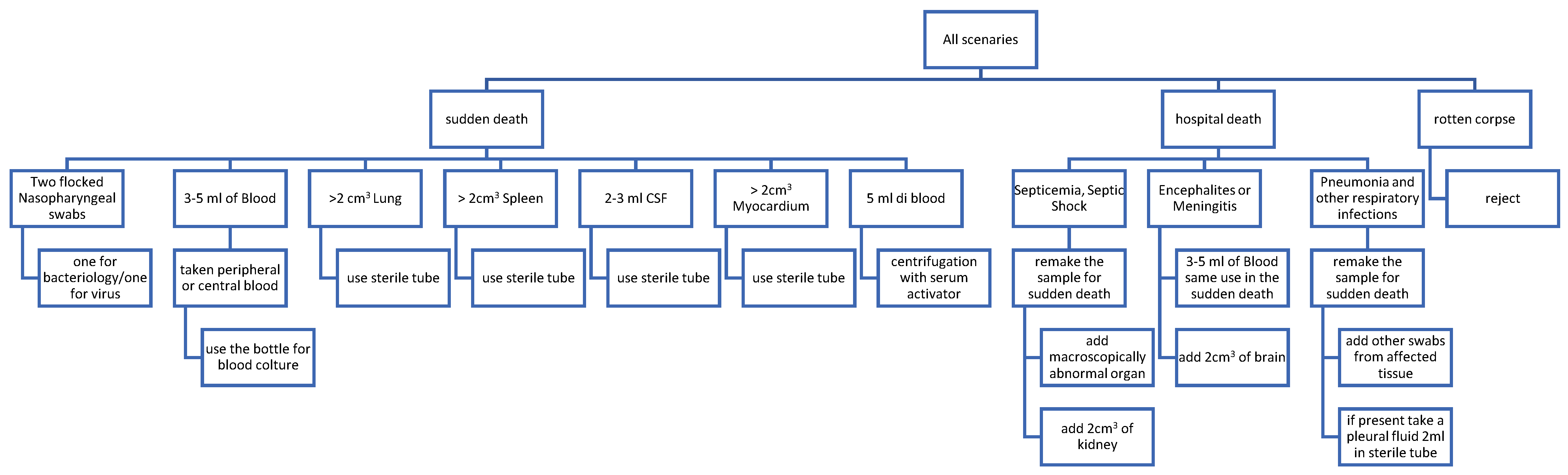
3. Microbiological Sampling after Death: When, Where and How; Pitfalls and Precautions
4. Next-Generation Sequencing (NGS) for Microbiological Post-Mortem Analysis
5. Main Applications
5.1. The Discriminatory Power of Forensic Microbiology
5.2. Sudden Death
5.3. Forensic Microbiology and the Deciphering of the Cause of Infectious Death in Outbreaks
5.4. Violence, Violent Death and Forensic Microbiology
6. Forensic Microbiology at the Border of Paleomicrobiology
7. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Budowle, B.; Schutzer, S.E.; Einseln, A.; Kelley, L.C.; Walsh, A.C.; Smith, J.A.; Marrone, B.L.; Robertson, J.; Campos, J. Building Microbial Forensics as a Response to Bioterrorism; American Association for the Advancement of Science: Washington, DC, USA, 2003; Volume 301, pp. 1852–1853. ISSN 0036-8075. [Google Scholar]
- Bazaj, A.; Turrina, S.; De Leo, D.; Cornaglia, G. Palaeomicrobiology Meets Forensic Medicine: Time as a Fourth-Dimension for the Crime Scene. New Microbes New Infect. 2015, 4, 5–6. [Google Scholar] [CrossRef]
- Spagnolo, E.V.; Mondello, C.; Stassi, C.; Baldino, G.; D’Aleo, F.; Conte, M.; Argo, A.; Zerbo, S. Forensic microbiology: A case series analysis. Euromediterranean Biomed. J. 2019, 14, 117–121. [Google Scholar]
- Fernández-Rodríguez, A.; Cohen, M.C.; Lucena, J.; Van de Voorde, W.; Angelini, A.; Ziyade, N.; Saegeman, V. How to Optimise the Yield of Forensic and Clinical Post-Mortem Microbiology with an Adequate Sampling: A Proposal for Standardisation. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1045–1057. [Google Scholar] [CrossRef]
- Burton, J.L.; Saegeman, V.; Arribi, A.; Rello, J.; Andreoletti, L.; Cohen, M.C.; Fernandez-Rodriguez, A.; ESGFOR Joint Working Group of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Study Group of Forensic and Postmortem Microbiology and the European Society of Pathology. Postmortem Microbiology Sampling Following Death in Hospital: An ESGFOR Task Force Consensus Statement. J. Clin. Pathol. 2019, 72, 329–336. [Google Scholar] [CrossRef]
- Schlipköter, U.; Flahault, A. Communicable Diseases: Achievements and Challenges for Public Health. Public Health Rev. 2010, 32, 90–119. [Google Scholar] [CrossRef]
- Schmedes, S.; Budowle, B. Microbial Forensics. Encycl. Microbiol. 2019, 134–145. [Google Scholar] [CrossRef]
- Castro, A.E.; De Ungria, M.C.A. Methods Used in Microbial Forensics and Epidemiological Investigations for Stronger Health Systems. Forensic Sci. Res. 2022, 7, 650–661. [Google Scholar] [CrossRef]
- Blondeau, L.D.; Rubin, J.E.; Deneer, H.; Kanthan, R.; Sanche, S.; Hamula, C.; Blondeau, J.M. Forensic, Investigative and Diagnostic Microbiology: Similar Technologies but Different Priorities. Future Microbiol. 2019, 14, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Piret, J.; Boivin, G. Pandemics throughout History. Front. Microbiol. 2021, 11, 631736. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, M. Was Famed Poet Pablo Neruda Poisoned? Scientists Warn Case Not Closed. Nature 2023, 615, 18–19. [Google Scholar] [CrossRef] [PubMed]
- Javan, G.T.; Finley, S.J.; Abidin, Z.; Mulle, J.G. The Thanatomicrobiome: A Missing Piece of the Microbial Puzzle of Death. Front. Microbiol. 2016, 7, 225. [Google Scholar] [CrossRef] [PubMed]
- Javan, G.T.; Finley, S.J.; Smith, T.; Miller, J.; Wilkinson, J.E. Cadaver Thanatomicrobiome Signatures: The Ubiquitous Nature of Clostridium Species in Human Decomposition. Front. Microbiol. 2017, 8, 2096. [Google Scholar] [CrossRef] [PubMed]
- Tambuzzi, S.; Maciocco, F.; Gentile, G.; Boracchi, M.; Faraone, C.; Andreola, S.; Zoja, R. Utility and Diagnostic Value of Postmortem Microbiology Associated with Histology for Forensic Purposes. Forensic Sci. Int. 2023, 342, 111534. [Google Scholar] [CrossRef] [PubMed]
- Cláudia-Ferreira, A.; Barbosa, D.J.; Saegeman, V.; Fernández-Rodríguez, A.; Dinis-Oliveira, R.J.; Freitas, A.R.; ESCMID Study Group of Forensic and Post-Mortem Microbiology (ESGFOR). The Future Is Now: Unraveling the Expanding Potential of Human (Necro) Microbiome in Forensic Investigations. Microorganisms 2023, 11, 2509. [Google Scholar] [CrossRef]
- Burcham, Z.M.; Belk, A.D.; McGivern, B.B.; Bouslimani, A.; Ghadermazi, P.; Martino, C.; Shenhav, L.; Zhang, A.R.; Shi, P.; Emmons, A. A Conserved Interdomain Microbial Network Underpins Cadaver Decomposition despite Environmental Variables. Nat. Microbiol. 2024, 9, 595–613. [Google Scholar] [CrossRef] [PubMed]
- FAQ: Human Microbiome. American Academy of Microbiology FAQ Reports; American Society for Microbiology: Washington, DC, USA, 2013.
- Martino, C.; Dilmore, A.H.; Burcham, Z.M.; Metcalf, J.L.; Jeste, D.; Knight, R. Microbiota Succession throughout Life from the Cradle to the Grave. Nat. Rev. Microbiol. 2022, 20, 707–720. [Google Scholar] [CrossRef] [PubMed]
- Melvin, J.J.; Cronholm, L.S.; Simson, L.J.; Isaacs, A.M. Bacterial Transmigration as an Indicator of Time of Death. J. Forensic Sci. 1984, 29, 412–417. [Google Scholar] [CrossRef]
- Metcalf, J.L. Estimating the Postmortem Interval Using Microbes: Knowledge Gaps and a Path to Technology Adoption. Forensic Sci. Int. Genet. 2019, 38, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Tomo, S.; Purohit, P.; Setia, P. Microbiome in Death and Beyond: Current Vistas and Future Trends. Front. Ecol. Evol. 2021, 9, 630397. [Google Scholar] [CrossRef]
- Dash, H.R.; Das, S. Thanatomicrobiome and Epinecrotic Community Signatures for Estimation of Post-Mortem Time Interval in Human Cadaver. Appl. Microbiol. Biotechnol. 2020, 104, 9497–9512. [Google Scholar] [CrossRef]
- Deel, H.; Bucheli, S.; Belk, A.; Ogden, S.; Lynne, A.; Carter, D.O.; Knight, R.; Metcalf, J.L. Chapter 12—Using Microbiome Tools for Estimating the Postmortem Interval. In Microbial Forensics, 3rd ed.; Budowle, B., Schutzer, S., Morse, S., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 171–191. ISBN 978-0-12-815379-6. [Google Scholar]
- Javan, G.T.; Finley, S.J.; Tuomisto, S.; Hall, A.; Benbow, M.E.; Mills, D. An Interdisciplinary Review of the Thanatomicrobiome in Human Decomposition. Forensic Sci. Med. Pathol. 2019, 15, 75–83. [Google Scholar] [CrossRef]
- Schotsmans, E.M.J.; Márquez-Grant, N.; Forbes, S.L. Taphonomy of Human Remains: Forensic Analysis of the Dead and the Depositional Environment; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 978-1-118-95332-7. [Google Scholar]
- Gunn, A.; Pitt, S. Microbes as Forensic Indicators. Trop. Biomed. 2012, 29, 311–330. [Google Scholar]
- Lutz, H.; Vangelatos, A.; Gottel, N.; Osculati, A.; Visona, S.; Finley, S.J.; Gilbert, J.A.; Javan, G.T. Effects of Extended Postmortem Interval on Microbial Communities in Organs of the Human Cadaver. Front. Microbiol. 2020, 11, 569630. [Google Scholar] [CrossRef]
- Vass, A.A.; Barshick, S.-A.; Sega, G.; Caton, J.; Skeen, J.T.; Love, J.C.; Synstelien, J.A. Decomposition Chemistry of Human Remains: A New Methodology for Determining the Postmortem Interval. J. Forensic Sci. 2002, 47, 542–553. [Google Scholar] [CrossRef] [PubMed]
- Lee Goff, M. Early Post-Mortem Changes and Stages of Decomposition in Exposed Cadavers. Exp. Appl. Acarol. 2009, 49, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Can, I.; Javan, G.T.; Pozhitkov, A.E.; Noble, P.A. Distinctive Thanatomicrobiome Signatures Found in the Blood and Internal Organs of Humans. J. Microbiol. Methods 2014, 106, 1–7. [Google Scholar] [CrossRef]
- Adserias-Garriga, J.; Quijada, N.M.; Hernandez, M.; Rodríguez Lázaro, D.; Steadman, D.; Garcia-Gil, L.J. Dynamics of the Oral Microbiota as a Tool to Estimate Time since Death. Mol. Oral Microbiol. 2017, 32, 511–516. [Google Scholar] [CrossRef] [PubMed]
- Damann, F.E.; Williams, D.E.; Layton, A.C. Potential Use of Bacterial Community Succession in Decaying Human Bone for Estimating Postmortem Interval. J. Forensic Sci. 2015, 60, 844–850. [Google Scholar] [CrossRef]
- Campobasso, C.P.; Mastroianni, G.; Feola, A.; Mascolo, P.; Carfora, A.; Liguori, B.; Zangani, P.; Dell’Annunziata, F.; Folliero, V.; Petrillo, A.; et al. MALDI-TOF Mass Spectrometry Analysis and Human Post-Mortem Microbial Community: A Pilot Study. Int. J. Environ. Res. Public Health 2022, 19, 4354. [Google Scholar] [CrossRef]
- Pechal, J.L.; Schmidt, C.J.; Jordan, H.R.; Benbow, M.E. A Large-Scale Survey of the Postmortem Human Microbiome, and Its Potential to Provide Insight into the Living Health Condition. Sci. Rep. 2018, 8, 5724. [Google Scholar] [CrossRef]
- Kodama, W.A.; Xu, Z.; Metcalf, J.L.; Song, S.J.; Harrison, N.; Knight, R.; Carter, D.O.; Happy, C.B. Trace Evidence Potential in Postmortem Skin Microbiomes: From Death Scene to Morgue. J. Forensic Sci. 2019, 64, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Xu, Z.Z.; Weiss, S.; Lax, S.; Van Treuren, W.; Hyde, E.R.; Song, S.J.; Amir, A.; Larsen, P.; Sangwan, N.; et al. Microbial Community Assembly and Metabolic Function during Mammalian Corpse Decomposition. Science 2016, 351, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Hyde, E.R.; Haarmann, D.P.; Petrosino, J.F.; Lynne, A.M.; Bucheli, S.R. Initial Insights into Bacterial Succession during Human Decomposition. Int. J. Leg. Med. 2015, 129, 661–671. [Google Scholar] [CrossRef] [PubMed]
- DeBruyn, J.M.; Hauther, K.A. Postmortem Succession of Gut Microbial Communities in Deceased Human Subjects. PeerJ 2017, 5, e3437. [Google Scholar] [CrossRef] [PubMed]
- Hauther, K.A.; Cobaugh, K.L.; Jantz, L.M.; Sparer, T.E.; DeBruyn, J.M. Estimating Time Since Death from Postmortem Human Gut Microbial Communities. J. Forensic Sci. 2015, 60, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- Tuomisto, S.; Karhunen, P.J.; Vuento, R.; Aittoniemi, J.; Pessi, T. Evaluation of Postmortem Bacterial Migration Using Culturing and Real-Time Quantitative PCR. J. Forensic Sci. 2013, 58, 910–916. [Google Scholar] [CrossRef] [PubMed]
- García, M.G.; Pérez-Cárceles, M.D.; Osuna, E.; Legaz, I. Impact of the Human Microbiome in Forensic Sciences: A Systematic Review. Appl. Environ. Microbiol. 2020, 86, e01451-20. [Google Scholar] [CrossRef] [PubMed]
- Javan, G.T.; Finley, S.J.; Can, I.; Wilkinson, J.E.; Hanson, J.D.; Tarone, A.M. Human Thanatomicrobiome Succession and Time Since Death. Sci. Rep. 2016, 6, 29598. [Google Scholar] [CrossRef]
- Bell, C.R.; Wilkinson, J.E.; Robertson, B.K.; Javan, G.T. Sex-related Differences in the Thanatomicrobiome in Postmortem Heart Samples Using Bacterial Gene Regions V1-2 and V4. Lett. Appl. Microbiol. 2018, 67, 144–153. [Google Scholar] [CrossRef]
- Tarone, A.M.; Mann, A.E.; Zhang, Y.; Zascavage, R.R.; Mitchell, E.A.; Morales, E.; Rusch, T.W.; Allen, M.S. The Devil Is in the Details: Variable Impacts of Season, BMI, Sampling Site Temperature, and Presence of Insects on the Post-Mortem Microbiome. Front. Microbiol. 2022, 13, 1064904. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, W.; Simayijiang, H.; Hu, P.; Yan, J. Application of Microbiome in Forensics. Genom. Proteom. Bioinform. 2022, 21, 97–107. [Google Scholar] [CrossRef]
- Johnson, H.R.; Trinidad, D.D.; Guzman, S.; Khan, Z.; Parziale, J.V.; DeBruyn, J.M.; Lents, N.H. A Machine Learning Approach for Using the Postmortem Skin Microbiome to Estimate the Postmortem Interval. PLoS ONE 2016, 11, e0167370. [Google Scholar] [CrossRef]
- Tsokos, M.; Püschel, K. Postmortem Bacteriology in Forensic Pathology: Diagnostic Value and Interpretation. Leg. Med. 2001, 3, 15–22. [Google Scholar] [CrossRef]
- Dobay, A.; Haas, C.; Fucile, G.; Downey, N.; Morrison, H.G.; Kratzer, A.; Arora, N. Microbiome-Based Body Fluid Identification of Samples Exposed to Indoor Conditions. Forensic Sci. Int. Genet. 2019, 40, 105–113. [Google Scholar] [CrossRef]
- Fleming, R.I.; Harbison, S. The Use of Bacteria for the Identification of Vaginal Secretions. Forensic Sci. Int. Genet. 2010, 4, 311–315. [Google Scholar] [CrossRef]
- Nields, H.; Kessler, S.C.; Boisot, S.; Evans, R. Streptococcal Toxic Shock Syndrome Presenting as Suspected Child Abuse. Am. J. Forensic Med. Pathol. 1998, 19, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Byard, R.W. “Shaken Baby Syndrome” and Forensic Pathology: An Uneasy Interface. Forensic Sci. Med. Pathol. 2014, 10, 239–241. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Principe, L.; Comandatore, F.; Perini, M.; Meroni, E.; Mattioni Marchetti, V.; Migliavacca, R.; Luzzaro, F. Whole-Genome Sequencing Investigation of a Large Nosocomial Outbreak Caused by ST131 H30Rx KPC-Producing Escherichia Coli. in Italy. Antibiotics 2021, 10, 718. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Amorim, A. Microbial Forensics: New Breakthroughs and Future Prospects. Appl. Microbiol. Biotechnol. 2018, 102, 10377–10391. [Google Scholar] [CrossRef]
- Burcham, Z.M.; Jordan, H.R. History, Current, and Future Use of Microorganisms as Physical Evidence. Forensic Microbiol. 2017, 25–55. [Google Scholar] [CrossRef]
- Cho, H.-W.; Eom, Y.-B. Forensic Analysis of Human Microbiome in Skin and Body Fluids Based on Geographic Location. Front. Cell. Infect. Microbiol. 2021, 11, 695191. [Google Scholar] [CrossRef] [PubMed]
- Habtom, H.; Demanèche, S.; Dawson, L.; Azulay, C.; Matan, O.; Robe, P.; Gafny, R.; Simonet, P.; Jurkevitch, E.; Pasternak, Z. Soil Characterisation by Bacterial Community Analysis for Forensic Applications: A Quantitative Comparison of Environmental Technologies. Forensic Sci. Int. Genet. 2017, 26, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lauber, C.L.; Zhou, N.; McDonald, D.; Costello, E.K.; Knight, R. Forensic Identification Using Skin Bacterial Communities. Proc. Natl. Acad. Sci. USA 2010, 107, 6477–6481. [Google Scholar] [CrossRef] [PubMed]
- Woerner, A.E.; Novroski, N.M.; Wendt, F.R.; Ambers, A.; Wiley, R.; Schmedes, S.E.; Budowle, B. Forensic Human Identification with Targeted Microbiome Markers Using Nearest Neighbor Classification. Forensic Sci. Int. Genet. 2019, 38, 130–139. [Google Scholar] [CrossRef] [PubMed]
- Phan, K.; Barash, M.; Spindler, X.; Gunn, P.; Roux, C. Retrieving Forensic Information about the Donor through Bacterial Profiling. Int. J. Leg. Med. 2020, 134, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Black, C.M.; Driebe, E.M.; Howard, L.A.; Fajman, N.N.; Sawyer, M.K.; Girardet, R.G.; Sautter, R.L.; Greenwald, E.; Beck-Sague, C.M.; Unger, E.R.; et al. Multicenter Study of Nucleic Acid Amplification Tests for Detection of Chlamydia Trachomatis and Neisseria Gonorrhoeae in Children Being Evaluated for Sexual Abuse. Pediatr. Infect. Dis. J. 2009, 28, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Lowe, S.M.; Rahman, N.; Forster, G. Chain of Evidence in Sexual Assault Cases. Int. J. STD AIDS 2009, 20, 799–800. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller, B.M.; Schuman, M.; Wennig, R. Was a Child Poisoned by Ethanol? Discrimination between Ante-Mortem Consumption and Post-Mortem Formation. Int. J. Leg. Med. 2008, 122, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.; Lowe, P.; Symonds, A. The Possible Influence of Micro-Organisms and Putrefaction in the Production of GHB in Post-Mortem Biological Fluid. Forensic Sci. Int. 2004, 139, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Clarke, T.H.; Gomez, A.; Singh, H.; Nelson, K.E.; Brinkac, L.M. Integrating the Microbiome as a Resource in the Forensics Toolkit. Forensic Sci. Int. Genet. 2017, 30, 141–147. [Google Scholar] [CrossRef]
- Gouello, A.; Dunyach-Remy, C.; Siatka, C.; Lavigne, J.-P. Analysis of Microbial Communities: An Emerging Tool in Forensic Sciences. Diagnostics 2021, 12, 1. [Google Scholar] [CrossRef]
- Riedel, S. The Value of Postmortem Microbiology Cultures. J. Clin. Microbiol. 2014, 52, 1028–1033. [Google Scholar] [CrossRef] [PubMed]
- Neagu, O.; Fernandez-Rodríguez, A.; Callon, D.; Andréoletti, L.; Cohen, M.C. Myocarditis Presenting as Sudden Death in Infants and Children: A Single Centre Analysis by ESGFOR Study Group. Pediatr. Dev. Pathol. 2021, 24, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Saegeman, V.; Cohen, M.C.; Burton, J.L.; Martinez, M.J.; Rakislova, N.; Offiah, A.C.; Fernandez-Rodriguez, A. Microbiology in Minimally Invasive Autopsy: Best Techniques to Detect Infection. ESGFOR (ESCMID Study Group of Forensic and Post-Mortem Microbiology) Guidelines. Forensic Sci. Med. Pathol. 2021, 17, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Christoffersen, S. The Importance of Microbiological Testing for Establishing Cause of Death in 42 Forensic Autopsies. Forensic Sci. Int. 2015, 250, 27–32. [Google Scholar] [CrossRef]
- Kruger, M.M.; Martin, L.J.; Maistry, S.; Heathfield, L.J. A Systematic Review Exploring the Relationship between Infection and Sudden Unexpected Death between 2000 and 2016: A Forensic Perspective. Forensic Sci. Int. 2018, 289, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Maujean, G.; Malicier, D.; Fanton, L. Air, Water, and Surface Bacterial Contamination in a University-Hospital Autopsy Room*. J. Forensic Sci. 2012, 57, 381–385. [Google Scholar] [CrossRef]
- d’Aleo, F.; Arghittu, M.; Bandi, C.; Conte, M.; Creti, R.; Farina, C. Guidance to Post-Mortem Collection and Storage of Biological Specimens for the Diagnosis of COVID-19 Infection. Microbiol. Medica 2020, 244–256. [Google Scholar] [CrossRef]
- Dell’Aquila, M.; Cattani, P.; Fantoni, M.; Marchetti, S.; Aquila, I.; Stigliano, E.; Carbone, A.; Oliva, A.; Arena, V. Postmortem Swabs in the Severe Acute Respiratory Syndrome Coronavirus 2 Pandemic: Report on 12 Complete Clinical Autopsy Cases. Arch. Pathol. Lab. Med. 2020, 144, 1298–1302. [Google Scholar] [CrossRef]
- Kuiper, I. Microbial Forensics: Next-Generation Sequencing as Catalyst: The Use of New Sequencing Technologies to Analyze Whole Microbial Communities Could Become a Powerful Tool for Forensic and Criminal Investigations. EMBO Rep. 2016, 17, 1085–1087. [Google Scholar] [CrossRef]
- Robinson, J.M.; Pasternak, Z.; Mason, C.E.; Elhaik, E. Forensic Applications of Microbiomics: A Review. Front. Microbiol. 2020, 11, 608101. [Google Scholar] [CrossRef]
- Yang, Y.; Xie, B.; Yan, J. Application of Next-Generation Sequencing Technology in Forensic Science. Genom. Proteom. Bioinform. 2014, 12, 190–197. [Google Scholar] [CrossRef]
- Shendure, J.; Ji, H. Next-Generation DNA Sequencing. Nat. Biotechnol. 2008, 26, 1135–1145. [Google Scholar] [CrossRef]
- Kanzi, A.M.; San, J.E.; Chimukangara, B.; Wilkinson, E.; Fish, M.; Ramsuran, V.; de Oliveira, T. Next Generation Sequencing and Bioinformatics Analysis of Family Genetic Inheritance. Front. Genet. 2020, 11, 544162. [Google Scholar] [CrossRef]
- Gunasekera, S.; Abraham, S.; Stegger, M.; Pang, S.; Wang, P.; Sahibzada, S.; O’Dea, M. Evaluating Coverage Bias in Next-Generation Sequencing of Escherichia coli. PLoS ONE 2021, 16, e0253440. [Google Scholar] [CrossRef]
- De Coster, W.; Van Broeckhoven, C. Newest Methods for Detecting Structural Variations. Trends Biotechnol. 2019, 37, 973–982. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun Metagenomics, from Sampling to Analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Zubakov, D.; Kokmeijer, I.; Ralf, A.; Rajagopalan, N.; Calandro, L.; Wootton, S.; Langit, R.; Chang, C.; Lagace, R.; Kayser, M. Towards Simultaneous Individual and Tissue Identification: A Proof-of-Principle Study on Parallel Sequencing of STRs, Amelogenin, and mRNAs with the Ion Torrent PGM. Forensic Sci. Int. Genet. 2015, 17, 122–128. [Google Scholar] [CrossRef]
- Galindo-González, L.; Pinzón-Latorre, D.; Bergen, E.A.; Jensen, D.C.; Deyholos, M.K. Ion Torrent Sequencing as a Tool for Mutation Discovery in the Flax (Linum usitatissimum L.) Genome. Plant Methods 2015, 11, 19. [Google Scholar] [CrossRef]
- Salipante, S.J.; Kawashima, T.; Rosenthal, C.; Hoogestraat, D.R.; Cummings, L.A.; Sengupta, D.J.; Harkins, T.T.; Cookson, B.T.; Hoffman, N.G. Performance Comparison of Illumina and Ion Torrent Next-Generation Sequencing Platforms for 16S rRNA-Based Bacterial Community Profiling. Appl. Environ. Microbiol. 2014, 80, 7583–7591. [Google Scholar] [CrossRef]
- Buermans, H.P.J.; den Dunnen, J.T. Next Generation Sequencing Technology: Advances and Applications. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the Effects of PCR Amplification and Sequencing Artifacts on 16S rRNA-Based Studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef] [PubMed]
- Aird, D.; Ross, M.G.; Chen, W.-S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and Minimizing PCR Amplification Bias in Illumina Sequencing Libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef] [PubMed]
- Quail, M.A.; Otto, T.D.; Gu, Y.; Harris, S.R.; Skelly, T.F.; McQuillan, J.A.; Swerdlow, H.P.; Oyola, S.O. Optimal Enzymes for Amplifying Sequencing Libraries. Nat. Methods 2012, 9, 10–11. [Google Scholar] [CrossRef]
- Quail, M.A.; Smith, M.; Coupland, P.; Otto, T.D.; Harris, S.R.; Connor, T.R.; Bertoni, A.; Swerdlow, H.P.; Gu, Y. A Tale of Three next Generation Sequencing Platforms: Comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq Sequencers. BMC Genom. 2012, 13, 341. [Google Scholar] [CrossRef]
- Cornelis, S.; Gansemans, Y.; Deleye, L.; Deforce, D.; Van Nieuwerburgh, F. Forensic SNP Genotyping Using Nanopore MinION Sequencing. Sci. Rep. 2017, 7, 41759. [Google Scholar] [CrossRef]
- Soneson, C.; Yao, Y.; Bratus-Neuenschwander, A.; Patrignani, A.; Robinson, M.D.; Hussain, S. A Comprehensive Examination of Nanopore Native RNA Sequencing for Characterization of Complex Transcriptomes. Nat. Commun. 2019, 10, 3359. [Google Scholar] [CrossRef]
- Rang, F.J.; Kloosterman, W.P.; de Ridder, J. From Squiggle to Basepair: Computational Approaches for Improving Nanopore Sequencing Read Accuracy. Genome Biol. 2018, 19, 90. [Google Scholar] [CrossRef]
- Delahaye, C.; Nicolas, J. Sequencing DNA with Nanopores: Troubles and Biases. PLoS ONE 2021, 16, e0257521. [Google Scholar] [CrossRef]
- Ogden, R.; Vasiljevic, N.; Prost, S. Nanopore Sequencing in Non-Human Forensic Genetics. Emerg. Top. Life Sci. 2021, 5, 465–473. [Google Scholar] [CrossRef]
- Zhou, W.; Bian, Y. Thanatomicrobiome Composition Profiling as a Tool for Forensic Investigation. Forensic Sci. Res. 2018, 3, 105–110. [Google Scholar] [CrossRef]
- Speruda, M.; Piecuch, A.; Borzęcka, J.; Kadej, M.; Ogórek, R. Microbial Traces and Their Role in Forensic Science. J. Appl. Microbiol. 2022, 132, 2547–2557. [Google Scholar] [CrossRef] [PubMed]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and Overcoming the Pitfalls and Biases of Next-Generation Sequencing (NGS) Methods for Use in the Routine Clinical Microbiological Diagnostic Laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef]
- Malla, M.A.; Dubey, A.; Kumar, A.; Yadav, S.; Hashem, A.; Abd_Allah, E.F. Exploring the Human Microbiome: The Potential Future Role of Next-Generation Sequencing in Disease Diagnosis and Treatment. Front. Immunol. 2019, 9, 2868. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Xu, Z.Z.; Bouslimani, A.; Dorrestein, P.; Carter, D.O.; Knight, R. Microbiome Tools for Forensic Science. Trends Biotechnol. 2017, 35, 814–823. [Google Scholar] [CrossRef]
- Ashe, E.C.; Comeau, A.M.; Zejdlik, K.; O’Connell, S.P. Characterization of Bacterial Community Dynamics of the Human Mouth Throughout Decomposition via Metagenomic, Metatranscriptomic, and Culturing Techniques. Front. Microbiol. 2021, 12, 689493. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Zhang, K.; Li, H.; Sun, Q.; Wei, X.; Li, H.; Zhang, S.; Fan, S.; Wang, Z. Dissecting the Microbial Community Structure of Internal Organs during the Early Postmortem Period in a Murine Corpse Model. BMC Microbiol. 2023, 23, 38. [Google Scholar] [CrossRef]
- Wang, L.-L.; Zhang, F.-Y.; Dong, W.-W.; Wang, C.-L.; Liang, X.-Y.; Suo, L.-L.; Cheng, J.; Zhang, M.; Guo, X.-S.; Jiang, P.-H.; et al. A Novel Approach for the Forensic Diagnosis of Drowning by Microbiological Analysis with Next-Generation Sequencing and Unweighted UniFrac-Based PCoA. Int. J. Leg. Med. 2020, 134, 2149–2159. [Google Scholar] [CrossRef]
- Richardson, M.; Gottel, N.; Gilbert, J.A.; Lax, S. Microbial Similarity between Students in a Common Dormitory Environment Reveals the Forensic Potential of Individual Microbial Signatures. mBio 2019, 10, e01054-19. [Google Scholar] [CrossRef] [PubMed]
- Khodakova, A.S.; Smith, R.J.; Burgoyne, L.; Abarno, D.; Linacre, A. Random Whole Metagenomic Sequencing for Forensic Discrimination of Soils. PLoS ONE 2014, 9, e104996. [Google Scholar] [CrossRef]
- Ben Zakour, N.L.; Venturini, C.; Beatson, S.A.; Walker, M.J. Analysis of a Streptococcus Pyogenes Puerperal Sepsis Cluster by Use of Whole-Genome Sequencing. J. Clin. Microbiol. 2012, 50, 2224–2228. [Google Scholar] [CrossRef]
- Mellmann, A.; Bletz, S.; Böking, T.; Kipp, F.; Becker, K.; Schultes, A.; Prior, K.; Harmsen, D. Real-Time Genome Sequencing of Resistant Bacteria Provides Precision Infection Control in an Institutional Setting. J. Clin. Microbiol. 2016, 54, 2874–2881. [Google Scholar] [CrossRef]
- Quainoo, S.; Coolen, J.P.M.; van Hijum, S.A.F.T.; Huynen, M.A.; Melchers, W.J.G.; van Schaik, W.; Wertheim, H.F.L. Whole-Genome Sequencing of Bacterial Pathogens: The Future of Nosocomial Outbreak Analysis. Clin. Microbiol. Rev. 2017, 30, 1015–1063. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; van der Plaat, D.A.; Liu, Y.; Wurmbach, E. Analyzing Degraded DNA and Challenging Samples Using the ForenSeqTM DNA Signature Prep Kit. Sci. Justice 2020, 60, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M. Recent Advances in Forensic Biology and Forensic DNA Typing: INTERPOL Review 2019–2022. Forensic Sci. Int. Synerg. 2023, 6, 100311. [Google Scholar] [CrossRef] [PubMed]
- Shadoff, R.; Panoyan, M.A.; Novroski, N. Microbial Forensics: A Present to Future Perspective on Genomic Targets, Bioinformatic Challenges, and Applications. Forensic Genom. 2022, 2, 42–64. [Google Scholar] [CrossRef]
- McCord, B.; Opel, K.; Funes, M.; Zoppis, S.; Meadows Jantz, L. An Investigation of the Effect of DNA Degradation and Inhibition on PCR Amplification of Single Source and Mixed Forensic Samples. In National Archive of Criminal Justice Data; US Department of Justice: Washington, DC, USA, 2011; pp. 1–65. Available online: http://www.icpsr.umich.edu/NACJD (accessed on 10 May 2024).
- Danko, D.; Bezdan, D.; Afshin, E.E.; Ahsanuddin, S.; Bhattacharya, C.; Butler, D.J.; Chng, K.R.; Donnellan, D.; Hecht, J.; Jackson, K.; et al. A Global Metagenomic Map of Urban Microbiomes and Antimicrobial Resistance. Cell 2021, 184, 3376–3393.e17. [Google Scholar] [CrossRef]
- Poinar, H.N. The Top 10 List: Criteria of Authenticity for DNA from Ancient and Forensic Samples. Int. Congr. Ser. 2003, 1239, 575–579. [Google Scholar] [CrossRef]
- Rivera-Perez, J.I.; Santiago-Rodriguez, T.M.; Toranzos, G.A. Paleomicrobiology: A Snapshot of Ancient Microbes and Approaches to Forensic Microbiology. Microbiol. Spectr. 2016, 4, 63–90. [Google Scholar] [CrossRef]
- Haas, C.; Neubauer, J.; Salzmann, A.P.; Hanson, E.; Ballantyne, J. Forensic Transcriptome Analysis Using Massively Parallel Sequencing. Forensic Sci. Int. Genet. 2021, 52, 102486. [Google Scholar] [CrossRef]
- Salzmann, A.P.; Russo, G.; Aluri, S.; Haas, C. Transcription and Microbial Profiling of Body Fluids Using a Massively Parallel Sequencing Approach. Forensic Sci. Int. Genet. 2019, 43, 102149. [Google Scholar] [CrossRef]
- Sijen, T. Molecular Approaches for Forensic Cell Type Identification: On mRNA, miRNA, DNA Methylation and Microbial Markers. Forensic Sci. Int. Genet. 2015, 18, 21–32. [Google Scholar] [CrossRef]
- Merkley, E.D. Proteomics for Microbial Forensics. In Applications in Forensic Proteomics: Protein Identification and Profiling; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2019; Volume 1339, pp. 143–160. ISBN 978-0-8412-3649-3. [Google Scholar]
- Lasch, P.; Schneider, A.; Blumenscheit, C.; Doellinger, J. Identification of Microorganisms by Liquid Chromatography-Mass Spectrometry (LC-MS1) and in Silico Peptide Mass Libraries. Mol. Cell Proteom. 2020, 19, 2125–2138. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, E.M.; Levin, E.; Braakman, R.; Prodan, A.; van Leeuwen, H.C.; Paauw, A. Untargeted Accurate Identification of Highly Pathogenic Bacteria Directly from Blood Culture Flasks. Int. J. Med. Microbiol. 2020, 310, 151376. [Google Scholar] [CrossRef]
- Murr, A.; Pink, C.; Hammer, E.; Michalik, S.; Dhople, V.M.; Holtfreter, B.; Völker, U.; Kocher, T.; Gesell Salazar, M. Cross-Sectional Association of Salivary Proteins with Age, Sex, Body Mass Index, Smoking, and Education. J. Proteome Res. 2017, 16, 2273–2281. [Google Scholar] [CrossRef] [PubMed]
- Clowers, B.H.; Wunschel, D.S.; Kreuzer, H.W.; Engelmann, H.E.; Valentine, N.; Wahl, K.L. Characterization of Residual Medium Peptides from Yersinia Pestis Cultures. Anal. Chem. 2013, 85, 3933–3939. [Google Scholar] [CrossRef]
- Wunschel, D.; Tulman, E.; Engelmann, H.; Clowers, B.H.; Geary, S.; Robinson, A.; Liao, X. Forensic Proteomics of Poxvirus Production. Analyst 2013, 138, 6385–6397. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D. Population-Level Analysis of Gut Microbiome Variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Flandroy, L.; Poutahidis, T.; Berg, G.; Clarke, G.; Dao, M.-C.; Decaestecker, E.; Furman, E.; Haahtela, T.; Massart, S.; Plovier, H.; et al. The Impact of Human Activities and Lifestyles on the Interlinked Microbiota and Health of Humans and of Ecosystems. Sci. Total Environ. 2018, 627, 1018–1038. [Google Scholar] [CrossRef]
- Zheng, Y.; Shi, J.; Chen, Q.; Deng, C.; Yang, F.; Wang, Y. Identifying Individual-Specific Microbial DNA Fingerprints from Skin Microbiomes. Front. Microbiol. 2022, 13, 960043. [Google Scholar] [CrossRef]
- Franceschetti, L.; Lodetti, G.; Blandino, A.; Amadasi, A.; Bugelli, V. Exploring the Role of the Human Microbiome in Forensic Identification: Opportunities and Challenges. Int. J. Leg. Med. 2024; epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Parfrey, L.W.; Zaneveld, J.; Lozupone, C.; Knight, R. Human-Associated Microbial Signatures: Examining Their Predictive Value. Cell Host Microbe 2011, 10, 292–296. [Google Scholar] [CrossRef] [PubMed]
- Moitas, B.; Caldas, I.M.; Sampaio-Maia, B. Forensic Microbiology and Geographical Location: A Systematic Review. Aust. J. Forensic Sci. 2023, 1–16. [Google Scholar] [CrossRef]
- Flores, G.E.; Bates, S.T.; Knights, D.; Lauber, C.L.; Stombaugh, J.; Knight, R.; Fierer, N. Microbial Biogeography of Public Restroom Surfaces. PLoS ONE 2011, 6, e28132. [Google Scholar] [CrossRef]
- Flores, G.E.; Bates, S.T.; Caporaso, J.G.; Lauber, C.L.; Leff, J.W.; Knight, R.; Fierer, N. Diversity, Distribution and Sources of Bacteria in Residential Kitchens. Environ. Microbiol. 2013, 15, 588–596. [Google Scholar] [CrossRef]
- Meadow, J.F.; Altrichter, A.E.; Green, J.L. Mobile Phones Carry the Personal Microbiome of Their Owners. PeerJ 2014, 2, e447. [Google Scholar] [CrossRef] [PubMed]
- Lax, S.; Hampton-Marcell, J.T.; Gibbons, S.M.; Colares, G.B.; Smith, D.; Eisen, J.A.; Gilbert, J.A. Forensic Analysis of the Microbiome of Phones and Shoes. Microbiome 2015, 3, 21. [Google Scholar] [CrossRef]
- Simon, L.M.; Flocco, C.; Burkart, F.; Methner, A.; Henke, D.; Rauer, L.; Müller, C.L.; Vogel, J.; Quaisser, C.; Overmann, J.; et al. Microbial Fingerprints Reveal Interaction between Museum Objects, Curators, and Visitors. iScience 2023, 26, 107578. [Google Scholar] [CrossRef]
- Jesmok, E.M.; Hopkins, J.M.; Foran, D.R. Next-Generation Sequencing of the Bacterial 16S rRNA Gene for Forensic Soil Comparison: A Feasibility Study. J. Forensic Sci. 2016, 61, 607–617. [Google Scholar] [CrossRef]
- Su, Q.; Yang, C.; Chen, L.; She, Y.; Xu, Q.; Zhao, J.; Liu, C.; Sun, H. Inference of Drowning Sites Using Bacterial Composition and Random Forest Algorithm. Front. Microbiol. 2023, 14, 1213271. [Google Scholar] [CrossRef] [PubMed]
- Gerber, G.K. The Dynamic Microbiome. FEBS Lett. 2014, 588, 4131–4139. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting Family Members Share Microbiota with One Another and with Their Dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Dill-McFarland, K.A.; Tang, Z.-Z.; Kemis, J.H.; Kerby, R.L.; Chen, G.; Palloni, A.; Sorenson, T.; Rey, F.E.; Herd, P. Close Social Relationships Correlate with Human Gut Microbiota Composition. Sci. Rep. 2019, 9, 703. [Google Scholar] [CrossRef] [PubMed]
- Thiene, G. Autopsy and Sudden Death. Eur. Heart J. Suppl. 2023, 25, C118–C129. [Google Scholar] [CrossRef]
- Dada, M.A.; Lazarus, N.G. SUDDEN NATURAL DEATH|Infectious Diseases. Encycl. Forensic Leg. Med. 2005, 229–236. [Google Scholar] [CrossRef]
- Krous, H.F.; Beckwith, J.B.; Byard, R.W.; Rognum, T.O.; Bajanowski, T.; Corey, T.; Cutz, E.; Hanzlick, R.; Keens, T.G.; Mitchell, E.A. Sudden Infant Death Syndrome and Unclassified Sudden Infant Deaths: A Definitional and Diagnostic Approach. Pediatrics 2004, 114, 234–238. [Google Scholar] [CrossRef]
- Prtak, L.; Al-Adnani, M.; Fenton, P.; Kudesia, G.; Cohen, M.C. Contribution of Bacteriology and Virology in Sudden Unexpected Death in Infancy. Arch. Dis. Child. 2010, 95, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Lafuente, R.; Aguilera, B.; Suárez-Mier, M.A.P.; Morentin, B.; Vallejo, G.; Gómez, J.; Fernández-Rodríguez, A. Detection of Human Herpesvirus-6, Epstein-Barr Virus and Cytomegalovirus in Formalin-Fixed Tissues from Sudden Infant Death: A Study with Quantitative Real-Time PCR. Forensic Sci. Int. 2008, 178, 106–111. [Google Scholar] [CrossRef]
- Leong, L.E.X.; Taylor, S.L.; Shivasami, A.; Goldwater, P.N.; Rogers, G.B. Intestinal Microbiota Composition in Sudden Infant Death Syndrome and Age-Matched Controls. J. Pediatr. 2017, 191, 63–68.e1. [Google Scholar] [CrossRef]
- Beebeejaun, K.; Parikh, S.R.; Campbell, H.; Gray, S.; Borrow, R.; Ramsay, M.E.; Ladhani, S.N. Invasive Meningococcal Disease: Timing and Cause of Death in England, 2008–2015. J. Infect. 2020, 80, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Ambrosio, L.; Neri, A.; Fazio, C.; Rossolini, G.M.; Vacca, P.; Riccobono, E.; Voller, F.; Miglietta, A.; Stefanelli, P. Genomic Analysis of Neisseria Meningitidis Carriage Isolates during an Outbreak of Serogroup C Clonal Complex 11, Tuscany, Italy. PLoS ONE 2019, 14, e0217500. [Google Scholar] [CrossRef]
- Fazio, C.; Daprai, L.; Neri, A.; Tirani, M.; Vacca, P.; Arghittu, M.; Ambrosio, L.; Cereda, D.; Gramegna, M.; Palmieri, A.; et al. Reactive Vaccination as Control Strategy for an Outbreak of Invasive Meningococcal Disease Caused by Neisseria Meningitidis C:P1.5-1,10-8:F3-6:ST-11(Cc11), Bergamo Province, Italy, December 2019 to January 2020. Euro Surveill 2022, 27, 2100919. [Google Scholar] [CrossRef]
- Schürch, A.C.; Siezen, R.J. Genomic Tracing of Epidemics and Disease Outbreaks. Microb. Biotechnol. 2010, 3, 628–633. [Google Scholar] [CrossRef]
- Onori, R.; Gaiarsa, S.; Comandatore, F.; Pongolini, S.; Brisse, S.; Colombo, A.; Cassani, G.; Marone, P.; Grossi, P.; Minoja, G. Tracking Nosocomial Klebsiella Pneumoniae Infections and Outbreaks by Whole-Genome Analysis: Small-Scale Italian Scenario within a Single Hospital. J. Clin. Microbiol. 2015, 53, 2861–2868. [Google Scholar] [CrossRef] [PubMed]
- Szarvas, J.; Ahrenfeldt, J.; Cisneros, J.L.B.; Thomsen, M.C.F.; Aarestrup, F.M.; Lund, O. Large Scale Automated Phylogenomic Analysis of Bacterial Isolates and the Evergreen Online Platform. Commun. Biol. 2020, 3, 137. [Google Scholar] [CrossRef]
- Berbel Caban, A.; Pak, T.R.; Obla, A.; Dupper, A.C.; Chacko, K.I.; Fox, L.; Mills, A.; Ciferri, B.; Oussenko, I.; Beckford, C. PathoSPOT Genomic Epidemiology Reveals Under-the-Radar Nosocomial Outbreaks. Genome Med. 2020, 12, 96. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, C.; Corbella, M.; Gaiarsa, S.; Comandatore, F.; Scaltriti, E.; Bandi, C.; Cambieri, P.; Marone, P.; Sassera, D. Multiple Klebsiella Pneumoniae KPC Clones Contribute to an Extended Hospital Outbreak. Front. Microbiol. 2019, 10, 2767. [Google Scholar] [CrossRef]
- Collin, S.M.; Lamb, P.; Jauneikaite, E.; Le Doare, K.; Creti, R.; Berardi, A.; Heath, P.T.; Sriskandan, S.; Lamagni, T. Hospital Clusters of Invasive Group B Streptococcal Disease: A Systematic Review. J. Infect. 2019, 79, 521–527. [Google Scholar] [CrossRef]
- Ward, D.V.; Hoss, A.G.; Kolde, R.; van Aggelen, H.C.; Loving, J.; Smith, S.A.; Mack, D.A.; Kathirvel, R.; Halperin, J.A.; Buell, D.J. Integration of Genomic and Clinical Data Augments Surveillance of Healthcare-Acquired Infections. Infect. Control Hosp. Epidemiol. 2019, 40, 649–655. [Google Scholar] [CrossRef]
- Gilchrist, C.A.; Turner, S.D.; Riley, M.F.; Petri, W.A., Jr.; Hewlett, E.L. Whole-Genome Sequencing in Outbreak Analysis. Clin. Microbiol. Rev. 2015, 28, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Syrowatka, A.; Kuznetsova, M.; Alsubai, A.; Beckman, A.L.; Bain, P.A.; Craig, K.J.T.; Hu, J.; Jackson, G.P.; Rhee, K.; Bates, D.W. Leveraging Artificial Intelligence for Pandemic Preparedness and Response: A Scoping Review to Identify Key Use Cases. Npj Digit. Med. 2021, 4, 96. [Google Scholar] [CrossRef] [PubMed]
- Ventura Spagnolo, E.; Stassi, C.; Mondello, C.; Zerbo, S.; Milone, L.; Argo, A. Forensic Microbiology Applications: A Systematic Review. Leg. Med. 2019, 36, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Schmedes, S.E.; Woerner, A.E.; Novroski, N.M.M.; Wendt, F.R.; King, J.L.; Stephens, K.M.; Budowle, B. Targeted Sequencing of Clade-Specific Markers from Skin Microbiomes for Forensic Human Identification. Forensic Sci. Int. Genet. 2018, 32, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, D.; Leung, M.H.Y.; Lee, P.K.H. Microbiota Fingerprints Lose Individually Identifying Features over Time. Microbiome 2017, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Edmonds-Wilson, S.L.; Nurinova, N.I.; Zapka, C.A.; Fierer, N.; Wilson, M. Review of Human Hand Microbiome Research. J. Dermatol. Sci. 2015, 80, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Giampaoli, S.; Berti, A.; Valeriani, F.; Gianfranceschi, G.; Piccolella, A.; Buggiotti, L.; Rapone, C.; Valentini, A.; Ripani, L.; Romano Spica, V. Molecular Identification of Vaginal Fluid by Microbial Signature. Forensic Sci. Int. Genet. 2012, 6, 559–564. [Google Scholar] [CrossRef] [PubMed]
- Flowers, L.; Grice, E.A. The Skin Microbiota: Balancing Risk and Reward. Cell Host Microbe 2020, 28, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Nakamura, I.; Mizutani, S.; Kurokawa, Y.; Mori, H.; Kurokawa, K.; Yamada, T. Minor Taxa in Human Skin Microbiome Contribute to the Personal Identification. PLoS ONE 2018, 13, e0199947. [Google Scholar] [CrossRef] [PubMed]
- Neckovic, A.; van Oorschot, R.A.H.; Szkuta, B.; Durdle, A. Investigation of Direct and Indirect Transfer of Microbiomes between Individuals. Forensic Sci. Int. Genet. 2020, 45, 102212. [Google Scholar] [CrossRef]
- Leake, S.L.; Pagni, M.; Falquet, L.; Taroni, F.; Greub, G. The Salivary Microbiome for Differentiating Individuals: Proof of Principle. Microbes Infect. 2016, 18, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Doi, M.; Gamo, S.; Okiura, T.; Nishimukai, H.; Asano, M. A Simple Identification Method for Vaginal Secretions Using Relative Quantification of Lactobacillus DNA. Forensic Sci. Int. Genet. 2014, 12, 93–99. [Google Scholar] [CrossRef]
- Choi, A.; Shin, K.-J.; Yang, W.I.; Lee, H.Y. Body Fluid Identification by Integrated Analysis of DNA Methylation and Body Fluid-Specific Microbial DNA. Int. J. Leg. Med. 2014, 128, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Díez López, C.; Montiel González, D.; Haas, C.; Vidaki, A.; Kayser, M. Microbiome-Based Body Site of Origin Classification of Forensically Relevant Blood Traces. Forensic Sci. Int. Genet. 2020, 47, 102280. [Google Scholar] [CrossRef] [PubMed]
- Baud, D.; Pattaroni, C.; Vulliemoz, N.; Castella, V.; Marsland, B.J.; Stojanov, M. Sperm Microbiota and Its Impact on Semen Parameters. Front. Microbiol. 2019, 10, 234. [Google Scholar] [CrossRef]
- Ghemrawi, M.; Torres, A.R.; Duncan, G.; Colwell, R.; Dadlani, M.; McCord, B. The Genital Microbiome and Its Potential for Detecting Sexual Assault. Forensic Sci. Int. Genet. 2021, 51, 102432. [Google Scholar] [CrossRef]
- Tackmann, J.; Arora, N.; Schmidt, T.S.B.; Rodrigues, J.F.M.; von Mering, C. Ecologically Informed Microbial Biomarkers and Accurate Classification of Mixed and Unmixed Samples in an Extensive Cross-Study of Human Body Sites. Microbiome 2018, 6, 192. [Google Scholar] [CrossRef]
- Williams, D.W.; Gibson, G. Classification of Individuals and the Potential to Detect Sexual Contact Using the Microbiome of the Pubic Region. Forensic Sci. Int. Genet. 2019, 41, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Tridico, S.R.; Murray, D.C.; Bunce, M.; Kirkbride, K.P. DNA Profiling of Bacteria from Human Hair. In Forensic Microbiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2017; pp. 358–375. ISBN 978-1-119-06258-5. [Google Scholar]
- Haarkötter, C.; Saiz, M.; Gálvez, X.; Medina-Lozano, M.I.; Álvarez, J.C.; Lorente, J.A. Usefulness of Microbiome for Forensic Geolocation: A Review. Life 2021, 11, 1322. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Finley, S.J.; Pechal, J.L.; Benbow, M.E.; Robertson, B.K.; Javan, G.T. Microbial Signatures of Cadaver Gravesoil during Decomposition. Microb. Ecol. 2016, 71, 524–529. [Google Scholar] [CrossRef] [PubMed]
- Lucci, A.; Campobasso, C.P.; Cirnelli, A.; Lorenzini, G. A Promising Microbiological Test for the Diagnosis of Drowning. Forensic Sci. Int. 2008, 182, 20–26. [Google Scholar] [CrossRef]
- Kakizaki, E.; Kozawa, S.; Imamura, N.; Uchiyama, T.; Nishida, S.; Sakai, M.; Yukawa, N. Detection of Marine and Freshwater Bacterioplankton in Immersed Victims: Post-Mortem Bacterial Invasion Does Not Readily Occur. Forensic Sci. Int. 2011, 211, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xu, Q.; Xiao, C.; Li, H.; Wu, W.; Du, W.; Zhao, J.; Liu, H.; Wang, H.; Liu, C. SYBR Green Real-Time qPCR Method: Diagnose Drowning More Rapidly and Accurately. Forensic Sci. Int. 2021, 321, 110720. [Google Scholar] [CrossRef] [PubMed]
- Tsuneya, S.; Yoshida, M.; Hoshioka, Y.; Chiba, F.; Inokuchi, G.; Torimitsu, S.; Iwase, H. Relevance of Diatom Testing on Closed Organs of a Drowned Cadaver Who Died after Receiving Treatment for 10 Days: A Case Report. Leg. Med. 2023, 60, 102168. [Google Scholar] [CrossRef]
- Uchiyama, T.; Kakizaki, E.; Kozawa, S.; Nishida, S.; Imamura, N.; Yukawa, N. A New Molecular Approach to Help Conclude Drowning as a Cause of Death: Simultaneous Detection of Eight Bacterioplankton Species Using Real-Time PCR Assays with TaqMan Probes. Forensic Sci. Int. 2012, 222, 11–26. [Google Scholar] [CrossRef]
- Swayambhu, M.; Kümmerli, R.; Arora, N. Microbiome-Based Stain Analyses in Crime Scenes. Appl. Environ. Microbiol. 2023, 89, e01325-22. [Google Scholar] [CrossRef]
- Drancourt, M.; Raoult, D. Palaeomicrobiology: Current Issues and Perspectives. Nat. Rev. Microbiol. 2005, 3, 23–35. [Google Scholar] [CrossRef]
- Raoult, D.; Drancourt, M. Molecular Detection of Past Pathogens. In Paleomicrobiology: Past Human Infections; Raoult, D., Drancourt, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2008; pp. 55–68. ISBN 978-3-540-75855-6. [Google Scholar]
- Bos, K.I.; Kühnert, D.; Herbig, A.; Esquivel-Gomez, L.R.; Andrades Valtueña, A.; Barquera, R.; Giffin, K.; Kumar Lankapalli, A.; Nelson, E.A.; Sabin, S.; et al. Paleomicrobiology: Diagnosis and Evolution of Ancient Pathogens. Annu. Rev. Microbiol. 2019, 73, 639–666. [Google Scholar] [CrossRef]
- Gorgé, O.; Bennett, E.A.; Massilani, D.; Daligault, J.; Pruvost, M.; Geigl, E.-M.; Grange, T. Analysis of Ancient DNA in Microbial Ecology. Methods Mol. Biol. 2016, 1399, 289–315. [Google Scholar] [CrossRef]
- Weyrich, L.S.; Duchene, S.; Soubrier, J.; Arriola, L.; Llamas, B.; Breen, J.; Morris, A.G.; Alt, K.W.; Caramelli, D.; Dresely, V. Neanderthal Behaviour, Diet, and Disease Inferred from Ancient DNA in Dental Calculus. Nature 2017, 544, 357–361. [Google Scholar] [CrossRef] [PubMed]
- Nodari, R.; Drancourt, M.; Barbieri, R. Paleomicrobiology of the Human Digestive Tract: A Review. Microb. Pathog. 2021, 157, 104972. [Google Scholar] [CrossRef] [PubMed]
- Dabney, J.; Knapp, M.; Glocke, I.; Gansauge, M.-T.; Weihmann, A.; Nickel, B.; Valdiosera, C.; García, N.; Pääbo, S.; Arsuaga, J.-L. Complete Mitochondrial Genome Sequence of a Middle Pleistocene Cave Bear Reconstructed from Ultrashort DNA Fragments. Proc. Natl. Acad. Sci. USA 2013, 110, 15758–15763. [Google Scholar] [CrossRef] [PubMed]
- Hendy, J. Ancient Protein Analysis in Archaeology. Sci. Adv. 2021, 7, eabb9314. [Google Scholar] [CrossRef]
- Adler, C.J.; Haak, W.; Donlon, D.; Cooper, A. Survival and Recovery of DNA from Ancient Teeth and Bones. J. Archaeol. Sci. 2011, 38, 956–964. [Google Scholar] [CrossRef]
- Handt, O.; Krings, M.; Ward, R.H.; Pääbo, S. The Retrieval of Ancient Human DNA Sequences. Am. J. Hum. Genet. 1996, 59, 368–376. [Google Scholar] [PubMed]
- Krings, M.; Stone, A.; Schmitz, R.W.; Krainitzki, H.; Stoneking, M.; Pääbo, S. Neandertal DNA Sequences and the Origin of Modern Humans. Cell 1997, 90, 19–30. [Google Scholar] [CrossRef]
- Briggs, A.W.; Stenzel, U.; Meyer, M.; Krause, J.; Kircher, M.; Pääbo, S. Removal of Deaminated Cytosines and Detection of in Vivo Methylation in Ancient DNA. Nucleic Acids Res. 2010, 38, e87. [Google Scholar] [CrossRef]
- Rohland, N.; Harney, E.; Mallick, S.; Nordenfelt, S.; Reich, D. Partial Uracil-DNA-Glycosylase Treatment for Screening of Ancient DNA. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20130624. [Google Scholar] [CrossRef]
- Spyrou, M.A.; Bos, K.I.; Herbig, A.; Krause, J. Ancient Pathogen Genomics as an Emerging Tool for Infectious Disease Research. Nat. Rev. Genet. 2019, 20, 323–340. [Google Scholar] [CrossRef]
- Barbieri, R.; Mekni, R.; Levasseur, A.; Chabrière, E.; Signoli, M.; Tzortzis, S.; Aboudharam, G.; Drancourt, M. Paleoproteomics of the Dental Pulp: The Plague Paradigm. PLoS ONE 2017, 12, e0180552. [Google Scholar] [CrossRef] [PubMed]
- Demarchi, B.; Hall, S.; Roncal-Herrero, T.; Freeman, C.L.; Woolley, J.; Crisp, M.K.; Wilson, J.; Fotakis, A.; Fischer, R.; Kessler, B.M.; et al. Protein Sequences Bound to Mineral Surfaces Persist into Deep Time. eLife 2016, 5, e17092. [Google Scholar] [CrossRef] [PubMed]
- Shved, N.; Haas, C.; Papageorgopoulou, C.; Akguel, G.; Paulsen, K.; Bouwman, A.; Warinner, C.; Rühli, F. Post Mortem DNA Degradation of Human Tissue Experimentally Mummified in Salt. PLoS ONE 2014, 9, e110753. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Martin, P.; Albarrán, C.; Garcia, P.; Fernandez de Simon, L.; Jesús Iturralde, M.; Fernández-Rodriguez, A.; Atienza, I.; Capilla, J.; García-Hirschfeld, J.; et al. Challenges of DNA Profiling in Mass Disaster Investigations. Croat. Med. J. 2005, 46, 540–548. [Google Scholar]
- Lozano-Peral, D.; Rubio, L.; Santos, I.; Gaitán, M.J.; Viguera, E.; Martín-de-las-Heras, S. DNA Degradation in Human Teeth Exposed to Thermal Stress. Sci. Rep. 2021, 11, 12118. [Google Scholar] [CrossRef]
- Yang, D.Y.; Watt, K. Contamination Controls When Preparing Archaeological Remains for Ancient DNA Analysis. J. Archaeol. Sci. 2005, 32, 331–336. [Google Scholar] [CrossRef]
- Poinar, H.N.; Cooper, A. Ancient DNA: Do It Right or Not at All. Science 2000, 5482, 416. [Google Scholar]
- Jónsson, H.; Ginolhac, A.; Schubert, M.; Johnson, P.L.F.; Orlando, L. mapDamage2.0: Fast Approximate Bayesian Estimates of Ancient DNA Damage Parameters. Bioinformatics 2013, 29, 1682–1684. [Google Scholar] [CrossRef]
- Peyrégne, S.; Peter, B.M. AuthentiCT: A Model of Ancient DNA Damage to Estimate the Proportion of Present-Day DNA Contamination. Genome Biol. 2020, 21, 246. [Google Scholar] [CrossRef]
- Fernández-Rodríguez, A.; Burton, J.L.; Andreoletti, L.; Alberola, J.; Fornes, P.; Merino, I.; Martínez, M.J.; Castillo, P.; Sampaio-Maia, B.; Caldas, I.M. Post-Mortem Microbiology in Sudden Death: Sampling Protocols Proposed in Different Clinical Settings. Clin. Microbiol. Infect. 2019, 25, 570–579. [Google Scholar] [CrossRef]
- International Organization for Standardization. ISO/IEC 17025:2017. General Requirements for the Competence of Testing and Calibration Laboratories. Available online: https://www.iso.org/standard/66912.html (accessed on 10 May 2024).
- International Organization for Standardization. ISO 21043-2:2018. Forensic Sciences. Part 2: Recognition, Recording, Collecting, Transport and Storage of Items. Available online: https://www.iso.org/standard/72041.html (accessed on 10 May 2024).
| Main Application | Type of Samples | Method | Selected Refs |
|---|---|---|---|
| Determination of post-mortem interval (PMI) | Autopsy tissues | Molecular analyses | [37,46] |
| Determining the infectious causes of death | Autopsy tissues/Body fluids | Bacteriological, histological and molecular analyses | [14,47] |
| Determining the type of biological fluid | Swabs or samples of the biological trace | Molecular analyses | [48,49] |
| Differential diagnosis in shaken baby and child injuries | Autopsy tissues | Bacteriological analyses with histological examination and molecular analyses | [50,51] |
| Identifying deaths attributable to infectious diseases during outbreaks | Environmental/autopsy tissue/body fluids | Bacteriological and molecular analyses with bioinformatics | [52,53] |
| Identifying instances of medical malpractice and nosocomial infections | Swabs of specific body regions/body fluid/objects | Bacteriological and molecular analyses | [53,54] |
| Identifying soil types and specific locations | Soil/objects containing soil | Molecular analyses and comparing the results with the forensic microbiome database (FMD) | [55,56] |
| Identifying the touch residue left on objects by skin | Swabs on objects | Molecular analyses | [35,57] |
| Personal identification | Swabs of specific body regions | Molecular analyses | [58,59] |
| Sexual violence investigations | Swabs of specific body regions/objects | Bacteriological and molecular analyses with comparisons between samples | [60,61] |
| Post-mortem toxicological analyses | Autopsy tissues | Bacteriological, chromatographic and molecular analyses | [62,63] |
| Violent death investigations | Swabs of specific body regions/objects | Bacteriological and molecular analyses with comparisons between samples | [64,65] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nodari, R.; Arghittu, M.; Bailo, P.; Cattaneo, C.; Creti, R.; D’Aleo, F.; Saegeman, V.; Franceschetti, L.; Novati, S.; Fernández-Rodríguez, A.; et al. Forensic Microbiology: When, Where and How. Microorganisms 2024, 12, 988. https://doi.org/10.3390/microorganisms12050988
Nodari R, Arghittu M, Bailo P, Cattaneo C, Creti R, D’Aleo F, Saegeman V, Franceschetti L, Novati S, Fernández-Rodríguez A, et al. Forensic Microbiology: When, Where and How. Microorganisms. 2024; 12(5):988. https://doi.org/10.3390/microorganisms12050988
Chicago/Turabian StyleNodari, Riccardo, Milena Arghittu, Paolo Bailo, Cristina Cattaneo, Roberta Creti, Francesco D’Aleo, Veroniek Saegeman, Lorenzo Franceschetti, Stefano Novati, Amparo Fernández-Rodríguez, and et al. 2024. "Forensic Microbiology: When, Where and How" Microorganisms 12, no. 5: 988. https://doi.org/10.3390/microorganisms12050988
APA StyleNodari, R., Arghittu, M., Bailo, P., Cattaneo, C., Creti, R., D’Aleo, F., Saegeman, V., Franceschetti, L., Novati, S., Fernández-Rodríguez, A., Verzeletti, A., Farina, C., & Bandi, C., on behalf of the ESCMID Study Group of Forensic and Post-Mortem Microbiology (ESGFOR) and the AMCLI Forensic Microbiology Study Group (GLAMIFO) . (2024). Forensic Microbiology: When, Where and How. Microorganisms, 12(5), 988. https://doi.org/10.3390/microorganisms12050988








