Insights into Effects of Combined Capric and Lauric Acid on Rumen Bacterial Composition
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Compliance
2.2. Animals, Diets, and Experimental Design
2.3. Rumen Fluid Collection
2.4. DNA Extraction, Polymerase Chain Reaction (PCR), and Amplicon Sequence Variant (ASV) Analysis
2.5. Visualization, Assessment, and Statistical Analysis of the Bacterial Community
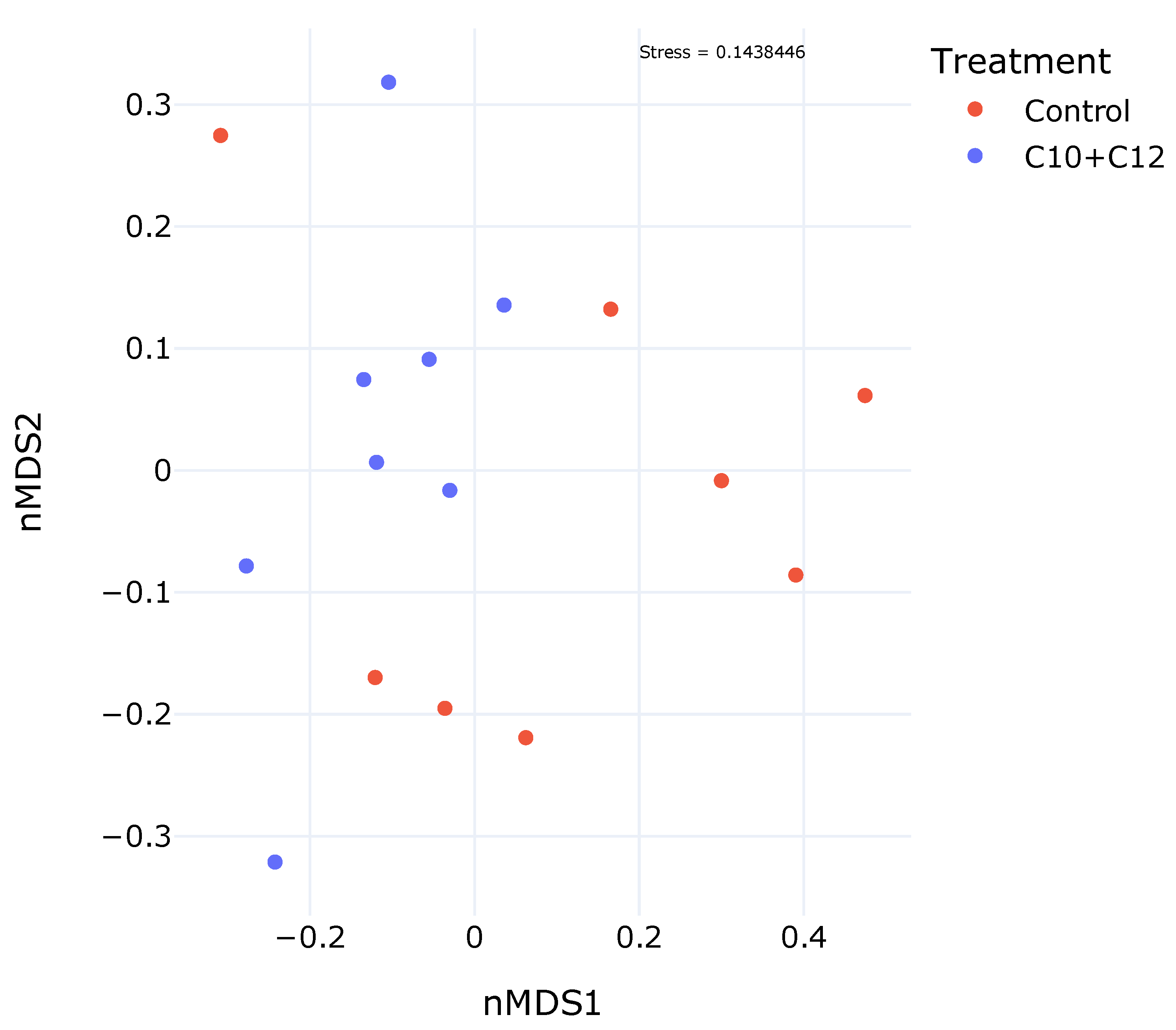
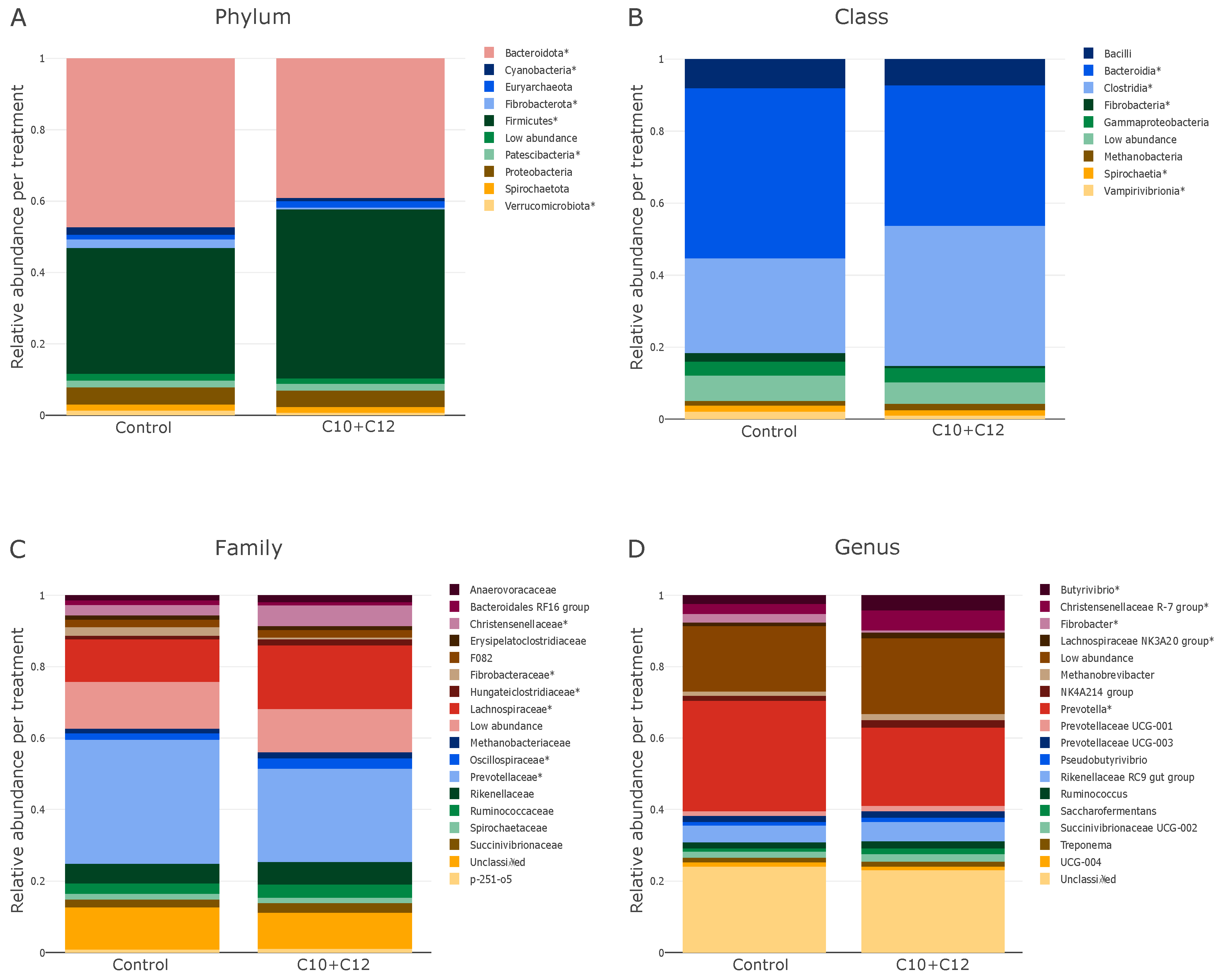
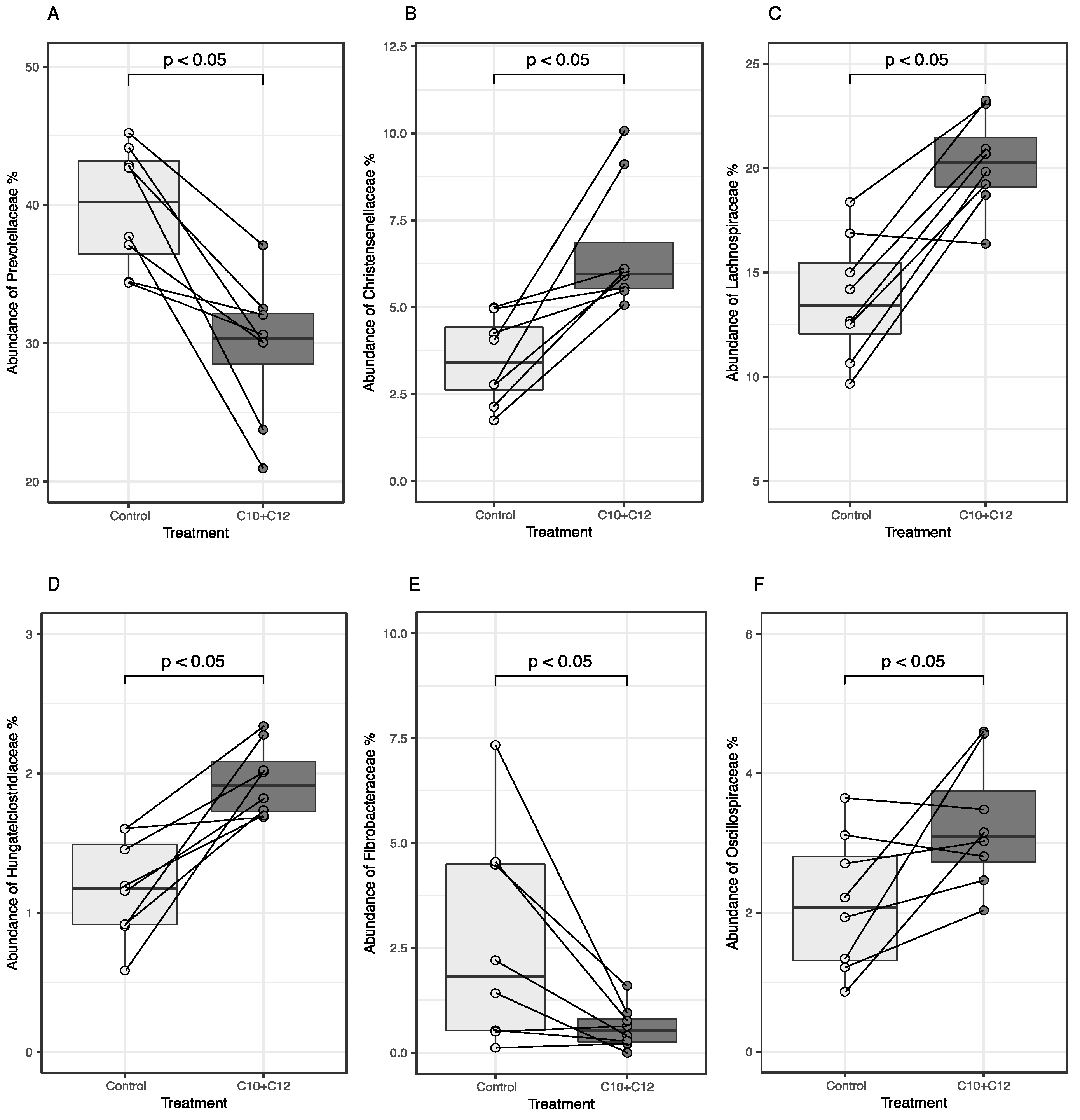
3. Results
4. Discussion
4.1. Bacterial Diversity, ASVs, and Indices
4.2. Microbial Relative Abundances
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamada, K.; Iwamae, K.; Suzuki, Y.; Koike, S.; Kobayashi, Y. Batch Culture Analysis to Identify Potent Organic Acids for Suppressing Ruminal Methane Production. Anim. Sci. J. 2023, 94, e13873. [Google Scholar] [CrossRef] [PubMed]
- Gruninger, R.J.; Zhang, X.M.; Smith, M.L.; Kung, L.; Vyas, D.; McGinn, S.M.; Kindermann, M.; Wang, M.; Tan, Z.L.; Beauchemin, K.A. Application of 3-Nitrooxypropanol and Canola Oil to Mitigate Enteric Methane Emissions of Beef Cattle Results in Distinctly Different Effects on the Rumen Microbial Community. Anim. Microbiome 2022, 4, 35. [Google Scholar] [CrossRef]
- Choi, Y.; Lee, S.J.; Kim, H.S.; Eom, J.S.; Jo, S.U.; Guan, L.L.; Seo, J.; Kim, H.; Lee, S.S.; Lee, S.S. Effects of Seaweed Extracts on in Vitro Rumen Fermentation Characteristics, Methane Production, and Microbial Abundance. Sci. Rep. 2021, 11, 24092. [Google Scholar] [CrossRef] [PubMed]
- Sikiru, A.; Michael, A.O.; John, M.O.; Egena, S.S.A.; Oleforuh-Okoleh, V.U.; Ambali, M.I.; Muhammad, I.R. Methane Emissions in Cattle Production: Biology, Measurement and Mitigation Strategies in Smallholder Farmer Systems. Environ. Dev. Sustain. 2024. [Google Scholar] [CrossRef]
- Tapio, I.; Snelling, T.J.; Strozzi, F.; Wallace, R.J. The Ruminal Microbiome Associated with Methane Emissions from Ruminant Livestock. J. Anim. Sci. Biotechnol. 2017, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- De Assis Lage, C.F.; Räisänen, S.E.; Melgar, A.; Nedelkov, K.; Chen, X.; Oh, J.; Fetter, M.E.; Indugu, N.; Bender, J.S.; Vecchiarelli, B.; et al. Comparison of Two Sampling Techniques for Evaluating Ruminal Fermentation and Microbiota in the Planktonic Phase of Rumen Digesta in Dairy Cows. Front. Microbiol. 2020, 11, 618032. [Google Scholar] [CrossRef] [PubMed]
- Honan, M.; Feng, X.; Tricarico, J.; Kebreab, E. Feed Additives as a Strategic Approach to Reduce Enteric Methane Production in Cattle: Modes of Action, Effectiveness and Safety. Anim. Prod. Sci. 2021, 62, 1303–1317. [Google Scholar] [CrossRef]
- Burdick, M.; Zhou, M.; Guan, L.; Oba, M. Effects of Medium-Chain Fatty Acid Supplementation on Performance and Rumen Fermentation of Lactating Holstein Dairy Cows. Animal 2022, 16, 100491. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, M.H.; Weisbjerg, M.R.; Larsen, M.; Højberg, O.; Ohlsson, C.; Walker, N.; Hellwing, A.L.F.; Lund, P. Gas Exchange, Rumen Hydrogen Sinks, and Nutrient Digestibility and Metabolism in Lactating Dairy Cows Fed 3-NOP and Cracked Rapeseed. J. Dairy Sci. 2023, 107, 2047–2065. [Google Scholar] [CrossRef] [PubMed]
- Dohme, F.; Machmüller, A.; Wasserfallen, A.; Kreuzer, M. Ruminal Methanogenesis as Influenced by Individual Fatty Acids Supplemented to Complete Ruminant Diets. Lett. Appl. Microbiol. 2001, 32, 47–51. [Google Scholar] [CrossRef]
- Machmüller, A. Medium-Chain Fatty Acids and Their Potential to Reduce Methanogenesis in Domestic Ruminants. Agric. Ecosyst. Environ. 2006, 112, 107–114. [Google Scholar] [CrossRef]
- Machmüller, A.; Kreuzer, M. Methane Suppression by Coconut Oil and Associated Effects on Nutrient and Energy Balance in Sheep. Can. J. Anim. Sci. 1999, 79, 65–72. [Google Scholar] [CrossRef]
- Machmüller, A.; Soliva, C.R.; Kreuzer, M. Methane-Suppressing Effect of Myristic Acid in Sheep as Affected by Dietary Calcium and Forage Proportion. Br. J. Nutr. 2003, 90, 529–540. [Google Scholar] [CrossRef] [PubMed]
- Joch, M.; Vadroňová, M.; Češpiva, M.; Zabloudilová, P.; Výborná, A.; Tyrolová, Y.; Kudrna, V.; Tichá, D.; Plachý, V.; Hroncová, Z. Capric and Lauric Acid Mixture Decreased Rumen Methane Production, While Combination with Nitrate Had No Further Benefit in Methane Reduction. Ann. Anim. Sci. 2023, 23, 799–808. [Google Scholar] [CrossRef]
- Muizelaar, W.; Bani, P.; Kuhla, B.; Tapio, I.; Yáñez-Ruiz, D.; van Gastelen, S. Rumen Fluid Sampling via Oral Stomach Tubing Method. In Methods in Cattle Physiology and Behaviour Research; Publisso: Cologne, Germany, 2020. [Google Scholar]
- Vadroňová, M.; Šťovíček, A.; Jochová, K.; Výborná, A.; Tyrolová, Y.; Tichá, D.; Homolka, P.; Joch, M. Combined Effects of Nitrate and Medium-Chain Fatty Acids on Methane Production, Rumen Fermentation, and Rumen Bacterial Populations In Vitro. Sci. Rep. 2023, 13, 21961. [Google Scholar] [CrossRef]
- Jiang, X.; Xu, H.J.; Cui, Z.Q.; Zhang, Y.G. Effects of Supplementation with Lactobacillus plantarum 299v on the Performance, Blood Metabolites, Rumen Fermentation and Bacterial Communities of Preweaning Calves. Livest. Sci. 2020, 239, 104120. [Google Scholar] [CrossRef]
- Cancino-Padilla, N.; Gajardo, F.; Neves, A.L.A.; Kholif, A.E.; Mele, M.; Huws, S.A.; Loor, J.J.; Romero, J.; Vargas-Bello-Pérez, E. Influence of Dietary Oils Rich in Omega-6 or Omega-3 Fatty Acids on Rumen Microbiome of Dairy Cows. Transl. Anim. Sci. 2023, 7, txad074. [Google Scholar] [CrossRef]
- Shabat, S.K.B.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific Microbiome-Dependent Mechanisms Underlie the Energy Harvest Efficiency of Ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The Rumen Microbiome: Balancing Food Security and Environmental Impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Díaz Carrasco, J.M.; Cabral, C.; Redondo, L.M.; Pin Viso, N.D.; Colombatto, D.; Farber, M.D.; Fernández Miyakawa, M.E. Impact of Chestnut and Quebracho Tannins on Rumen Microbiota of Bovines. BioMed Res. Int. 2017, 2017, 9610810. [Google Scholar] [CrossRef]
- Zhou, X.; Meile, L.; Kreuzer, M.; Zeitz, J.O. The Effect of Saturated Fatty Acids on Methanogenesis and Cell Viability of Methanobrevibacter ruminantium. Archaea 2013, 2013, e106916. [Google Scholar] [CrossRef] [PubMed]
- Yanza, Y.R.; Szumacher-Strabel, M.; Jayanegara, A.; Kasenta, A.M.; Gao, M.; Huang, H.; Patra, A.K.; Warzych, E.; Cieślak, A. The Effects of Dietary Medium-chain Fatty Acids on Ruminal Methanogenesis and Fermentation In Vitro and In Vivo: A Meta-analysis. J. Anim. Physiol. Anim. Nutr. 2020, 105, 874–889. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Q.; Liu, J.-X.; Lu, Y.; Zhu, W.Y.; Denman, S.E.; McSweeney, C.S. Effect of Tea Saponin on Methanogenesis, Microbial Community Structure and Expression of mcrA Gene, in Cultures of Rumen Micro-Organisms. Lett. Appl. Microbiol. 2008, 47, 421–426. [Google Scholar] [CrossRef]
- Danielsson, R.; Ramin, M.; Bertilsson, J.; Lund, P.; Huhtanen, P. Evaluation of a Gas In Vitro System for Predicting Methane Production In Vivo. J. Dairy Sci. 2017, 100, 8881–8894. [Google Scholar] [CrossRef]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen Methanogens and Mitigation of Methane Emission by Anti-Methanogenic Compounds and Substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Medrano, R.F.; Wang, M.; Beauchemin, K.A.; Ma, Z.; Wang, R.; Wen, J.; Bernard, L.A.; Tan, Z. Effects of Urea plus Nitrate Pretreated Rice Straw and Corn Oil Supplementation on Fiber Digestibility, Nitrogen Balance, Rumen Fermentation, Microbiota and Methane Emissions in Goats. J. Anim. Sci. Biotechnol. 2019, 10, 6. [Google Scholar] [CrossRef]
- Cancino-Padilla, N.; Catalán, N.; Siu-Ting, K.; Creevey, C.J.; Huws, S.A.; Romero, J.; Vargas-Bello-Pérez, E. Long-Term Effects of Dietary Supplementation with Olive Oil and Hydrogenated Vegetable Oil on the Rumen Microbiome of Dairy Cows. Microorganisms 2021, 9, 1121. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; White, B.A.; Mizrahi, I. Potential Role of the Bovine Rumen Microbiome in Modulating Milk Composition and Feed Efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Lima, F.S.; Oikonomou, G.; Lima, S.F.; Bicalho, M.L.S.; Ganda, E.K.; de Oliveira Filho, J.C.; Lorenzo, G.; Trojacanec, P.; Bicalho, R.C. Prepartum and Postpartum Rumen Fluid Microbiomes: Characterization and Correlation with Production Traits in Dairy Cows. Appl. Environ. Microbiol. 2015, 81, 1327–1337. [Google Scholar] [CrossRef]
- Zhang, X.M.; Smith, M.L.; Gruninger, R.J.; Kung, L., Jr.; Vyas, D.; McGinn, S.M.; Kindermann, M.; Wang, M.; Tan, Z.L.; Beauchemin, K.A. Combined Effects of 3-Nitrooxypropanol and Canola Oil Supplementation on Methane Emissions, Rumen Fermentation and Biohydrogenation, and Total Tract Digestibility in Beef Cattle. J. Anim. Sci. 2021, 99, skab081. [Google Scholar] [CrossRef]
- Jiao, P.; Wang, Z.; Yang, W. Effect of Clostridium Butyricum Supplementation on In Vitro Rumen Fermentation and Microbiota with High Grain Substrate Varying with Media pH Levels. Front. Microbiol. 2022, 13, 912042. [Google Scholar] [CrossRef]
- Huaiquipán, R.; Quiñones, J.; Díaz, R.; Velásquez, C.; Sepúlveda, G.; Velázquez, L.; Paz, E.A.; Tapia, D.; Cancino, D.; Sepúlveda, N. Review: Effect of Experimental Diets on the Microbiome of Productive Animals. Microorganisms 2023, 11, 2219. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Hassan, F.; Li, M.; Xie, H.; Peng, L.; Tang, Z.; Yang, C. Effect of Sodium Nitrate and Cysteamine on In Vitro Ruminal Fermentation, Amino Acid Metabolism and Microbiota in Buffalo. Microorganisms 2022, 10, 2038. [Google Scholar] [CrossRef]
- Ungerfeld, E.M.; Beauchemin, K.A.; Muñoz, C. Current Perspectives on Achieving Pronounced Enteric Methane Mitigation From Ruminant Production. Front. Anim. Sci. 2022, 2, 795200. [Google Scholar] [CrossRef]
- Hristov, A.N.; Oh, J.; Giallongo, F.; Frederick, T.W.; Harper, M.T.; Weeks, H.L.; Branco, A.F.; Moate, P.J.; Deighton, M.H.; Williams, S.R.O. An Inhibitor Persistently Decreased Enteric Methane Emission from Dairy Cows with No Negative Effect on Milk Production. Proc. Natl. Acad. Sci. USA 2015, 112, 10663–10668. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xiong, B.; Zhao, X. Could Propionate Formation Be Used to Reduce Enteric Methane Emission in Ruminants? Sci. Total Environ. 2023, 855, 158867. [Google Scholar] [CrossRef] [PubMed]
- Guyader, J.; Tavendale, M.; Martin, C.; Muetzel, S. Dose-Response Effect of Nitrate on Hydrogen Distribution between Rumen Fermentation End Products: An In Vitro Approach. Anim. Prod. Sci. 2016, 56, 224–230. [Google Scholar] [CrossRef]
- Tan, P.; Liu, H.; Zhao, J.; Gu, X.; Wei, X.; Zhang, X.; Ma, N.; Johnston, L.J.; Bai, Y.; Zhang, W.; et al. Amino Acids Metabolism by Rumen Microorganisms: Nutrition and Ecology Strategies to Reduce Nitrogen Emissions from the inside to the Outside. Sci. Total Environ. 2021, 800, 149596. [Google Scholar] [CrossRef] [PubMed]
- Millen, D.D.; Arrigoni, M.D.B.; Pacheco, R.D.L. Rumenology; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 3-319-30533-6. [Google Scholar]
- Zhang, Z.; Wei, W.; Yang, S.; Huang, Z.; Li, C.; Yu, X.; Qi, R.; Liu, W.; Loor, J.J.; Wang, M.; et al. Regulation of Dietary Protein Solubility Improves Ruminal Nitrogen Metabolism In Vitro: Role of Bacteria–Protozoa Interactions. Nutrients 2022, 14, 2972. [Google Scholar] [CrossRef]
- Pitta, D.; Indugu, N.; Narayan, K.; Hennessy, M. Symposium Review: Understanding the Role of the Rumen Microbiome in Enteric Methane Mitigation and Productivity in Dairy Cows. J. Dairy Sci. 2022, 105, 8569–8585. [Google Scholar] [CrossRef]
- Weinroth, M.D.; Belk, A.D.; Dean, C.; Noyes, N.; Dittoe, D.K.; Rothrock, M.J.; Ricke, S.C.; Myer, P.R.; Henniger, M.T.; Ramírez, G.A.; et al. Considerations and Best Practices in Animal Science 16S Ribosomal RNA Gene Sequencing Microbiome Studies. J. Anim. Sci. 2022, 100, skab346. [Google Scholar] [CrossRef] [PubMed]

| Treatment | Shannon Diversity Index | Simpson Diversity Index | Chao1 Richness Index | Pielou Evenness Index | ASV Count | Archaeal Relative Abundance |
|---|---|---|---|---|---|---|
| Control | 9.925562 | 0.995680 | 5569 | 0.797338 | 5589 | 0.013248 |
| C10 + C12 | 9.982964 | 0.996355 | 5830 | 0.806313 | 5334 | 0.017648 |
| SEM | 0.05372 | 0.0004 | 22.26162 | 0.00391 | 21.45958 | 0.16 |
| p-value | 0.2828 | 0.2918 | 0.8492 | 0.1208 | 0.8956 | 0.195312 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vadroňová, M.; Šťovíček, A.; Výborná, A.; Tyrolová, Y.; Tichá, D.; Joch, M. Insights into Effects of Combined Capric and Lauric Acid on Rumen Bacterial Composition. Microorganisms 2024, 12, 1085. https://doi.org/10.3390/microorganisms12061085
Vadroňová M, Šťovíček A, Výborná A, Tyrolová Y, Tichá D, Joch M. Insights into Effects of Combined Capric and Lauric Acid on Rumen Bacterial Composition. Microorganisms. 2024; 12(6):1085. https://doi.org/10.3390/microorganisms12061085
Chicago/Turabian StyleVadroňová, Mariana, Adam Šťovíček, Alena Výborná, Yvona Tyrolová, Denisa Tichá, and Miroslav Joch. 2024. "Insights into Effects of Combined Capric and Lauric Acid on Rumen Bacterial Composition" Microorganisms 12, no. 6: 1085. https://doi.org/10.3390/microorganisms12061085
APA StyleVadroňová, M., Šťovíček, A., Výborná, A., Tyrolová, Y., Tichá, D., & Joch, M. (2024). Insights into Effects of Combined Capric and Lauric Acid on Rumen Bacterial Composition. Microorganisms, 12(6), 1085. https://doi.org/10.3390/microorganisms12061085






