Bacillus amyloliquefaciens LM-1 Affects Multiple Cell Biological Processes in Magnaporthe oryzae to Suppress Rice Blast
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Identification of B. amyloliquefaciens LM-1
2.3. Enzyme Activity Detection
2.4. Genome Annotation of B. amyloliquefaciens LM-1
2.5. Sensitivity Detection for Mycelial Growth of M. oryzae and B. oryzae with B. amyloliquefaciens LM-1
2.6. Pathogenicity Detection
2.7. Microscopy Observation
2.8. Effects of Strain LM-1 on Growth and Development of Rice
3. Results
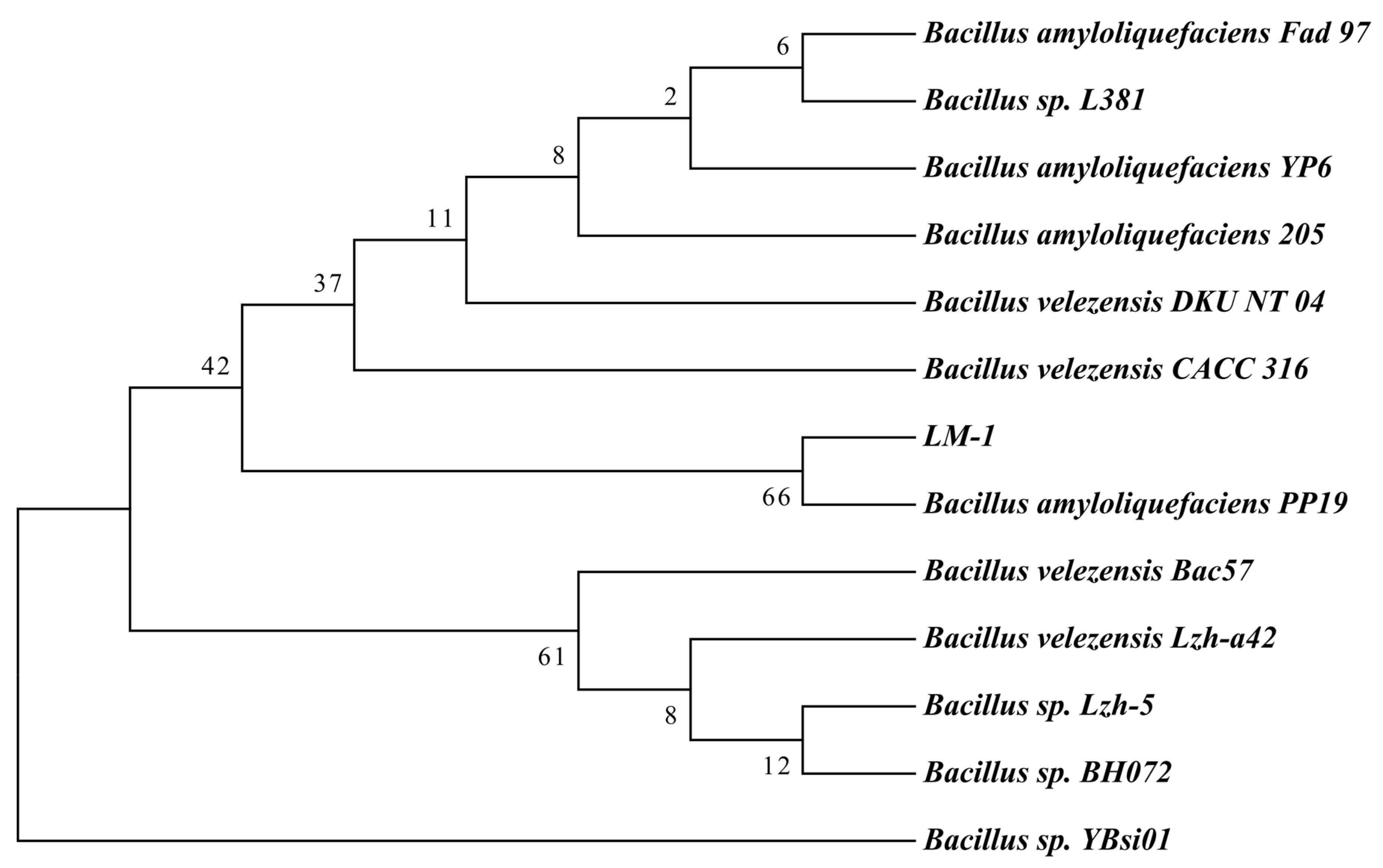
3.1. Isolation and Identification of Bacillus amyloliquefaciens LM-1
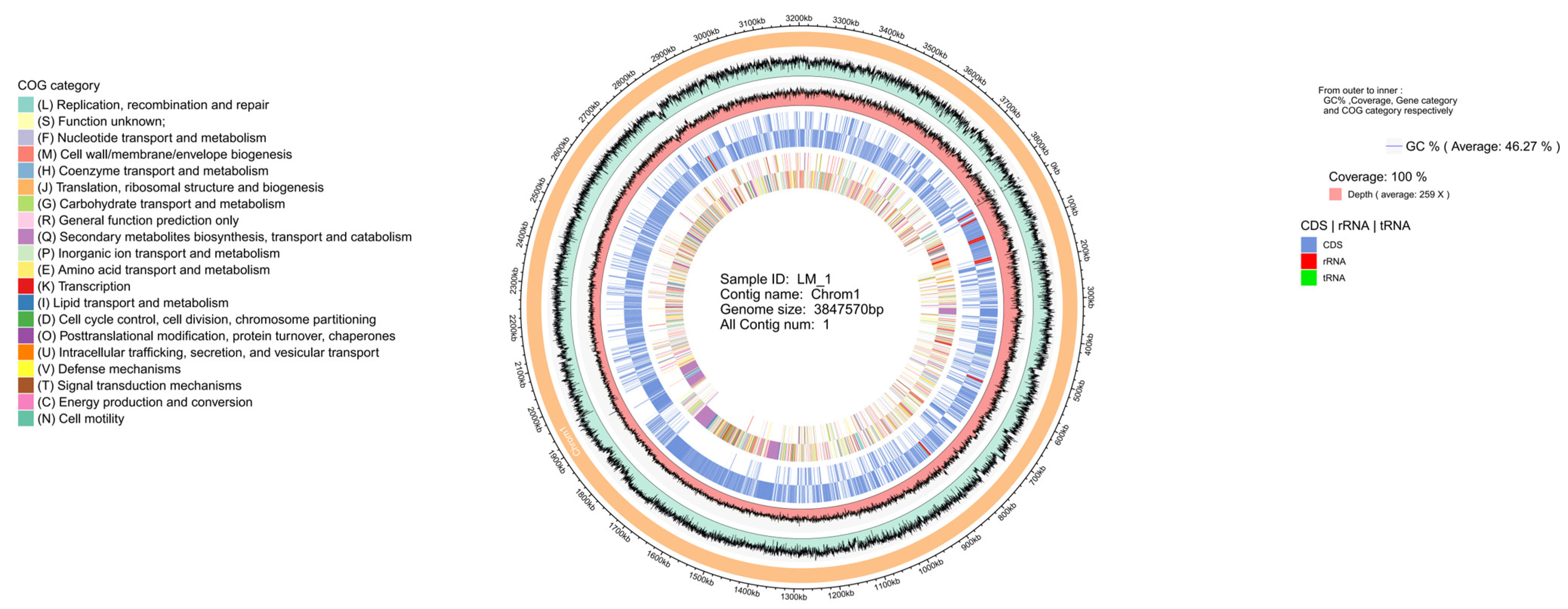
3.2. Genomic Features of B. amyloliquefaciens LM-1
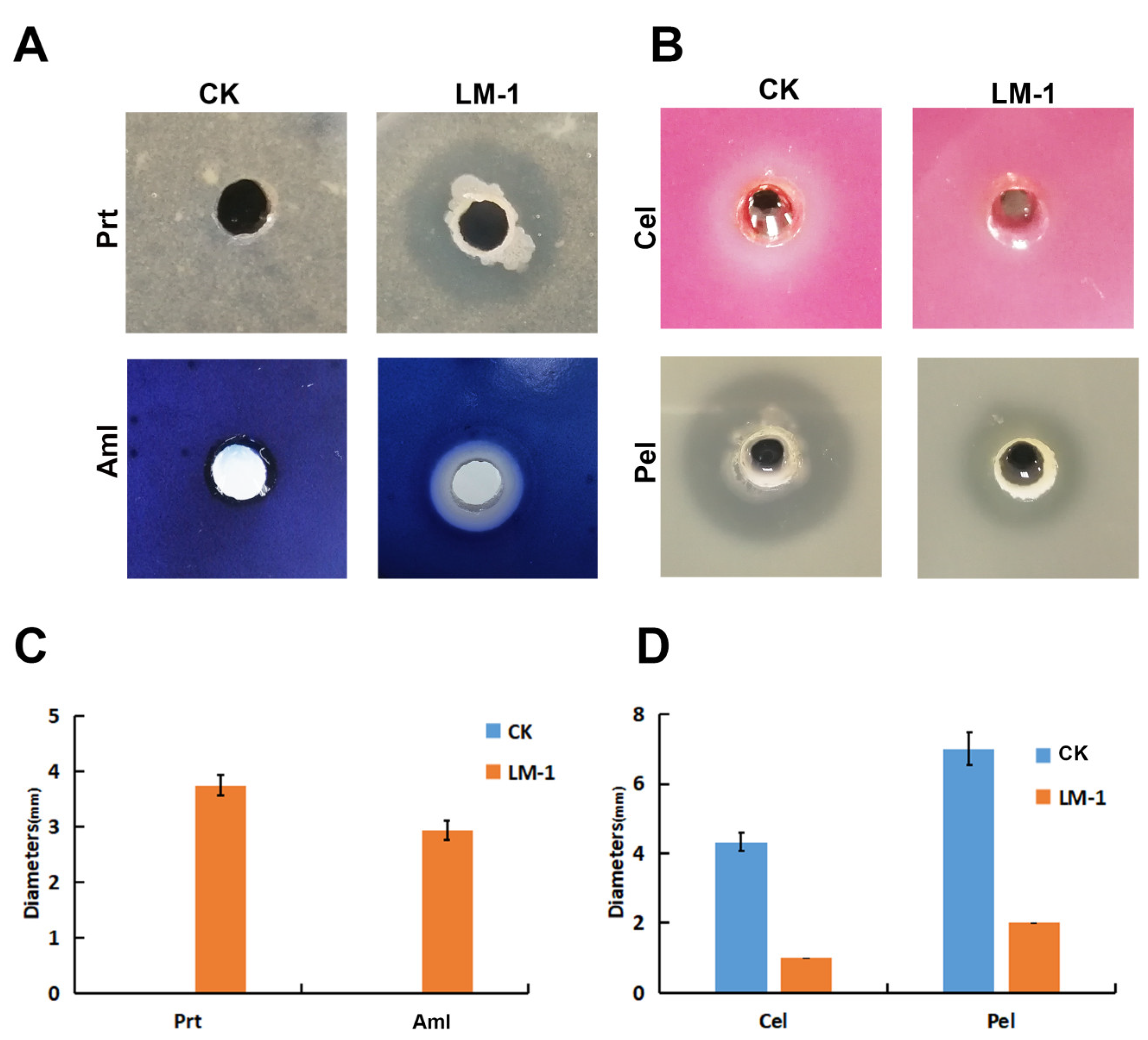
3.3. Enzyme Activity Detection of B. amyloliquefaciens LM-1
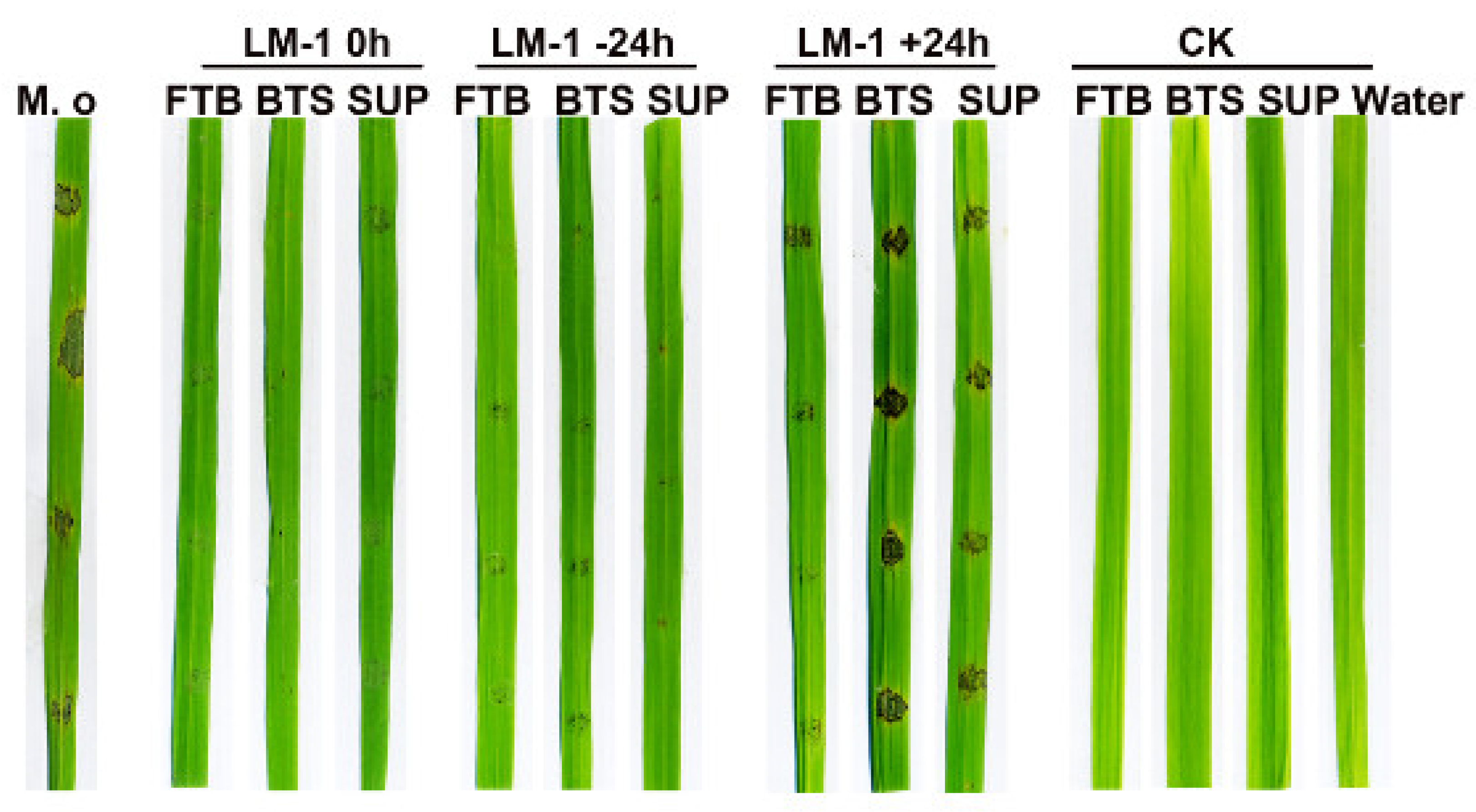
3.4. The Rice Fungal Diseases Can Be Controlled by B. amyloliquefaciens LM-1
3.5. LM-1 Inhibits Sporulation and Damages the Integrity of the Cell Wall in M. oryzae
3.6. LM-1 Suppresses Appressorium Formation and Conidial Cell Death of M. oryzae
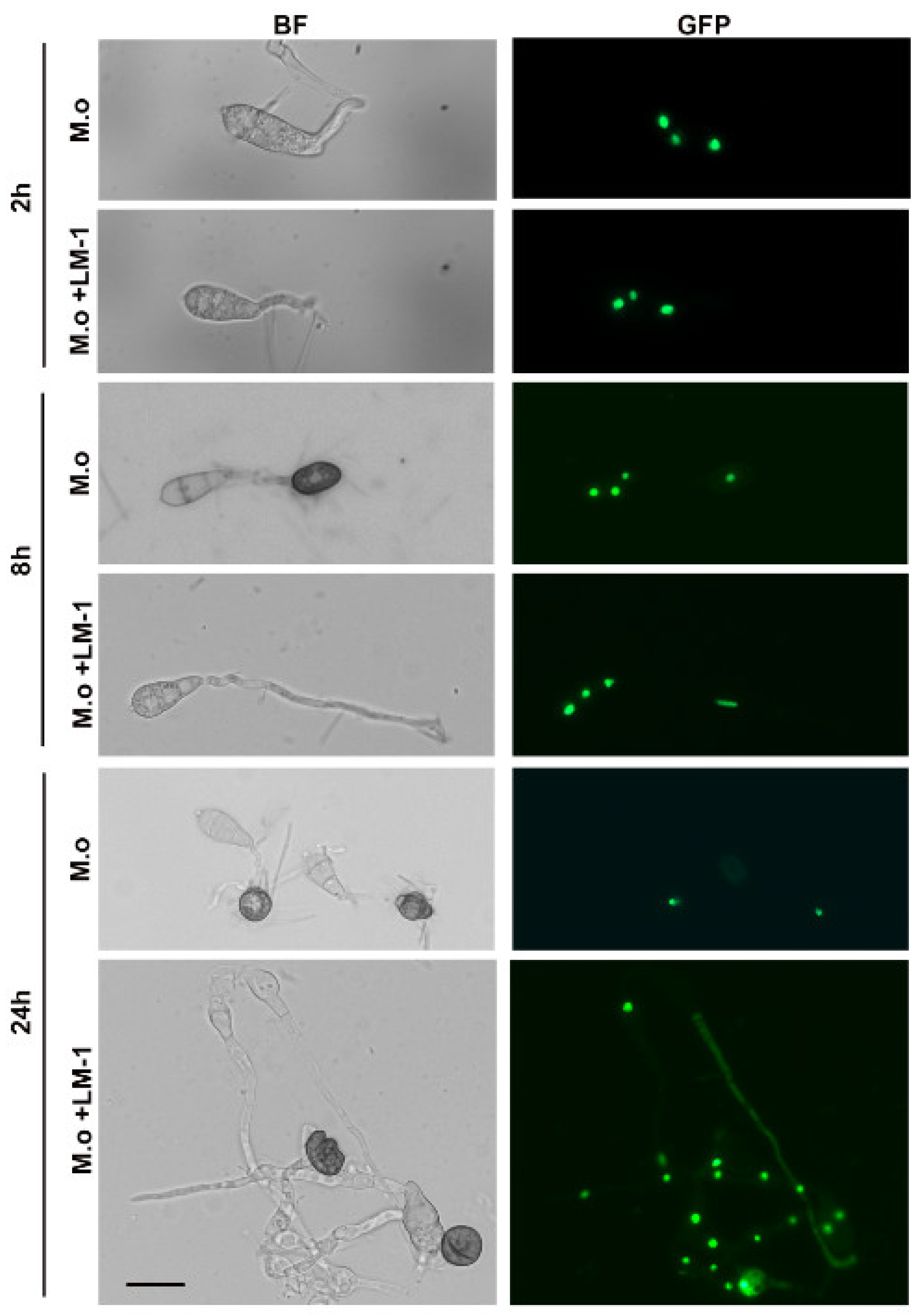
3.7. Effects of LM-1 on Autophagy during Conidial Germination in M. oryzae
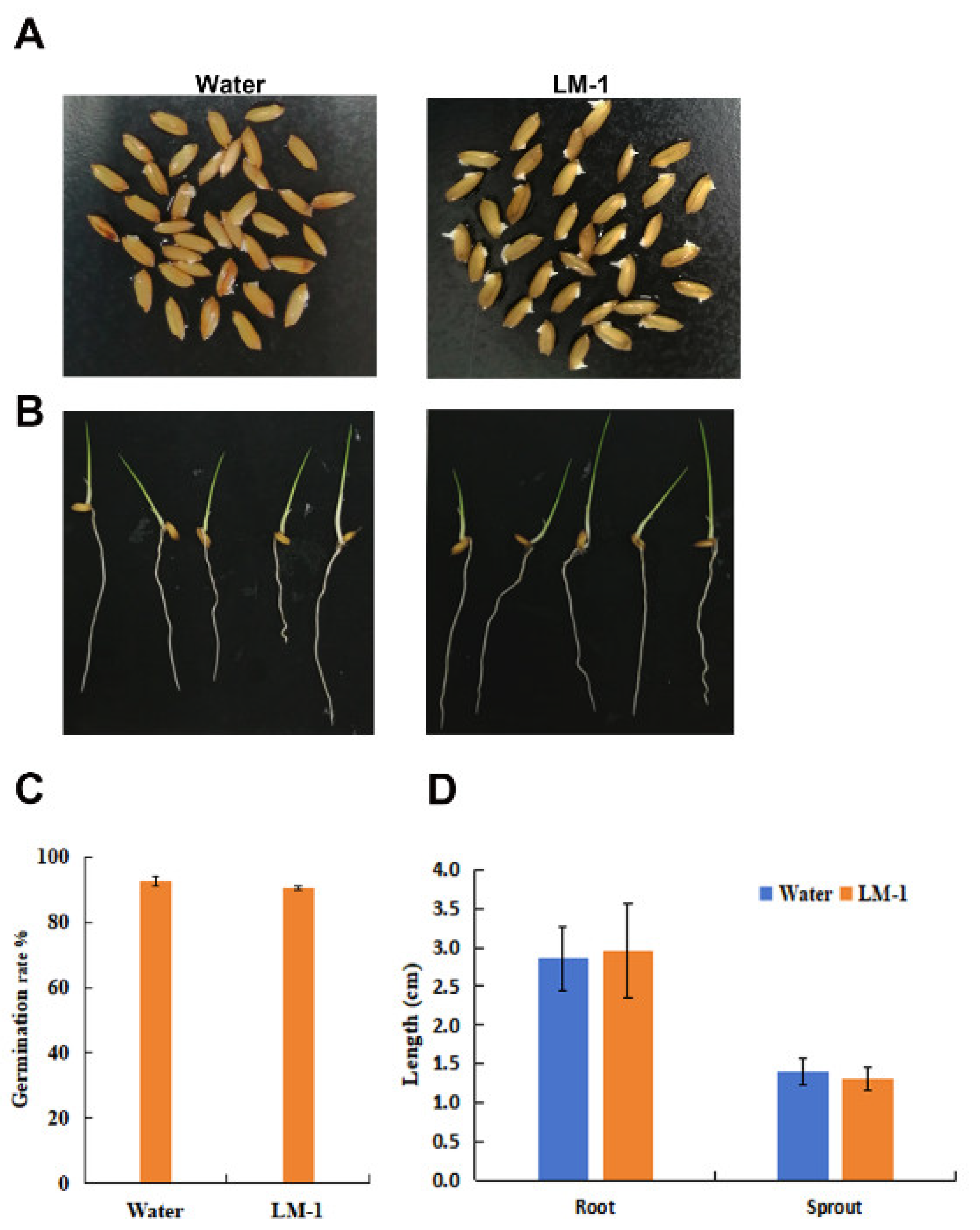
3.8. Effects of Strain LM-1 on Growth and Development of Rice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant. Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.T.; Croll, D.; Gladieux, P.; Soanes, D.M.; Persoons, A.; Bhattacharjee, P.; Hossain, M.S.; Gupta, D.R.; Rahman, M.M.; Mahboob, M.G.; et al. Emergence of wheat blast in Bangladesh was caused by a South American lineage of Magnaporthe oryzae. BMC Biol. 2016, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; Mahmud, N.U.; Ullah, C.; Rahman, M.; Islam, T. Biological and biorational management of blast diseases in cereals caused by Magnaporthe oryzae. Crit. Rev. Biotechnol. 2021, 41, 994–1022. [Google Scholar] [CrossRef]
- Howard, R.J.; Valent, B. Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 1996, 50, 491–512. [Google Scholar] [CrossRef]
- Talbot, N.J. Appressoria. Curr. Biol. 2019, 29, R144–R146. [Google Scholar] [CrossRef] [PubMed]
- Veneault-Fourrey, C.; Barooah, M.; Egan, M.; Wakley, G.; Talbot, N.J. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science 2006, 312, 580–583. [Google Scholar] [CrossRef] [PubMed]
- Castroagudin, V.L.; Ceresini, P.C.; Oliveira, S.C.; Reges, J.T.; Maciel, J.L.; Bonato, A.L.; Dorigan, A.F.; McDonald, B.A. Resistance to QoI fungicides is widespread in Brazilian populations of the wheat blast pathogen Magnaporthe oryzae. Phytopathology 2015, 105, 284–294. [Google Scholar] [CrossRef]
- Vaezi, A.; Fakhim, H.; Javidnia, J.; Khodavaisy, S.; Abtahian, Z.; Vojoodi, M.; Nourbakhsh, F.; Badali, H. Pesticide behavior in paddy fields and development of azole-resistant Aspergillus fumigatus: Should we be concerned? J. Mycol. Med. 2018, 28, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, Y.; Wang, J.; Liu, L.; Zhao, Y.; Jiang, Z.; Wang, Y.; Li, Z.; Fu, L.; Cui, Z. Biocontrol potential of Burkholderia sp. BV6 against the rice blast fungus Magnaporthe oryzae. J. Appl. Microbiol. 2022, 133, 883–897. [Google Scholar]
- Surovy, M.Z.; Rahman, S.; Rostas, M.; Islam, T.; von Tiedemann, A. Suppressive effects of volatile compounds from Bacillus spp. on Magnaporthe oryzae Triticum (MoT) pathotype, causal agent of wheat blast. Microorganisms 2023, 11, 1291. [Google Scholar] [CrossRef]
- Wang, S.Y.; Herrera-Balandrano, D.D.; Wang, Y.X.; Shi, X.C.; Chen, X.; Jin, Y.; Liu, F.Q.; Laborda, P. Biocontrol ability of the Bacillus amyloliquefaciens Group, B. amyloliquefaciens, B. velezensis, B. nakamurai, and B. siamensis, for the management of fungal postharvest diseases: A Review. J. Agric. Food Chem. 2022, 70, 6591–6616. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, A.T. Bacillus subtilis. Trends Microbiol. 2019, 27, 724–725. [Google Scholar] [CrossRef] [PubMed]
- Boottanun, P.; Potisap, C.; Hurdle, J.G.; Sermswan, R.W. Secondary metabolites from Bacillus amyloliquefaciens isolated from soil can kill Burkholderia pseudomallei. AMB Express 2017, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Amruta, N.; Prasanna, K.M.K.; Puneeth, M.E.; Sarika, G.; Kandikattu, H.K.; Vishwanath, K.; Narayanaswamy, S. Exploring the potentiality of novel Rhizospheric bacterial strains against the rice blast fungus Magnaporthe oryzae. Plant Pathol. J. 2018, 34, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Ben, A.D.; Frikha-Gargouri, O.; Tounsi, S. Bacillus amyloliquefaciens strain 32a as a source of lipopeptides for biocontrol of Agrobacterium tumefaciens strains. J. Appl. Microbiol. 2015, 119, 196–207. [Google Scholar]
- Alvarez, F.; Castro, M.; Principe, A.; Borioli, G.; Fischer, S.; Mori, G.; Jofre, E. The plant-associated Bacillus amyloliquefaciens strains MEP2 18 and ARP2 3 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J. Appl. Microbiol. 2012, 112, 159–174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, Y.; Harvey, P.R.; Stummer, B.E.; Yang, J.; Ji, Z. Wheat rhizosphere colonization by Bacillus amyloliquefaciens W10 and Pseudomonas protegens FD6 suppress soil and in planta abundance of the sharp eyespot pathogen Rhizoctonia cerealis. J. Appl. Microbiol. 2023, 134, lxad101. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Uddin, W.; Wenner, N.G. Induced systemic resistance responses in perennial ryegrass against Magnaporthe oryzae elicited by semi-purified surfactin lipopeptides and live cells of Bacillus amyloliquefaciens. Mol. Plant Pathol. 2015, 16, 546–558. [Google Scholar] [CrossRef]
- Liang, M.; Ye, H.; Shen, Q.; Jiang, X.; Cui, G.; Gu, W.; Zhang, L.H.; Naqvi, N.I.; Deng, Y.Z. Tangeretin inhibits fungal ferroptosis to suppress rice blast. J. Integr. Plant Biol. 2021, 63, 2136–2149. [Google Scholar] [CrossRef]
- Shen, Q.; Liang, M.; Yang, F.; Deng, Y.Z.; Naqvi, N.I. Ferroptosis contributes to developmental cell death in rice blast. New Phytol. 2020, 227, 1831–1846. [Google Scholar] [CrossRef]
- Feng, A.; Zhu, X.; Wang, C.; Chen, K.; Feng, J.; Yang, J.; Lin, X.; Chen, S. Efficacy of thirteen gungicides for controlling rice brown spot disease. Plant Prot. 2022, 48, 352–360. [Google Scholar]
- Zhou, J.; Xie, Y.; Liao, Y.; Li, X.; Li, Y.; Li, S.; Ma, X.; Lei, S.; Lin, F.; Jiang, W.; et al. Characterization of a Bacillus velezensis strain isolated from Bolbostemmatis Rhizoma displaying strong antagonistic activities against a variety of rice pathogens. Front. Microbiol. 2022, 13, 983781. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Xu, T.; Peng, J.; Zhou, K.; Zhu, Y.; Zhou, W.; Cheng, H.; Zhou, H. Overexpression of an endogenous raw starch digesting mesophilic alpha-amylase gene in Bacillus amyloliquefaciens Z3 by in vitro methylation protocol. J. Sci. Food Agric. 2020, 100, 3013–3023. [Google Scholar] [CrossRef] [PubMed]
- Bekli, S.; Aktas, B.; Gencer, D.; Aslim, B. Biochemical and molecular characterizations of a novel pH- and temperature-stable ppectate lyase from Bacillus amyloliquefaciens S6 for industrial application. Mol. Biotechnol. 2019, 61, 681–693. [Google Scholar] [CrossRef] [PubMed]
- Dangol, S.; Chen, Y.; Hwang, B.K.; Jwa, N.S. Iron- and reactive oxygen species-dependent ferroptotic cell death in rice-Magnaporthe oryzae interactions. Plant Cell 2019, 31, 189–209. [Google Scholar] [CrossRef] [PubMed]
- Lv, M.; Chen, Y.; Hu, M.; Yu, Q.; Duan, C.; Ye, S.; Ling, J.; Zhou, J.; Zhou, X.; Zhang, L. OhrR is a central transcriptional regulator of virulence in Dickeya zeae. Mol. Plant Pathol. 2022, 23, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A. Magnaporthe oryzae. Trends Microbiol. 2021, 29, 663–664. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.C.; Dagdas, Y.F.; Gupta, Y.K.; Mentlak, T.A.; Yi, M.; Martinez-Rocha, A.L.; Saitoh, H.; Terauchi, R.; Talbot, N.J.; Valent, B. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun. 2013, 4, 1996. [Google Scholar] [CrossRef] [PubMed]
- Fira, D.; Dimkic, I.; Beric, T.; Lozo, J.; Stankovic, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Syed, A.R.S.; Pieterse, C.M.J.; Schenk, P.M. Emerging microbial biocontrol strategies for plant pathogens. Plant Sci. 2018, 267, 102–111. [Google Scholar] [CrossRef]
- Yang, P.; Yuan, P.; Liu, W.; Zhao, Z.; Bernier, M.C.; Zhang, C.; Adhikari, A.; Opiyo, S.O.; Zhao, L.; Banks, F.; et al. Plant growth promotion and plant disease suppression induced by Bacillus amyloliquefaciens Strain GD4a. Plants 2024, 13, 672. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Dai, Z.; Zhang, J.; Li, S.; Ahmed, A.; Munir, S.; Liu, Q.; Tan, Y.; Ji, G.; Zhao, Z. Plant-microbe interaction: Mining the impact of native Bacillus amyloliquefaciens WS-10 on tobacco bacterial wilt disease and rhizosphere microbial communities. Microbiol. Spectr. 2022, 10, e0147122. [Google Scholar] [CrossRef] [PubMed]
- Zalila-Kolsi, I.; Ben-Mahmoud, A.; Al-Barazie, R. Bacillus amyloliquefaciens: Harnessing its potential for industrial, medical, and agricultural applications-a comprehensive review. Microorganisms 2023, 11, 2215. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Zeng, Q.; Sui, S. Screening and application of Bacillus strains isolated from nonrhizospheric rice Soil for the biocontrol of rice blast. Plant Pathol. J. 2020, 36, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.Y.; Hou, Z.J.; Ding, M.Z.; Gao, G.R.; Qiao, B.; Wei, S.Y.; Cheng, J.S. Integrated biofilm modification and transcriptional analysis for improving fengycin production in Bacillus amyloliquefaciens. Probiotics Antimicrob. Proteins 2024. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Yoshino, K.; Dagdas, G.; Ryder, L.S.; Bielska, E.; Steinberg, G.; Talbot, N.J. Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 2012, 336, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, K.; Chen, R.; Jiang, J. Bacillus subtilis KLBMPGC81 suppresses appressorium-mediated plant infection by altering the cell wall integrity signaling pathway and multiple cell biological processes in Magnaporthe oryzae. Front. Cell Infect. Microbiol. 2022, 12, 983757. [Google Scholar] [CrossRef] [PubMed]
- Kgosi, V.T.; Tingting, B.; Ying, Z.; Liu, H. Anti-fungal analysis of Bacillus subtilis DL76 on conidiation, appressorium formation, growth, multiple stress response, and pathogenicity in Magnaporthe oryzae. Int. J. Mol. Sci. 2022, 23, 5314. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.A.; Talbot, N.J. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat. Rev. Microbiol. 2009, 7, 185–195. [Google Scholar] [CrossRef]
- Kershaw, M.J.; Talbot, N.J. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc. Natl. Acad. Sci. USA 2009, 106, 15967–15972. [Google Scholar] [CrossRef]
- Eseola, A.B.; Oses-Ruiz, M.; Findlay, K.; Yan, X.; Cruz-Mireles, N.; Molinari, C.; Garduno-Rosales, M.; Talbot, N.J. Investigating the cell and developmental biology of plant infection by the rice blast fungus Magnaporthe oryzae. Fungal Genet. Biol. 2021, 154, 103562. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhao, M.; Chen, W.; Yu, H.; Jia, W.; Pan, H.; Zhang, X. Multi-omics techniques for analysis antifungal mechanisms of lipopeptides produced by Bacillus velezensis GS-1 against Magnaporthe oryzae in vitro. Int. J. Mol. Sci. 2022, 23, 3762. [Google Scholar] [CrossRef] [PubMed]
- Sha, Y.; Wang, Q.; Li, Y. Suppression of Magnaporthe oryzae and interaction between Bacillus subtilis and rice plants in the control of rice blast. SpringerPlus 2016, 5, 1238. [Google Scholar] [CrossRef] [PubMed]
- Thepbandit, W.; Srisuwan, A.; Siriwong, S.; Nawong, S.; Athinuwat, D. Bacillus vallismortis TU-Orga21 blocks rice blast through both direct effect and stimulation of plant defense. Front Plant Sci. 2023, 14, 1103487. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ren, Z.H.; Zu, X.; Yu, X.Y.; Zhu, H.J.; Li, X.J.; Zhong, J.; Liu, E.M. Efficacy of plant growth-promoting bacteria Bacillus cereus YN917 for biocontrol of rice blast. Front. Microbiol. 2021, 12, 684888. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.J.; Wang, Z.Y.; Jones, M.A.; Smirnoff, N.; Talbot, N.J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA 2007, 104, 11772–11777. [Google Scholar] [CrossRef]
- Sanchez-Sanuy, F.; Mateluna-Cuadra, R.; Tomita, K.; Okada, K.; Sacchi, G.A.; Campo, S.; San, S.B. Iron induces resistance against the rice blast fungus Magnaporthe oryzae through potentiation of immune responses. Rice 2022, 15, 68. [Google Scholar] [CrossRef]





| Treatment Group | Disease Grade % | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| M.o | 43.3% ± 9.81 | 43.3% ± 7.2 | 13.3% ± 2.72 | |||||
| M.o+LM-1 | 50.0% ± 4.7 | 30.0% ± 4.7 | 20.0% ± 4.7 | |||||
| B.o | 10.0% ± 8.2 | 10.0% ± 8.2 | 36.7% ± 9.8 | 43.3% ± 16.6 | ||||
| B.o+LM-1 | 33.3% ± 5.4 | 30.0% ± 8.2 | 26.7% ± 10.9 | 6.7% ± 5.4 | 3.3% ± 2.7 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, M.; Feng, A.; Wang, C.; Zhu, X.; Su, J.; Xu, Z.; Yang, J.; Wang, W.; Chen, K.; Chen, B.; et al. Bacillus amyloliquefaciens LM-1 Affects Multiple Cell Biological Processes in Magnaporthe oryzae to Suppress Rice Blast. Microorganisms 2024, 12, 1246. https://doi.org/10.3390/microorganisms12061246
Liang M, Feng A, Wang C, Zhu X, Su J, Xu Z, Yang J, Wang W, Chen K, Chen B, et al. Bacillus amyloliquefaciens LM-1 Affects Multiple Cell Biological Processes in Magnaporthe oryzae to Suppress Rice Blast. Microorganisms. 2024; 12(6):1246. https://doi.org/10.3390/microorganisms12061246
Chicago/Turabian StyleLiang, Meiling, Aiqing Feng, Congying Wang, Xiaoyuan Zhu, Jing Su, Zihan Xu, Jianyuan Yang, Wenjuan Wang, Kailing Chen, Bing Chen, and et al. 2024. "Bacillus amyloliquefaciens LM-1 Affects Multiple Cell Biological Processes in Magnaporthe oryzae to Suppress Rice Blast" Microorganisms 12, no. 6: 1246. https://doi.org/10.3390/microorganisms12061246
APA StyleLiang, M., Feng, A., Wang, C., Zhu, X., Su, J., Xu, Z., Yang, J., Wang, W., Chen, K., Chen, B., Lin, X., Feng, J., & Chen, S. (2024). Bacillus amyloliquefaciens LM-1 Affects Multiple Cell Biological Processes in Magnaporthe oryzae to Suppress Rice Blast. Microorganisms, 12(6), 1246. https://doi.org/10.3390/microorganisms12061246





