Amazonian Bacteria from River Sediments as a Biocontrol Solution against Ralstonia solanacearum
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. In Vitro Antimicrobial Activity against Ralstonia Solanacearum
2.3. Biocontrol Evaluation under Greenhouse Conditions
2.4. Analysis of R. solanacearum Suppression in Soil
2.5. DNA Extraction, Sequencing, and Genome Assembly
2.6. Phylogenomic Identification
2.7. Production of Extracellular Enzymes
2.8. Production of In Vitro Growth Promotion Inducers
2.8.1. Phosphate (P) and Zinc (Zn) Solubilization
2.8.2. Siderophore
2.8.3. Indole Acetic Acid (IAA)
2.8.4. Ammonia
2.9. Statistical Analysis
3. Results
3.1. In Vitro Antimicrobial Activity
3.2. Phylogenomic Identification
3.3. Production of Growth Promotion Inducers and Enzymes
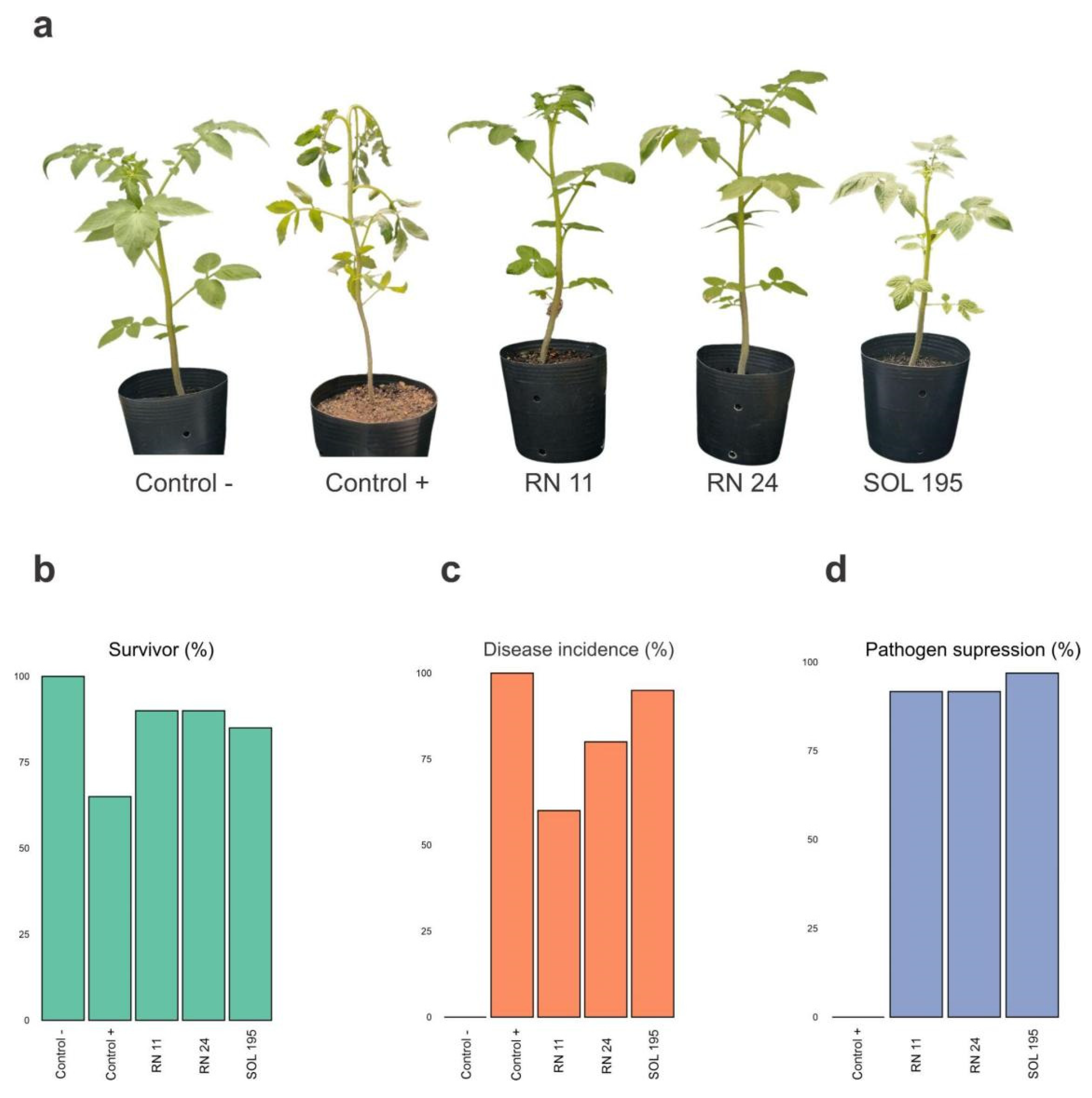
3.4. Biological Control under Greenhouse Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Assefa, W.; Tesfaye, B.; Dessalegn, L. Influence of Inter-Intra Row Spacing on Yield Losses of Tomato Cultivars. Ethiop. J. Agric. Sci. 2015, 25, 15–28. [Google Scholar]
- Elnaggar, S.; Mohamed, A.M.; Bakeer, A.; Osman, T.A. Current Status of Bacterial Wilt (Ralstonia solanacearum) Disease in Major Tomato (Solanum lycopersicum L.) Growing Areas in Egypt. Arch. Agric. Environ. Sci. 2018, 3, 399–406. [Google Scholar] [CrossRef]
- Kurabachew, H.; Ayana, G. Bacterial Wilt Caused by Ralstonia solanacearum in Ethiopia: StatusaAnd Management Approaches: A Review. Int. J. Phytopathol. 2016, 5, 107–119. [Google Scholar] [CrossRef]
- Coelho Netto, R.A.; Noda, H.; Boher, B. Melanthera discoidea: Um novo hospedeiro de Ralstonia solanacearum. Fitopatol. Bras. 2001, 26, 781. [Google Scholar] [CrossRef]
- Coelho Netto, R.A.; Pereira, B.G.; Noda, H.; Boher, B. Caracterização de isolados de Ralstonia solanacearum obtidos de tomateiros em várzea e em terra firme, no Estado do Amazonas. Fitopatol. Bras. 2003, 28, 362–366. [Google Scholar] [CrossRef]
- Coelho Netto, R.A.; Pereira, B.G.; Noda, H.; Boher, B. Murcha bacteriana no estado do Amazonas, Brasil. Fitopatol. Bras. 2004, 29, 17–23. [Google Scholar] [CrossRef]
- Lopes, C.A.; Rossato, M. History and Status of Selected Hosts of the Ralstonia solanacearum Species Complex Causing Bacterial Wilt in Brazil. Front. Microbiol. 2018, 9, 1228. [Google Scholar] [CrossRef] [PubMed]
- Vailleau, F.; Genin, S. Ralstonia solanacearum: An Arsenal of Virulence Strategies and Prospects for Resistance. Annu. Rev. Phytopathol. 2023, 61, 25–47. [Google Scholar] [CrossRef] [PubMed]
- Cellier, G.; Prior, P. Deciphering Phenotypic Diversity of Ralstonia solanacearum Strains Pathogenic to Potato. Phytopathology 2010, 100, 1250–1261. [Google Scholar] [CrossRef]
- Lebeau, A.; Daunay, M.C.; Frary, A.; Palloix, A.; Wang, J.F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial Wilt Resistance in Tomato, Pepper, and Eggplant: Genetic Resources Respond to Diverse Strains in the Ralstonia solanacearum Species Complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef]
- Romo, J.P.; Osorio, J.G.M.; Yepes, M.S. Identification of New Hosts for Ralstonia solanacearum (Smith) Race 2 from Colombia. Rev. Protección Veg. 2012, 27, 151–161. [Google Scholar]
- Wicker, E.; Lefeuvre, P.; De Cambiaire, J.-C.; Lemaire, C.; Poussier, S.; Prior, P. Contrasting Recombination Patterns and Demographic Histories of the Plant Pathogen Ralstonia solanacearum Inferred from MLSA. ISME J. 2012, 6, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Santiago, T.R.; Lopes, C.A.; Caetano-Anollés, G.; Mizubuti, E.S.G. Phylotype and Sequevar Variability of Ralstonia solanacearum in Brazil, an Ancient Centre of Diversity of the Pathogen. Plant Pathol. 2017, 66, 383–392. [Google Scholar] [CrossRef]
- Bergsma-Vlami, M.; Van De Bilt, J.L.J.; Tjou-Tam-Sin, N.N.A.; Westenberg, M.; Meekes, E.T.M.; Teunissen, H.A.S.; Van Vaerenbergh, J. Phylogenetic Assignment of Ralstonia pseudosolanacearum (Ralstonia solanacearum Phylotype I) Isolated from Rosa spp. Plant Dis. 2018, 102, 2258–2267. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, F.; Ana Carolina, P.N.; Daniel Diego, C.C.; Mylla Crysthyan, R. Induction of Tolerance to Bacterial Wilt in Hybrids of Tomatoes by Application of Gibberellin. Rev. Cienc. Agrovet. 2018, 17, 54–60. [Google Scholar] [CrossRef]
- Huet, G. Breeding for Resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [PubMed]
- Kaari, M.; Joseph, J.; Manikkam, R.; Sreenivasan, A.; Venugopal, G. Biological Control of Streptomyces sp. UT4A49 to Suppress Tomato Bacterial Wilt Disease and Its Metabolite Profiling. J. King Saud Univ.-Sci. 2022, 34, 101688. [Google Scholar] [CrossRef]
- Kaari, M.; Joseph, J.; Manikkam, R.; Sreenivasan, A.; Venugopal, G.; Alexander, B.; Krishnan, S. Anti-Biofilm Activity and Biocontrol Potential of Streptomyces Cultures Against Ralstonia solanacearum on Tomato Plants. Indian J. Microbiol. 2022, 62, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Genin, S.; Denny, T.P. Pathogenomics of the Ralstonia solanacearum Species Complex. Annu. Rev. Phytopathol. 2012, 50, 67–89. [Google Scholar] [CrossRef] [PubMed]
- Saputra, R.; Arwiyanto, T.; Wibowo, A. Biological Control of Ralstonia solanacearum Causes of Bacterial Wilt Disease with Pseudomonas Putida and Streptomyces spp. on Some Tomato Varieties. IOP Conf. Ser. Earth Environ. Sci. 2020, 515, 012007. [Google Scholar] [CrossRef]
- Noda, H.; Pahlen, A.V.; Der Silva Filho, D.F. Avaliação Da Resistência de Progênies de Tomate à Murcha Bacteriana Em Solo Naturalmente Infestado Por Pseudomonas solanacearum (Smith) Dows. Rev. Bras. Genética 1986, 9, 55–66. [Google Scholar]
- Costa, K.D.D.S.; Santos, P.R.; Santos, A.M.M.; Silva, A.M.F.; Chagas, J.T.B.; Carvalho Filho, J.L.S.; Pereira, J.W.D.L.; Silva, M.D.O.; Silva, J.R.; Menezes, D. Genetic Control of Tomato Resistance to Ralstonia solanacearum. Euphytica 2019, 215, 136. [Google Scholar] [CrossRef]
- Marques, M.J.; Vizú, J.D.F.; Silva Filho, D.F.; Ticona-Benavente, C.A. Tomato Progenies Selection in Rondônia, Brazil. Hortic. Bras. 2019, 37, 106–111. [Google Scholar] [CrossRef]
- Ishikawa, R.; Fujimori, K.; Matsuura, K. Antibacterial Activity of Validamycin A against Pseudomonas Solanacearum and Its Efficacy against Tomato Bacterial Wilt. Jpn. J. Phytopathol. 1996, 62, 478–482. [Google Scholar] [CrossRef][Green Version]
- Sing’ombe Ombiro, G.; Sawai, T.; Noutoshi, Y.; Nishina, Y.; Matsui, H.; Yamamoto, M.; Toyoda, K.; Ichinose, Y. Specific Growth Inhibitors of Ralstonia Solanacearum, Xanthomonas oryzae Pv. Oryzae, X. campestris Pv. Campestris, and Clavibacter michiganensis Subsp. michiganensis. Microbiol. Res. 2018, 215, 29–35. [Google Scholar] [CrossRef]
- Yuliar; Nion, Y.A.; Toyota, K. Recent Trends in Control Methods for Bacterial Wilt Diseases Caused by Ralstonia solanacearum. Microbes Environ. 2015, 30, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, J.; Liu, B.; Zhu, Y.; Xiao, R.; Yang, W.; Ge, C.; Chen, Z. Biocontrol of Tomato Bacterial Wilt by the New Strain Bacillus velezensis FJAT-46737 and Its Lipopeptides. BMC Microbiol. 2020, 20, 160. [Google Scholar] [CrossRef] [PubMed]
- Boukaew, S.; Chuenchit, S.; Petcharat, V. Evaluation of Streptomyces spp. for Biological Control of Sclerotium Root and Stem Rot and Ralstonia Wilt of Chili Pepper. BioControl 2011, 56, 365–374. [Google Scholar] [CrossRef]
- Elsayed, T.R.; Jacquiod, S.; Nour, E.H.; Sørensen, S.J.; Smalla, K. Biocontrol of Bacterial Wilt Disease Through Complex Interaction between Tomato Plant, Antagonists, the Indigenous Rhizosphere Microbiota, and Ralstonia solanacearum. Front. Microbiol. 2020, 10, 2835. [Google Scholar] [CrossRef]
- Ling, L.; Han, X.; Li, X.; Zhang, X.; Wang, H.; Zhang, L.; Cao, P.; Wu, Y.; Wang, X.; Zhao, J.; et al. A Streptomyces Sp. NEAU-HV9: Isolation, Identification, and Potential as a Biocontrol Agent against Ralstonia solanacearum of Tomato Plants. Microorganisms 2020, 8, 351. [Google Scholar] [CrossRef]
- Raza, W.; Ling, N.; Yang, L.; Huang, Q.; Shen, Q. Response of Tomato Wilt Pathogen Ralstonia Solanacearum to the Volatile Organic Compounds Produced by a Biocontrol Strain Bacillus amyloliquefaciens SQR-9. Sci. Rep. 2016, 6, 24856. [Google Scholar] [CrossRef]
- Tan, H.M.; Cao, L.X.; He, Z.F.; Su, G.J.; Lin, B.; Zhou, S.N. Isolation of Endophytic Actinomycetes from Different Cultivars of Tomato and Their Activities against Ralstonia Solanacearum in Vitro. World J. Microbiol. Biotechnol. 2006, 22, 1275–1280. [Google Scholar] [CrossRef]
- Tan, H.; Zhou, S.; Deng, Z.; He, M.; Cao, L. Ribosomal-Sequence-Directed Selection for Endophytic Streptomycete Strains Antagonistic to Ralstonia solanacearum to Control Tomato Bacterial Wilt. Biol. Control 2011, 59, 245–254. [Google Scholar] [CrossRef]
- Zhao, J.; Han, L.; Yu, M.; Cao, P.; Li, D.; Guo, X.; Liu, Y.; Wang, X.; Xiang, W. Characterization of Streptomyces sporangiiformans sp. nov., a Novel Soil Actinomycete with Antibacterial Activity against Ralstonia solanacearum. Microorganisms 2019, 7, 360. [Google Scholar] [CrossRef]
- Zhuang, X.; Gao, C.; Peng, C.; Wang, Z.; Zhao, J.; Shen, Y.; Liu, C. Characterization of a Novel Endophytic Actinomycete, Streptomyces physcomitrii sp. nov., and Its Biocontrol Potential against Ralstonia solanacearum on Tomato. Microorganisms 2020, 8, 2025. [Google Scholar] [CrossRef]
- Velho-Pereira, S.; Kamat, N.M. Antimicrobial Screening of Actinobacteria Using a Modified Cross-Streak Method. Indian J. Pharm. Sci. 2011, 73, 223–228. [Google Scholar] [CrossRef]
- Agarwal, H.; Dowarah, B.; Baruah, P.M.; Bordoloi, K.S.; Krishnatreya, D.B.; Agarwala, N. Endophytes from Gnetum gnemon L. Can Protect Seedlings against the Infection of Phytopathogenic Bacterium Ralstonia solanacearum as Well as Promote Plant Growth in Tomato. Microbiol. Res. 2020, 238, 126503. [Google Scholar] [CrossRef]
- Kim, S.G.; Hur, O.-S.; Ro, N.-Y.; Ko, H.-C.; Rhee, J.-H.; Sung, J.S.; Ryu, K.-Y.; Lee, S.-Y.; Baek, H.J. Evaluation of Resistance to Ralstonia solanacearum in Tomato Genetic Resources at Seedling Stage. Plant Pathol. J. 2016, 32, 58–64. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Lee, I.; Kim, Y.O.; Park, S.C.; Chun, J. OrthoANI: An Improved Algorithm and Software for Calculating Average Nucleotide Identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Öker, M.G. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Abd-Elhalem, B.T.; El-Sawy, M.; Gamal, R.F.; Abou-Taleb, K.A. Production of Amylases from Bacillus Amyloliquefaciens under Submerged Fermentation Using Some Agro-Industrial by-Products. Ann. Agric. Sci. 2015, 60, 193–202. [Google Scholar] [CrossRef]
- Kasana, R.; Salwan, R.; Dhar, H.; Dutt, S.; Gulati, A. A Rapid and Easy Method for the Detection of Microbial Cellulases on Agar Plates Using Gram’s Iodine. Curr. Microbiol. 2008, 57, 503–507. [Google Scholar] [CrossRef]
- Haba, E.; Bresco, O.; Ferrer, C.; Marqués, A.; Busquets, M.; Manresa, A. Isolation of Lipase-Secreting Bacteria by Deploying Used Frying Oil as Selective Substrate. Enzym. Microb. Technol. 2000, 26, 40–44. [Google Scholar] [CrossRef]
- Masi, C.; Gemechu, G.; Tafesse, M. Isolation, Screening, Characterization, and Identification of Alkaline Protease-Producing Bacteria from Leather Industry Effluent. Ann. Microbiol. 2021, 71, 24. [Google Scholar] [CrossRef]
- Souza, C.; Burbano-Rosero, E.; Almeida, B.; Martins, G.; Albertini, L.; Rivera, I. Culture Medium for Isolating Chitinolytic Bacteria from Seawater and Plankton. World J. Microbiol. Biotechnol. 2009, 25, 2079–2082. [Google Scholar] [CrossRef]
- Subba Rao, N.S. Soil Microorganisms and Plant Growth; Oxford and IBH Publishing Co.: Delhi, India, 1977. [Google Scholar]
- Saravanan, V.S.; Subramoniam, S.R.; Raj, S.A. Assessing in Vitro Solubilization Potential of Different Zinc Solubilizing Bacterial (Zsb) Isolates. Braz. J. Microbiol. 2004, 35, 121–125. [Google Scholar] [CrossRef]
- Thampi, A.; Bhai, R.S. Rhizosphere Actinobacteria for Combating Phytophthora capsici and Sclerotium rolfsii, the Major Soil Borne Pathogens of Black Pepper (Piper nigrum L.). Biol. Control 2017, 109, 1–13. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Cappucino, J.C.; Sherman, N. Microbiology: A Laboratory Manual; Benjamin/Cumming Pub., Co.: New York, NY, USA, 1992. [Google Scholar]
- Weisse, T. Biodiversity of Freshwater Microorganisms—Achievements, Problems, and Perspectives. Pol. J. Ecol. 2006, 54, 633–652. [Google Scholar]
- Barreto, E.S.; Torres, A.R.; Barreto, M.R.; Vasconcelos, A.T.R.; Astolfi-Filho, S.; Hungria, M. Diversity in Antifungal Activity of Strains of Chromobacterium violaceum from the Brazilian Amazon. J. Ind. Microbiol. Biotechnol. 2008, 35, 783–790. [Google Scholar] [CrossRef]
- Farjalla, V.F. Are the Mixing Zones between Aquatic Ecosystems Hot Spots of Bacterial Production in the Amazon River System? Hydrobiologia 2014, 728, 153–165. [Google Scholar] [CrossRef]
- Toyama, D.; Santos-Júnior, C.D.; Kishi, L.T.; Oliveira, T.C.S.; Garcia, J.W.; Sarmento, H.; Miranda, F.P.; Henrique-Silva, F. A Snapshot on Prokaryotic Diversity of the Solimões River Basin (Amazon, Brazil). Genet. Mol. Res. 2017, 16, gmr16029567. [Google Scholar] [CrossRef]
- Doherty, M.; Yager, P.L.; Moran, M.A.; Coles, V.J.; Fortunato, C.S.; Krusche, A.V.; Medeiros, P.M.; Payet, J.P.; Richey, J.E.; Satinsky, B.M.; et al. Bacterial Biogeography across the Amazon River-Ocean Continuum. Front. Microbiol. 2017, 8, 882. [Google Scholar] [CrossRef]
- Santos-Júnior, C.D.; Sarmento, H.; Miranda, F.P.; Henrique-Silva, F.; Logares, R. Uncovering the Genomic Potential of the Amazon River Microbiome to Degrade Rainforest Organic Matter. Microbiome 2020, 8, 151. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.P.; Oliveira, T.R.; Gama, A.R.; Alves, K.R.; Santos, R.K.; Vieira, J.D.G.; Pires, D.J.; Carneiro, L.C. Phenotypic Plasticity and Characterization of Chromobacterium Isolates from Aquatic Environment. Res. Soc. Dev. 2022, 11, e28111536821. [Google Scholar] [CrossRef]
- Motta, A.S.; Cladera-Olivera, F.; Brandelli, A. Screening for Antimicrobial Activity among Bacteria Isolated from the Amazon Basin. Braz. J. Microbiol. 2004, 35, 307–310. [Google Scholar] [CrossRef]
- Oliveira-Longatti, S.M.; Marra, L.M.; Lima Soares, B.; Bomfeti, C.A.; Silva, K.; Avelar Ferreira, P.A.; Souza Moreira, F.M. Bacteria Isolated from Soils of the Western Amazon and from Rehabilitated Bauxite-Mining Areas Have Potential as Plant Growth Promoters. World J. Microbiol. Biotechnol. 2014, 30, 1239–1250. [Google Scholar] [CrossRef]
- Pereira, J.O.; Souza, A.Q.L.; Souza, A.D.L.; Castro França, S.; Oliveira, L.A. Overview on Biodiversity, Chemistry, and Biotechnological Potential of Microorganisms from the Brazilian Amazon. In Diversity and Benefits of Microorganisms from the Tropics; De Azevedo, J.L., Quecine, M.C., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 71–103. [Google Scholar] [CrossRef]
- Do Nascimento Monte, C.; Correa Saldanha, E.; Costa, I.; Shinaigger Rocha Do Nascimento, T.; Santos Pereira, M.; Farias Batista, L.; Costa Pinheiro, D. The Physical-Chemical Characteristics of Surface Waters in the Management of Quality in Clearwater Rivers in the Brazilian Amazon. Water Policy 2021, 23, 1303–1313. [Google Scholar] [CrossRef]
- Pinheiro, L.A.; Borges, J.T. Avaliação hidroquímica qualitativa das aguas do baixo Rio Negro. RUnPetro 2013, 1, 26–32. [Google Scholar]
- Silva, H.A.; Araújo, N.B.; Santos, N.L.; Bolson, A.; Gontijo, E.S.J.; Junior, E.S. Concentração Letal (CL50) e Potencial de Substâncias Húmicas Aquáticas Do Rio Negro (Brasil) Na Redução de Toxicidade Aguda de Íons Cu2+. Sci. Amazon. 2020, 9, 63–75. [Google Scholar]
- Queiroz, M.M.A.; Horbe, A.M.C.; Seyler, P.; Moura, C.A.V. Hidroquímica do rio Solimões na região entre Manacapuru e Alvarães: Amazonas-Brasil. Acta Amaz. 2009, 39, 943–952. [Google Scholar] [CrossRef]
- Feliatra, F.; Mardalisa, M.; Setiadi, J.; Lukistyowaty, I.; Hutasoit, A.Y. Potential of Secondary Metabolite from Marine Heterotrophic Bacteria against Pathogenic Bacteria in Aquaculture. J. Phys. Conf. Ser. 2020, 1655, 012044. [Google Scholar] [CrossRef]
- Setiaji, J.; Feliatra, F.; Teruna, H.Y.; Lukistyowati, I.; Suharman, I.; Muchlisin, Z.A.; Johan, T.I. Antibacterial Activity in Secondary Metabolite Extracts of Heterotrophic Bacteria against Vibrio alginolyticus, Aeromonas hydrophila, and Pseudomonas aeruginosa. F1000Research 2020, 9, 1491. [Google Scholar] [CrossRef] [PubMed]
- Viju, N.; Punitha, S.M.J.; Satheesh, S. In Vitro and In Silico Antifouling Activity Analysis of Secondary Metabolites Extracted from the Marine Bacterium Vibrio alginolyticus. Thalassas 2024, 40, 225–235. [Google Scholar] [CrossRef]
- Srinivas, V.; Gopalakrishnan, S.; Kamidi, J.P.; Chander, G. Effect of plant growth-promoting Streptomyces sp. on plant growth and yield of tomato and chilli. Andhra Pradesh J. Agril. Sci. 2020, 6, 65–70. [Google Scholar]
- Javed, Z.; Tripathi, G.D.; Mishra, M.; Dashora, K. Actinomycetes—The Microbial Machinery for the Organic-Cycling, Plant Growth, and Sustainable Soil Health. Biocatal. Agric. Biotechnol. 2021, 31, 101893. [Google Scholar] [CrossRef]
- Dede, A.; Güven, K.; Şahin, N. Isolation, Plant Growth-Promoting Traits, Antagonistic Effects on Clinical and Plant Pathogenic Organisms and Identification of Actinomycetes from Olive Rhizosphere. Microb. Pathog. 2020, 143, 104134. [Google Scholar] [CrossRef]
- Chaurasia, A.; Meena, B.R.; Tripathi, A.N.; Pandey, K.K.; Rai, A.B.; Singh, B. Actinomycetes: An Unexplored Microorganisms for Plant Growth Promotion and Biocontrol in Vegetable Crops. World J. Microbiol. Biotechnol. 2018, 34, 132. [Google Scholar] [CrossRef]
- Cunha-Ferreira, I.C.; Vizzotto, C.S.; Freitas, M.A.M.; Peixoto, J.; Carvalho, L.S.; Tótola, M.R.; Thompson, F.L.; Krüger, R.H. Genomic and Physiological Characterization of Kitasatospora sp. Nov., an Actinobacterium with Potential for Biotechnological Application Isolated from Cerrado Soil. Braz. J. Microbiol. 2024, 55, 1099–1115. [Google Scholar] [CrossRef]
- Li, C.; Pan, D.; Li, M.; Wang, Y.; Song, L.; Yu, D.; Zuo, Y.; Wang, K.; Liu, Y.; Wei, Z.; et al. Aerobactin-Mediated Iron Acquisition Enhances Biofilm Formation, Oxidative Stress Resistance, and Virulence of Yersinia pseudotuberculosis. Front. Microbiol. 2021, 12, 699913. [Google Scholar] [CrossRef]
- Tu, C.K.; Wang, P.H.; Lee, M.H. Endophytic Bacterium Lysobacter Firmicutimachus Strain 5-7 Is a Promising Biocontrol Agent against Rice Seedling Disease Caused by Pythium arrhenomanes in Nursery Trays. Plant Dis. 2023, 107, 1075–1086. [Google Scholar] [CrossRef]
- Haesler, F.; Hagn, A.; Frommberger, M.; Hertkorn, N.; Schmitt-Kopplin, P.; Munch, J.C.; Schloter, M. In Vitro Antagonism of an Actinobacterial Kitasatospora Isolate against the Plant Pathogen Phytophthora citricola as Elucidated with Ultrahigh Resolution Mass Spectrometry. J. Microbiol. Methods 2008, 75, 188–195. [Google Scholar] [CrossRef]
- Pithakkit, S.; Petcharat, V.; Chuenchit, S.; Pornsuriya, C.; Sunpapao, A. Isolation of Antagonistic Actinomycetes Species from Rhizosphere as Effective Biocontrol against Oil Palm Fungal Diseases. Agric. Technol. Biol. Sci. Walailak J. Sci. Tech. 2015, 12, 481–490. [Google Scholar]
- Shrivastava, S.; D’souza, S.F.; Desai, P.D. Production of Indole-3-Acetic Acid by Immobilized Actinomycete (Kitasatospora sp.) for Soil Applications. Curr. Sci. 2008, 94, 1595–1604. [Google Scholar]
- Takahashi, Y. Genus Kitasatospora, Taxonomic Features and Diversity of Secondary Metabolites. J. Antibiot. 2017, 70, 506–513. [Google Scholar] [CrossRef]
- Viaene, T.; Langendries, S.; Beirinckx, S.; Maes, M.; Goormachtig, S. Streptomyces as a Plant’s Best Friend? FEMS Microbiol. Ecol. 2016, 92, fiw119. [Google Scholar] [CrossRef]
- Singh, D.P.; Patil, H.J.; Prabha, R.; Yandigeri, M.S.; Prasad, S.R. Chapter 2—Actinomycetes as Potential Plant Growth-Promoting Microbial Communities. In Crop Improvement through Microbial Biotechnology; Prasad, R., Gill, S.S., Tuteja, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 27–38. [Google Scholar] [CrossRef]
- Prihatiningsih, N.; Asnani, A.; Djatmiko, H.A. Extracellular Protease from Bacillus Subtilis B315 with Antagonistic Activity against Bacterial Wilt Pathogen (Ralstonia solanacearum) of Chili. Biodiversitas 2021, 22, 1291–1295. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J.M. Siderophores in Environmental Research: Roles and Applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Mumtaz, M.Z.; Ahmad, M.; Jamil, M.; Hussain, T. Zinc Solubilizing Bacillus Spp. Potential Candidates for Biofortification in Maize. Microbiol. Res. 2017, 202, 51–60. [Google Scholar] [CrossRef]
- Yadav, D.K.; Devappa, V.; Kashyap, A.S.; Kumar, N.; Rana, V.S.; Sunita, K.; Singh, D. Boosting the Biocontrol Efficacy of Bacillus amyloliquefaciens DSBA-11 through Physical and Chemical Mutagens to Control Bacterial Wilt Disease of Tomato Caused by Ralstonia solanacearum. Microorganisms 2023, 11, 1790. [Google Scholar] [CrossRef]
- Singh, D.; Kumar Yadav, D. Potential of Bacillus amyloliquefaciens for Biocontrol of Bacterial Wilt of Tomato Incited by Ralstonia solanacearum. J. Plant Pathol. Microbiol. 2016, 7, 1000327. [Google Scholar] [CrossRef]
- Tan, S.; Gu, Y.; Yang, C.; Dong, Y.; Mei, X.; Shen, Q.; Xu, Y. Bacillus amyloliquefaciens T-5 May Prevent Ralstonia solanacearum Infection through Competitive Exclusion. Biol. Fertil. Soils 2016, 52, 341–351. [Google Scholar] [CrossRef]
- Kang, B.R.; Park, J.S.; Jung, W.-J. Antifungal Evaluation of Fengycin Isoforms Isolated from Bacillus amyloliquefaciens PPL against Fusarium oxysporum f. Sp. Lycopersici. Microb. Pathog. 2020, 149, 104509. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst Phytohormones from Planta and PGPR under Biotic and Abiotic Stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Khan, M.; Salman, M.; Ahmad Jan, S.; Khan Shinwari, Z. Biological Control of Fungal Phytopathogens: A Comprehensive Review Based on Bacillus Species. MOJ Biol. Med. 2021, 6, 90–92. [Google Scholar] [CrossRef]
- Khan, A.R.; Mustafa, A.; Hyder, S.; Valipour, M.; Rizvi, Z.F.; Gondal, A.S.; Yousuf, Z.; Iqbal, R.; Daraz, U. Bacillus spp. as Bioagents: Uses and Application for Sustainable Agriculture. Biology 2022, 11, 1763. [Google Scholar] [CrossRef] [PubMed]
- Mousa, M.A.A.; Abo-Elyousr, K.A.M.; Abdel Alal, A.M.K.; Alshareef, N.O. Management Fusarium Wilt Disease in Tomato by Combinations of Bacillus amyloliquefaciens and Peppermint Oil. Agronomy 2021, 11, 2536. [Google Scholar] [CrossRef]
- Gupta, R.S.; Patel, S.; Saini, N.; Chen, S. Robust Demarcation of 17 Distinct Bacillus Species Clades, Proposed as Novel Bacillaceae Genera, by Phylogenomics and Comparative Genomic Analyses: Description of Robertmurraya kyonggiensis sp. Nov. and Proposal for an Emended Genus Bacillus Limiting It Only to the Members of the Subtilis and Cereus Clades of Species. Int. J. Syst. Evol. Microbiol. 2020, 70, 5753–5798. [Google Scholar] [CrossRef]
- Akram, W.; Waqar, S.; Hanif, S.; Anjum, T.; Aftab, Z.-E.-H.; Li, G.; Ali, B.; Rizwana, H.; Hassan, A.; Rehman, A.; et al. Comparative Effect of Seed Coating and Biopriming of Bacillus aryabhattai Z-48 on Seedling Growth, Growth Promotion, and Suppression of Fusarium Wilt Disease of Tomato Plants. Microorganisms 2024, 12, 792. [Google Scholar] [CrossRef]
- Yanti, Y.; Hamid, H.; Reflin. Development of the PGPR and Cyanobacteria Consortium for Growth Promotion and Control Ralstonia syzigii subsp. indonesiensis of Tomato. IOP Conf. Ser. Earth Environ. Sci. 2021, 709, 012085. [Google Scholar] [CrossRef]
| Isolate | Size (pb) | Scaffolds | Type Species | NCBI Accession | Specie | ANI (%) | dDDH2 (%) |
|---|---|---|---|---|---|---|---|
| RN 11 | 5.262.007 | 45 | Priestia aryabhattai | NZ_CP024035 | Priestia aryabhattai | 98.61 | 88.3 |
| RN 24 | 8.364.889 | 366 | Streptomyces ardesiascus | BEWC01000001.1 | Streptomyces sp. nov. | 92.29 | 46.8 |
| SOL 195 | 9.091.611 | 397 | Kitasatospora aureofaciens | CP020567.1 | Kitasatospora sp. nov. | 86.36 | 31.1 |
| Assay | P. aryabhattai RN 11 | Streptomyces sp. RN 24 | Kitasatospora sp. SOL 195 |
|---|---|---|---|
| Siderophore (mm) | 12 ± 1.6 | 0 | 5 ± 0.8 |
| IAA (µg/mL) | 26.1 ± 2 | 42.1 ± 1.8 | 47.8 ± 2.3 |
| Ammonia | + | ++ | +++ |
| P (mm) | 0 | 0 | 0 |
| ZnO (mm) | 11 ± 1.5 | 0 | 0 |
| ZnSO4 (mm) | 15 ± 2 | 0 | 0 |
| Test | Height (cm) | Stem Diameter (cm) | Root (cm) | Branch (unid) | Leaf (cm) | ADW (g) | RDW (g) |
|---|---|---|---|---|---|---|---|
| NC | 51.96 ± 5.9 a | 0.40 a | 14.77 ± 2.1 a | 8 ± 1 b | 7.05 ± 0.17 a | 0.618 ± 0.06 a | 0.116 ± 0.02 c |
| PC | 42.64 ± 7.2 b | 0.20 c | 10.69 ± 1.7 c | 6 ± 1 c | 3.77 ± 0.22 c | 0.309 ± 0.02 c | 0.094 ± 0.01 d |
| RN 11 | 46.21 ± 3.1 b | 0.36 ± 0.05 a | 15.46 ± 2.8 a | 8 ± 1 b | 7.04 ± 0.12 a | 0.525 ± 0.03 b | 0.151 ± 0.01 b |
| RN 24 | 50.81 ± 3.2 a | 0.32 ± 0.07 b | 13.37 ± 1.9 b | 8 ± 1 b | 6.81 ± 0.35 b | 0.509 ± 0.02 b | 0.142 ± 0.01 b |
| SOL 195 | 44.85 ± 3.3 b | 0.25 ± 0.05 c | 13.96 ± 1.6 b | 7 ± 1 b | 3.74 ± 0.36 c | 0.332 ± 0.01 c | 0.116 ± 0.01 c |
| Test | Height (cm) | Stem Diameter (cm) | Root (cm) | Branch (unid) | Leaf (cm) | ADW (g) | RDW (g) |
|---|---|---|---|---|---|---|---|
| NC | 37.69 ± 3.1 b | 0.3 ± 0.09 b | 27 ± 4.3 a | 7 ± 1 b | 5.71 ± 0.4 a | 1.1 ± 0.28 b | 0.266 ± 0.08 a |
| PC | 34.19 ± 2.5 c | 0.22 ± 0.02 c | 19.73 ± 3.6 b | 7 ± 1 b | 4.77 ± 0.9 b | 0.94 ± 0.11 b | 0.126 ± 0.03 b |
| RN 11 | 42.04 ± 4.8 a | 0.38 ± 0.03 a | 29.04 ± 3.6 a | 8 ± 1 a | 5.77 ± 0.5 a | 1.32 ± 0.23 a | 0.26 ± 0.07 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fonseca, J.S.d.; Sousa, T.F.; Almeida, S.V.R.d.; Silva, C.N.; Castro, G.d.S.; Yamagishi, M.E.B.; Koolen, H.H.F.; Hanada, R.E.; Silva, G.F.d. Amazonian Bacteria from River Sediments as a Biocontrol Solution against Ralstonia solanacearum. Microorganisms 2024, 12, 1364. https://doi.org/10.3390/microorganisms12071364
Fonseca JSd, Sousa TF, Almeida SVRd, Silva CN, Castro GdS, Yamagishi MEB, Koolen HHF, Hanada RE, Silva GFd. Amazonian Bacteria from River Sediments as a Biocontrol Solution against Ralstonia solanacearum. Microorganisms. 2024; 12(7):1364. https://doi.org/10.3390/microorganisms12071364
Chicago/Turabian StyleFonseca, Jennifer Salgado da, Thiago Fernandes Sousa, Suene Vanessa Reis de Almeida, Carina Nascimento Silva, Gleucinei dos Santos Castro, Michel Eduardo Beleza Yamagishi, Hector Henrique Ferreira Koolen, Rogério Eiji Hanada, and Gilvan Ferreira da Silva. 2024. "Amazonian Bacteria from River Sediments as a Biocontrol Solution against Ralstonia solanacearum" Microorganisms 12, no. 7: 1364. https://doi.org/10.3390/microorganisms12071364
APA StyleFonseca, J. S. d., Sousa, T. F., Almeida, S. V. R. d., Silva, C. N., Castro, G. d. S., Yamagishi, M. E. B., Koolen, H. H. F., Hanada, R. E., & Silva, G. F. d. (2024). Amazonian Bacteria from River Sediments as a Biocontrol Solution against Ralstonia solanacearum. Microorganisms, 12(7), 1364. https://doi.org/10.3390/microorganisms12071364









