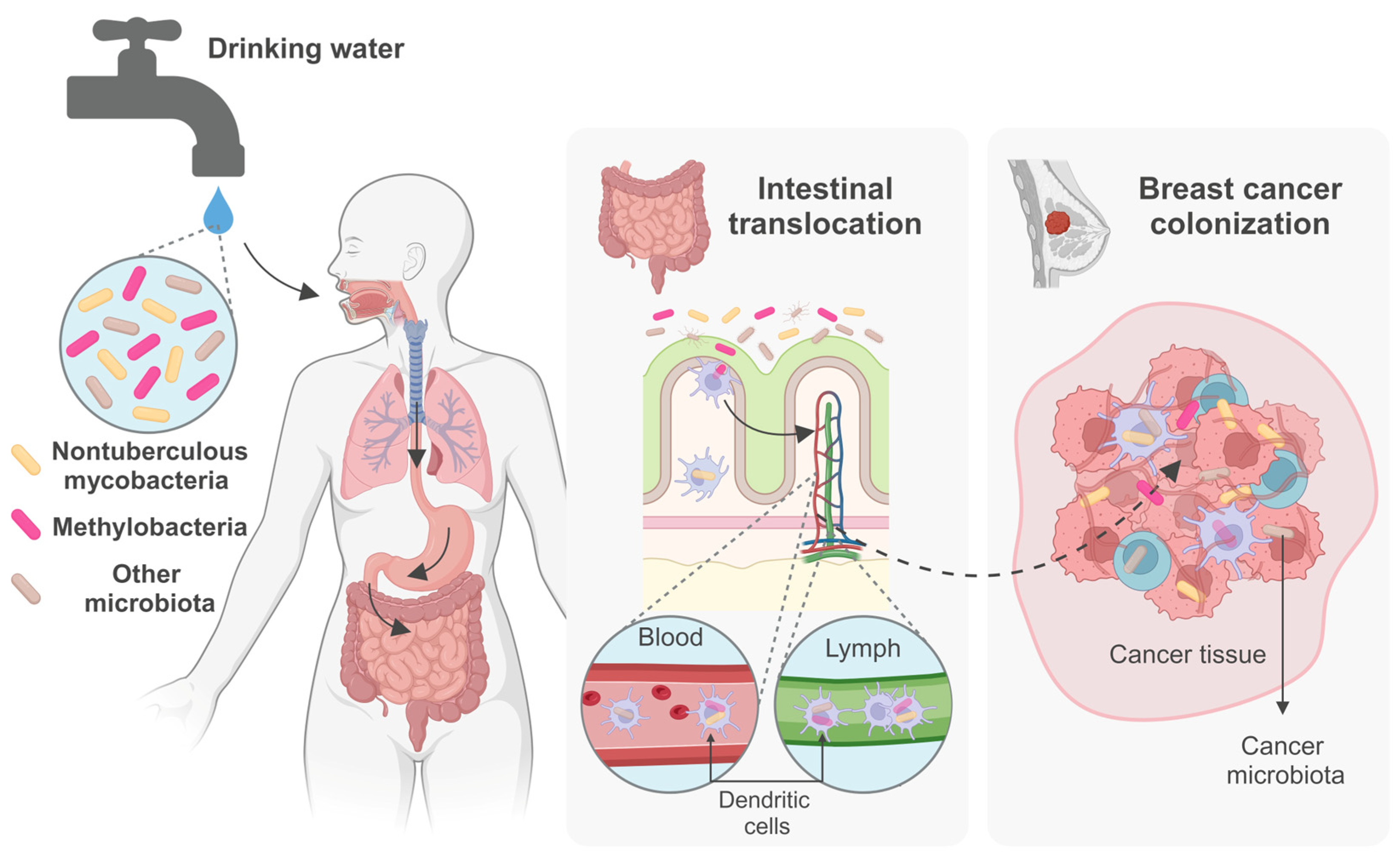
Drinking Water Microbiota, Entero-Mammary Pathways, and Breast Cancer: Focus on Nontuberculous Mycobacteria
Abstract
1. Microbiology of Drinking Water
2. Nontuberculous Mycobacteria: Environmentally Versatile Opportunistic Pathogens
3. Breast Cancer: Epidemiology, Biology, and Pathology
4. Bacteria and Cancer
5. Entero-Mammary Pathways and the Intratumoral Microbiome
6. Mycobacteria and Cancer: Focus on Breast Cancer
7. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- NRC. Indicators for Waterborne Pathogens; Committee on Indicators for Waterborne Pathogens, Board on Life Sciences; Water Science and Technology Board, Division on Earth and Life Studies, National Research Council (NRC): Washington, DC, USA, 2004. [Google Scholar]
- WHO. Guidelines for Drinking-Water Quality, 4th ed.; Incorporating the First and Second Addenda; WHO: Geneva, Switzerland, 2022. [Google Scholar]
- Manaia, C.M.; Aga, D.S.; Cytryn, E.; Gaze, W.H.; Graham, D.W.; Guo, J.; Leonard, A.F.C.; Li, L.; Murray, A.K.; Nunes, O.C.; et al. The Complex Interplay Between Antibiotic Resistance and Pharmaceutical and Personal Care Products in the Environment. Environ. Toxicol. Chem. 2004, 43, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Vanhaecke, T.; Bretin, O.; Poirel, M.; Tap, J. Drinking Water Source and Intake Are Associated with Distinct Gut Microbiota Signatures in US and UK Populations. J. Nutr. 2022, 152, 171–182. [Google Scholar] [CrossRef] [PubMed]
- IOM. Global Issues in Water, Sanitation, and Health: Workshop Summary; Institute of Medicine The National Academies Press: Washington, DC, USA, 2009; p. 328. [Google Scholar]
- Falkinham, J.O., III; Pruden, A.; Edwards, M. Opportunistic Premise Plumbing Pathogens: Increasingly Important Pathogens in Drinking Water. Pathogens 2015, 4, 373–386. [Google Scholar] [CrossRef]
- Abkar, L.; Moghaddam, H.S.; Fowler, S.J. Microbial ecology of drinking water from source to tap. Sci. Total Environ. 2023, 908, 168077. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Li, W.; Chen, J.; Zhou, Y.; Wei, Z.; Gong, L. Microbial diversity in full-scale water supply systems through sequencing technology: A review. RSC Adv. 2021, 11, 25484–25496. [Google Scholar] [CrossRef] [PubMed]
- Hammes, F.; Berney, M.; Wang, Y.; Vital, M.; Koster, O.; Egli, T. Flow-cytometric total bacterial cell counts as a descriptive microbiological parameter for drinking water treatment processes. Water Res. 2008, 42, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, Y.; van der Mark, E.; Magic-Knezev, A.; Pinto, A.; van den Bogert, B.; Liu, W.; van der Meer, W.; Medema, G. Assessing the origin of bacteria in tap water and distribution system in an unchlorinated drinking water system by SourceTracker using microbial community fingerprints. Water Res. 2018, 138, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Dinkla, I.J.T.; Muyzer, G. Microbial ecology of biofiltration used for producing safe drinking water. Appl. Microbiol. Biotechnol. 2022, 106, 4813–4829. [Google Scholar] [CrossRef] [PubMed]
- Pinto, A.J.; Xi, C.; Raskin, L. Bacterial community structure in the drinking water microbiome is governed by filtration processes. Environ. Sci. Technol. 2012, 46, 8851–8859. [Google Scholar] [CrossRef]
- Bruno, A.; Agostinetto, G.; Fumagalli, S.; Ghisleni, G.; Sandionigi, A. It’s a Long Way to the Tap: Microbiome and DNA-Based Omics at the Core of Drinking Water Quality. Int. J. Environ. Res. Public Health 2022, 19, 7940. [Google Scholar] [CrossRef]
- Thom, C.; Smith, C.J.; Moore, G.; Weir, P.; Ijaz, U.Z. Microbiomes in drinking water treatment and distribution: A meta-analysis from source to tap. Water Res. 2022, 212, 118106. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Sevillano-Rivera, M.C.; Calus, S.T.; Bautista-de Los Santos, Q.M.; Eren, A.M.; van der Wielen, P.; Ijaz, U.Z.; Pinto, A.J. Disinfection exhibits systematic impacts on the drinking water microbiome. Microbiome 2020, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Delafont, V.; Brouke, A.; Bouchon, D.; Moulin, L.; Hechard, Y. Microbiome of free-living amoebae isolated from drinking water. Water Res. 2013, 47, 6958–6965. [Google Scholar] [CrossRef] [PubMed]
- Ling, F.; Hwang, C.; LeChevallier, M.W.; Andersen, G.L.; Liu, W.T. Core-satellite populations and seasonality of water meter biofilms in a metropolitan drinking water distribution system. ISME J. 2016, 10, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Revetta, R.P.; Gomez-Alvarez, V.; Gerke, T.L.; Curioso, C.; Santo Domingo, J.W.; Ashbolt, N.J. Establishment and early succession of bacterial communities in monochloramine-treated drinking water biofilms. FEMS Microbiol. Ecol. 2013, 86, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Revetta, R.P.; Gomez-Alvarez, V.; Gerke, T.L.; Santo Domingo, J.W.; Ashbolt, N.J. Changes in bacterial composition of biofilm in a metropolitan drinking water distribution system. J. Appl. Microbiol. 2016, 121, 294–305. [Google Scholar] [CrossRef]
- Vaz-Moreira, I.; Nunes, O.C.; Manaia, C.M. Diversity and Antibiotic Resistance Patterns of Sphingomonadaceae Isolates from Drinking Water. Appl. Environ. Microbiol. 2011, 77, 5697–5706. [Google Scholar] [CrossRef]
- Ren, H.; Wang, W.; Liu, Y.; Liu, S.; Lou, L.; Cheng, D.; He, X.; Zhou, X.; Qiu, S.; Fu, L.; et al. Pyrosequencing analysis of bacterial communities in biofilms from different pipe materials in a city drinking water distribution system of East China. Appl. Microbiol. Biotechnol. 2015, 99, 10713–10724. [Google Scholar] [CrossRef]
- Aggarwal, S.; Gomez-Smith, C.K.; Jeon, Y.; LaPara, T.M.; Waak, M.B.; Hozalski, R.M. Effects of Chloramine and Coupon Material on Biofilm Abundance and Community Composition in Bench-Scale Simulated Water Distribution Systems and Comparison with Full-Scale Water Mains. Environ. Sci. Technol. 2018, 52, 13077–13088. [Google Scholar] [CrossRef]
- Waak, M.B.; Hozalski, R.M.; Hallé, C.; LaPara, T.M. Comparison of the microbiomes of two drinking water distribution systems—With and without residual chloramine disinfection. Microbiome 2019, 7, 87. [Google Scholar] [CrossRef]
- Bautista-de los Santos, Q.M.; Schroeder, J.L.; Sevillano-Rivera, M.C.; Sungthong, R.; Ijaz, U.Z.; Sloan, W.T.; Pinto, A.J. Emerging investigators series: Microbial communities in full-scale drinking water distribution systems—A meta-analysis. Environ. Sci. Water Res. Technol. 2016, 2, 631–644. [Google Scholar] [CrossRef]
- Chiao, T.-H.; Clancy, T.M.; Pinto, A.; Xi, C.; Raskin, L. Differential Resistance of Drinking Water Bacterial Populations to Monochloramine Disinfection. Environ. Sci. Technol. 2014, 48, 4038–4047. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Bédard, E.; Prévost, M.; Camper, A.K.; Hill, V.R.; Pruden, A. Methodological approaches for monitoring opportunistic pathogens in premise plumbing: A review. Water Res. 2017, 117, 68–86. [Google Scholar] [CrossRef] [PubMed]
- Feazel, L.M.; Baumgartner, L.K.; Peterson, K.L.; Frank, D.N.; Harris, J.K.; Pace, N.R. Opportunistic pathogens enriched in showerhead biofilms. Proc. Natl. Acad. Sci. USA 2009, 106, 16393–16399. [Google Scholar] [CrossRef] [PubMed]
- Hayward, C.; Ross, K.E.; Brown, M.H.; Bentham, R.; Whiley, H. The Presence of Opportunistic Premise Plumbing Pathogens in Residential Buildings: A Literature Review. Water 2022, 14, 1129. [Google Scholar] [CrossRef]
- De Sotto, R.; Tang, R.; Bae, S. Biofilms in premise plumbing systems as a double-edged sword: Microbial community composition and functional profiling of biofilms in a tropical region. J. Water Health 2020, 18, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Hayward, C.; Brown, M.H.; Whiley, H. Hospital water as the source of healthcare-associated infection and antimicrobial-resistant organisms. Curr. Opin. Infect. Dis. 2022, 35, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Logan-Jackson, A.R.; Batista, M.D.; Healy, W.; Ullah, T.; Whelton, A.J.; Bartrand, T.A.; Proctor, C. A Critical Review on the Factors that Influence Opportunistic Premise Plumbing Pathogens: From Building Entry to Fixtures in Residences. Environ. Sci. Technol. 2023, 57, 6360–6372. [Google Scholar] [CrossRef]
- Shen, Y.; Haig, S.-J.; Prussin, A.J., II; LiPuma, J.J.; Marr, L.C.; Raskin, L. Shower water contributes viable nontuberculous mycobacteria to indoor air. PNAS Nexus 2022, 1, pgac145. [Google Scholar] [CrossRef]
- Gebert, M.J.; Delgado-Baquerizo, M.; Oliverio, A.M.; Webster, T.M.; Nichols, L.M.; Honda, J.R.; Chan, E.D.; Adjemian, J.; Dunn, R.R.; Fierer, N. Ecological Analyses of Mycobacteria in Showerhead Biofilms and Their Relevance to Human Health. mBio 2018, 9, e01614-18. [Google Scholar] [CrossRef]
- Thomson, R.M.; Tolson, C.; Carter, R.; Coulter, C.; Huygens, F.; Hargreaves, M. Isolation of Nontuberculous Mycobacteria (NTM) from Household Water and Shower Aerosols in Patients with Pulmonary Disease Caused by NTM. J. Clin. Microbiol. 2013, 51, 3006–3011. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Pfyffer, G.E.; Parrish, N. Mycobacterium: General Characteristics, Laboratory Processing, Staining, Isolation and Detection Procedures. In Manual of Clinical Microbiology, 12th ed.; Carroll, K.C., Pfaller, M.A., Eds.; ASM Press: Washington, DC, USA, 2023. [Google Scholar]
- Etymologia: Mycobacterium. Emerg. Infect. Dis. 2008, 14, 377. [CrossRef]
- Fedrizzi, T.; Meehan, C.J.; Grottola, A.; Giacobazzi, E.; Fregni Serpini, G.; Tagliazucchi, S.; Fabio, A.; Bettua, C.; Bertorelli, R.; De Sanctis, V.; et al. Genomic characterization of Nontuberculous Mycobacteria. Sci. Rep. 2017, 7, 45258. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S.; Lo, B.; Son, J. Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 2018, 9, 67. [Google Scholar] [CrossRef]
- Meehan, C.J.; Barco, R.A.; Loh, Y.-H.E.; Cogneau, S.; Rigouts, L. Reconstituting the genus Mycobacterium. Int. J. Syst. Evol. Microbiol. 2021, 71, 004922. [Google Scholar] [CrossRef] [PubMed]
- Falkinham III, J.O. Nontuberculous mycobacteria in the environment. Tuberculosis 2022, 137, 102267. [Google Scholar] [CrossRef]
- Magee, J.G.; Ward, A.C. Mycobacterium. In Bergey’s Manual of Systematics of Archaea and Bacteria; John Wiley & Sons. Inc.: Hoboken, NJ, USA, 2015; pp. 1–84. [Google Scholar]
- Drapal, M.; Wheeler, P.R.; Fraser, P.D. Metabolite analysis of Mycobacterium species under aerobic and hypoxic conditions reveals common metabolic traits. Microbiology 2016, 162, 1456–1467. [Google Scholar] [CrossRef]
- Kalia, N.P.; Singh, S.; Hards, K.; Cheung, C.-Y.; Sviriaeva, E.; Banaei-Esfahani, A.; Aebersold, R.; Berney, M.; Cook, G.M.; Pethe, K. M. tuberculosis relies on trace oxygen to maintain energy homeostasis and survive in hypoxic environments. Cell Rep. 2023, 42, 112444. [Google Scholar] [CrossRef]
- Ashenafi, S.; Brighenti, S. Reinventing the human tuberculosis (TB) granuloma: Learning from the cancer field. Front. Immunol. 2022, 13, 1059725. [Google Scholar] [CrossRef]
- Shi, Y.; Queller, D.C.; Tian, Y.; Zhang, S.; Yan, Q.; He, Z.; He, Z.; Wu, C.; Wang, C.; Shu, L. The Ecology and Evolution of Amoeba-Bacterium Interactions. Appl. Environ. Microbiol. 2021, 87, e01866-20. [Google Scholar] [CrossRef]
- Pereira, A.C.; Ramos, B.; Reis, A.C.; Cunha, M.V. Non-Tuberculous Mycobacteria: Molecular and Physiological Bases of Virulence and Adaptation to Ecological Niches. Microorganisms 2020, 8, 1380. [Google Scholar] [CrossRef] [PubMed]
- Blanc, S.M.; Robinson, D.; Fahrenfeld, N.L. Potential for nontuberculous mycobacteria proliferation in natural and engineered water systems due to climate change: A literature review. City Environ. Interact. 2021, 11, 100070. [Google Scholar] [CrossRef]
- Brown-Elliott, B.A.; Philley, J.V. Rapidly Growing Mycobacteria. Microbiol. Spectr. 2017, 5, 703–723. [Google Scholar] [CrossRef] [PubMed]
- Holt, M.R.; Kasperbauer, S. Management of Extrapulmonary Nontuberculous Mycobacterial Infections. Semin. Respir. Crit. Care Med. 2018, 39, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Costa, D.; Alarico, S.; Dalcolmo, M.P.; Correia-Neves, M.; Empadinhas, N. The looming tide of nontuberculous mycobacterial infections in Portugal and Brazil. Tuberculosis 2016, 96, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Omori, K.; Kitagawa, H.; Yamaguchi, K.; Sakamoto, S.; Horimasu, Y.; Masuda, T.; Miyamoto, S.; Nakashima, T.; Iwamoto, H.; Fujitaka, K.; et al. Clinical characteristics of extrapulmonary nontuberculous mycobacteria infections in comparison with pulmonary infections: A single-center, retrospective study in Japan. J. Infect. Chemother. 2023, 29, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Grigg, C.; Jackson, K.A.; Barter, D.; Czaja, C.A.; Johnston, H.; Lynfield, R.; Vagnone, P.S.; Tourdot, L.; Spina, N.; Dumyati, G.; et al. Epidemiology of Pulmonary and Extrapulmonary Nontuberculous Mycobacteria Infections at 4 US Emerging Infections Program Sites: A 6-Month Pilot. Clin. Infect. Dis. 2023, 77, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, P.M.; Hedberg, K.; Saulson, A.; McNelly, E.; Winthrop, K.L. Nontuberculous Mycobacterial Disease Prevalence and Risk Factors: A Changing Epidemiology. Clin. Infect. Dis. 2009, 49, e124–e129. [Google Scholar] [CrossRef] [PubMed]
- Prevots, D.R.; Marshall, J.E.; Wagner, D.; Morimoto, K. Global Epidemiology of Nontuberculous Mycobacterial Pulmonary Disease: A Review. Clin. Chest Med. 2023, 44, 675–721. [Google Scholar] [CrossRef]
- Ricotta, E.; Adjemian, J.; Blakney, R.; Lai, Y.L.; Kadri, S.; Prevots, D.R. Extrapulmonary Nontuberculous Mycobacteria Infections in Hospitalized Patients, United States, 2009–2014. Emerg. Infect. Dis. 2021, 27, 845. [Google Scholar] [CrossRef]
- Griffith, D.E.; Aksamit, T.R. Diagnostic Criteria and the Decision to Treat Nontuberculous Mycobacterial Pulmonary Disease. Clin. Chest Med. 2023, 44, 757–769. [Google Scholar] [CrossRef]
- Sawka, A.; Burke, A. Medications and Monitoring in Treatment of Nontuberculous Mycobacterial Pulmonary Disease. Clin. Chest Med. 2023, 44, 815–828. [Google Scholar] [CrossRef]
- Gleeson, L.E.; Waterer, G. Beyond antibiotics: Recent developments in the diagnosis and management of nontuberculous mycobacterial infection. Breathe 2022, 18, 210171. [Google Scholar] [CrossRef]
- Alffenaar, J.-W.; Märtson, A.-G.; Heysell, S.K.; Cho, J.-G.; Patanwala, A.; Burch, G.; Kim, H.Y.; Sturkenboom, M.G.G.; Byrne, A.; Marriott, D.; et al. Therapeutic Drug Monitoring in Non-Tuberculosis Mycobacteria Infections. Clin. Pharmacokinet. 2021, 60, 711–725. [Google Scholar] [CrossRef]
- Honda, J.R.; Virdi, R.; Chan, E.D. Global Environmental Nontuberculous Mycobacteria and Their Contemporaneous Man-Made and Natural Niches. Front. Microbiol. 2018, 9, 2029. [Google Scholar] [CrossRef]
- Chan, E.D.; Iseman, M.D. Slender, Older Women Appear to Be More Susceptible to Nontuberculous Mycobacterial Lung Disease. Gend. Med. 2010, 7, 5–18. [Google Scholar] [CrossRef] [PubMed]
- Park, S.C.; Kang, M.J.; Han, C.H.; Lee, S.M.; Kim, C.J.; Lee, J.M.; Kang, Y.A. Prevalence, incidence, and mortality of nontuberculous mycobacterial infection in Korea: A nationwide population-based study. BMC Pulm. Med. 2019, 19, 140. [Google Scholar] [CrossRef] [PubMed]
- Weathered, C.; Wei, N.; Pienaar, E. Reduced macrophage killing of M. avium drives infection risk in post-menopausal patients. Tuberculosis 2023, 139, 102304. [Google Scholar] [CrossRef] [PubMed]
- Namkoong, H.; Holland, S.M. Host Susceptibility to Nontuberculous Mycobacterial Pulmonary Disease. Clin. Chest Med. 2023, 44, 723–730. [Google Scholar] [CrossRef]
- Namkoong, H.; Omae, Y.; Asakura, T.; Ishii, M.; Suzuki, S.; Morimoto, K.; Kawai, Y.; Emoto, K.; Oler, A.J.; Szymanski, E.P.; et al. Genome-wide association study in patients with pulmonary Mycobacterium avium complex disease. Eur. Respir. J. 2021, 58, 1902269. [Google Scholar] [CrossRef]
- van Ingen, J.; Aksamit, T.; Andrejak, C.; Böttger, E.C.; Cambau, E.; Daley, C.L.; Griffith, D.E.; Guglielmetti, L.; Holland, S.M.; Huitt, G.A.; et al. Treatment outcome definitions in nontuberculous mycobacterial pulmonary disease: An NTM-NET consensus statement. Eur. Respir. J. 2018, 51, 1800170. [Google Scholar] [CrossRef]
- Winthrop, K.L.; Marras, T.K.; Adjemian, J.; Zhang, H.; Wang, P.; Zhang, Q. Incidence and Prevalence of Nontuberculous Mycobacterial Lung Disease in a Large U.S. Managed Care Health Plan, 2008–2015. Ann. Am. Thorac. Soc. 2020, 17, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Schildkraut, J.A.; Gallagher, J.; Morimoto, K.; Lange, C.; Haworth, C.; Floto, R.A.; Hoefsloot, W.; Griffith, D.E.; Wagner, D.; Ingen, J.v. Epidemiology of nontuberculous mycobacterial pulmonary disease in Europe and Japan by Delphi estimation. Respir. Med. 2020, 173, 106164. [Google Scholar] [CrossRef]
- Zhou, Y.; Mu, W.; Zhang, J.; Wen, S.W.; Pakhale, S. Global prevalence of non-tuberculous mycobacteria in adults with non-cystic fibrosis bronchiectasis 2006–2021: A systematic review and meta-analysis. BMJ Open 2022, 12, e055672. [Google Scholar] [CrossRef]
- De Groote, M.A.; Pace, N.R.; Fulton, K.; Falkinham III, J.O. Relationships between Mycobacterium Isolates from Patients with Pulmonary Mycobacterial Infection and Potting Soils. Appl. Environ. Microbiol. 2006, 72, 7602–7606. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, K.; Ito, Y.; Hirai, T.; Kubo, T.; Imai, S.; Tatsumi, S.; Fujita, K.; Takakura, S.; Niimi, A.; Iinuma, Y.; et al. Environmental Risk Factors for Pulmonary Mycobacterium avium-intracellulare Complex Disease. Chest 2011, 140, 723–729. [Google Scholar] [CrossRef]
- Honda, J.R. Environmental Sources and Transmission of Nontuberculous Mycobacteria. Clin. Chest Med. 2023, 44, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Nishiuchi, Y.; Maekura, R.; Kitada, S.; Tamaru, A.; Taguri, T.; Kira, Y.; Hiraga, T.; Hirotani, A.; Yoshimura, K.; Miki, M.; et al. The Recovery of Mycobacterium avium-intracellulare Complex (MAC) from the Residential Bathrooms of Patients with Pulmonary MAC. Clin. Infect. Dis. 2007, 45, 347–351. [Google Scholar] [CrossRef] [PubMed]
- Tzou, C.L.; Dirac, M.A.; Becker, A.L.; Beck, N.K.; Weigel, K.M.; Meschke, J.S.; Cangelosi, G.A. Association between Mycobacterium avium Complex Pulmonary Disease and Mycobacteria in Home Water and Soil. Ann. Am. Thorac. Soc. 2020, 17, 57–62. [Google Scholar] [CrossRef]
- Lande, L.; Alexander, D.C.; Wallace, R.J.; Kwait, R.; Iakhiaeva, E.; Williams, M.; Cameron, A.D.S.; Olshefsky, S.; Devon, R.; Vasireddy, R.; et al. Mycobacterium avium in Community and Household Water, Suburban Philadelphia, Pennsylvania, USA, 2010–2012. Emerg. Infect. Dis. 2019, 25, 473–481. [Google Scholar] [CrossRef]
- Griffin, I.; Schmitz, A.; Oliver, C.; Pritchard, S.; Zhang, G.; Rico, E.; Davenport, E.; Llau, A.; Moore, E.; Fernandez, D.; et al. Outbreak of Tattoo-associated Nontuberculous Mycobacterial Skin Infections. Clin. Infect. Dis. 2019, 69, 949–955. [Google Scholar] [CrossRef] [PubMed]
- CDC. Tattoo-associated nontuberculous mycobacterial skin infections--multiple states, 2011–2012. MMWR Morb. Mortal. Wkly. Rep. 2012, 61, 653–656. [Google Scholar]
- Fjällbrant, H.; Akerstrom, M.; Svensson, E.; Andersson, E. Hot tub lung: An occupational hazard. Eur. Respir. Rev. 2013, 22, 88–90. [Google Scholar] [CrossRef] [PubMed]
- Prevots, D.R.; Adjemian, J.; Fernandez, A.G.; Knowles, M.R.; Olivier, K.N. Environmental Risks for Nontuberculous Mycobacteria. Individual Exposures and Climatic Factors in the Cystic Fibrosis Population. Ann. Am. Thorac. Soc. 2014, 11, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Kwak, S.H.; Yong, S.H.; Lee, S.H.; Leem, A.Y.; Kim, S.Y.; Lee, S.H.; Chung, K.; Kim, E.Y.; Jung, J.Y.; et al. The Association between Behavioral Risk Factors and Nontuberculous Mycobacterial Pulmonary Disease. Yonsei Med. J. 2021, 62, 702–707. [Google Scholar] [CrossRef] [PubMed]
- Lyman, M.M.; Grigg, C.; Kinsey, C.B.; Keckler, M.S.; Moulton-Meissner, H.; Cooper, E.; Soe, M.M.; Noble-Wang, J.; Longenberger, A.; Walker, S.R.; et al. Invasive Nontuberculous Mycobacterial Infections among Cardiothoracic Surgical Patients Exposed to Heater-Cooler Devices. Emerg. Infect. Dis. 2017, 23, 796–805. [Google Scholar] [CrossRef] [PubMed]
- Peralta, G.; Tobin-D’Angelo, M.; Parham, A.; Edison, L.; Lorentzson, L.; Smith, C.; Drenzek, C. Notes from the Field: Mycobacterium abscessus Infections Among Patients of a Pediatric Dentistry Practice--Georgia, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 355–356. [Google Scholar] [CrossRef] [PubMed]
- Padoveze, M.C.; Fortaleza, C.M.C.B.; Freire, M.P.; Assis, D.B.d.; Madalosso, G.; Pellini, A.C.G.; César, M.L.V.; Neto, V.P.; Beltramelli, M.M.; Chimara, E.; et al. Outbreak of surgical infection caused by non-tuberculous mycobacteria in breast implants in Brazil. J. Hosp. Infect. 2007, 67, 161–167. [Google Scholar] [CrossRef]
- Daniau, C.; Lecorche, E.; Mougari, F.; Benmansour, H.; Bernet, C.; Blanchard, H.; Robert, J.; Berger-Carbonne, A.; Cambau, E. Association of Healthcare and Aesthetic Procedures with Infections Caused by Nontuberculous Mycobacteria, France, 2012–2020. Emerg. Infect. Dis. 2022, 28, 518–526. [Google Scholar] [CrossRef]
- Shapiro, K.; Cross, S.J.; Morton, T.H.; Inaba, H.; Holland, A.; Fasipe, F.R.; Adderson, E.E. Healthcare-Associated Infections Caused by Mycolicibacterium neoaurum. Emerg. Infect. Dis. 2023, 29, 1516–1523. [Google Scholar] [CrossRef]
- van der Wielen, P.W.J.J.; van der Kooij, D. Nontuberculous Mycobacteria, Fungi, and Opportunistic Pathogens in Unchlorinated Drinking Water in the Netherlands. Appl. Environ. Microbiol. 2013, 79, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.M.; Carter, R.; Tolson, C.; Coulter, C.; Huygens, F.; Hargreaves, M. Factors associated with the isolation of Nontuberculous mycobacteria (NTM) from a large municipal water system in Brisbane, Australia. BMC Microbiol. 2013, 13, 89. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.M.; Furuya-Kanamori, L.; Coffey, C.; Bell, S.C.; Knibbs, L.D.; Lau, C.L. Influence of climate variables on the rising incidence of nontuberculous mycobacterial (NTM) infections in Queensland, Australia 2001–2016. Sci. Total Environ. 2020, 740, 139796. [Google Scholar] [CrossRef] [PubMed]
- Adjemian, J.; Olivier, K.N.; Seitz, A.E.; Falkinham III, J.O.; Holland, S.M.; Prevots, D.R. Spatial Clusters of Nontuberculous Mycobacterial Lung Disease in the United States. Am. J. Respir. Crit. Care Med. 2012, 186, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Lipner, E.M.; Crooks, J.L.; French, J.; Strong, M.; Nick, J.A.; Prevots, D.R. Nontuberculous mycobacterial infection and environmental molybdenum in persons with cystic fibrosis: A case–control study in Colorado. J. Expo. Sci. Environ. Epidemiol. 2022, 32, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Lipner, E.M.; French, J.P.; Falkinham III, J.O.; Crooks, J.L.; Mercaldo, R.A.; Henkle, E.; Prevots, D.R. Nontuberculous Mycobacteria Infection Risk and Trace Metals in Surface Water: A Population-based Ecologic Epidemiologic Study in Oregon. Ann. Am. Thorac. Soc. 2022, 19, 543–550. [Google Scholar] [CrossRef] [PubMed]
- DeFlorio-Barker, S.; Egorov, A.; Smith, G.S.; Murphy, M.S.; Stout, J.E.; Ghio, A.J.; Hudgens, E.E.; Messier, K.P.; Maillard, J.-M.; Hilborn, E.D. Environmental risk factors associated with pulmonary isolation of nontuberculous mycobacteria, a population-based study in the southeastern United States. Sci. Total Environ. 2021, 763, 144552. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Ponnuswamy, A.; Capstick, T.G.D.; Chen, C.; McCabe, D.; Hurst, R.; Morrison, L.; Moore, F.; Gallardo, M.; Keane, J.; et al. Non-tuberculous mycobacterial pulmonary disease (NTM-PD): Epidemiology, diagnosis and multidisciplinary management. Clin. Med. 2024, 24, 100017. [Google Scholar] [CrossRef] [PubMed]
- Falkinham III, J.O.; Norton, C.D.; LeChevallier, M.W. Factors Influencing Numbers of Mycobacterium avium, Mycobacterium intracellulare, and Other Mycobacteria in Drinking Water Distribution Systems. Appl. Environ. Microbiol. 2001, 67, 1225–1231. [Google Scholar] [CrossRef]
- Donohue, M.J.; Mistry, J.H.; Donohue, J.M.; O’Connell, K.; King, D.; Byran, J.; Covert, T.; Pfaller, S. Increased Frequency of Nontuberculous Mycobacteria Detection at Potable Water Taps within the United States. Environ. Sci. Technol. 2015, 49, 6127–6133. [Google Scholar] [CrossRef]
- Torvinen, E.; Suomalainen, S.; Lehtola, M.J.; Miettinen, I.T.; Zacheus, O.; Paulin, L.; Katila, M.-L.; Martikainen, P.J. Mycobacteria in Water and Loose Deposits of Drinking Water Distribution Systems in Finland. Appl. Environ. Microbiol. 2004, 70, 1973–1981. [Google Scholar] [CrossRef] [PubMed]
- Dantec, C.L.; Duguet, J.-P.; Montiel, A.; Dumoutier, N.; Dubrou, S.; Vincent, V. Occurrence of Mycobacteria in Water Treatment Lines and in Water Distribution Systems. Appl. Environ. Microbiol. 2002, 68, 5318–5325. [Google Scholar] [CrossRef] [PubMed]
- Loret, J.-F.; Dumoutier, N. Non-tuberculous mycobacteria in drinking water systems: A review of prevalence data and control means. Int. J. Hyg. Environ. Health 2019, 222, 628–634. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Liu, T.; Men, Y.; Li, S.; Graham, N.; Yu, W. Understanding point-of-use tap water quality: From instrument measurement to intelligent analysis using sample filtration. Water Res. 2022, 225, 119205. [Google Scholar] [CrossRef] [PubMed]
- Norton, G.J.; Williams, M.; Falkinham III, J.O.; Honda, J.R. Physical Measures to Reduce Exposure to Tap Water–Associated Nontuberculous Mycobacteria. Front. Public Health 2020, 8, 190. [Google Scholar] [CrossRef]
- Säve-Söderbergh, M.; Toljander, J.; Mattisson, I.; Åkesson, A.; Simonsson, M. Drinking water consumption patterns among adults—SMS as a novel tool for collection of repeated self-reported water consumption. J. Expo. Sci. Environ. Epidemiol. 2018, 28, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Rosinger, A.Y.; Herrick, K.A.; Wutich, A.Y.; Yoder, J.S.; Ogden, C.L. Disparities in plain, tap and bottled water consumption among US adults: National Health and Nutrition Examination Survey (NHANES) 2007–2014. Public Health Nutr. 2018, 21, 1455–1464. [Google Scholar] [CrossRef] [PubMed]
- Chongwe, G.; Michelo, C.; Kelly, P. Diagnostic yield of nontuberculous mycobacteria in patients booked for endoscopy at the University Teaching Hospital, Lusaka. BMC Res. Notes 2017, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Falony, G.; Joossens, M.; Vieira-Silva, S.; Wang, J.; Darzi, Y.; Faust, K.; Kurilshikov, A.; Bonder, M.J.; Valles-Colomer, M.; Vandeputte, D.; et al. Population-level analysis of gut microbiome variation. Science 2016, 352, 560–564. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Luengo-Fernandez, R.; Leal, J.; Gray, A.; Sullivan, R. Economic burden of cancer across the European Union: A population-based cost analysis. Lancet Oncol. 2013, 14, 1165–1174. [Google Scholar] [CrossRef] [PubMed]
- Allemani, C.; Weir, H.K.; Carreira, H.; Harewood, R.; Spika, D.; Wang, X.-S.; Bannon, F.; Ahn, J.V.; Johnson, C.J.; Bonaventure, A.; et al. Global surveillance of cancer survival 1995–2009: Analysis of individual data for 25 676 887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet 2015, 385, 977–1010. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, O.; Bray, F.; Coleman, M.P.; Vanderpuye, V.; Eniu, A.; Kotha, S.R.; Sarker, M.; Huong, T.T.; Allemani, C.; Dvaladze, A.; et al. The global burden of women’s cancers: A grand challenge in global health. Lancet 2017, 389, 847–860. [Google Scholar] [CrossRef]
- Chen, S.; Parmigiani, G. Meta-analysis of BRCA1 and BRCA2 penetrance. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2007, 25, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Nur, U.; El Reda, D.; Hashim, D.; Weiderpass, E. A prospective investigation of oral contraceptive use and breast cancer mortality: Findings from the Swedish women’s lifestyle and health cohort. BMC Cancer 2019, 19, 807. [Google Scholar] [CrossRef] [PubMed]
- Mørch, L.S.; Skovlund, C.W.; Hannaford, P.C.; Iversen, L.; Fielding, S.; Lidegaard, Ø. Contemporary Hormonal Contraception and the Risk of Breast Cancer. N. Engl. J. Med. 2017, 377, 2228–2239. [Google Scholar] [CrossRef] [PubMed]
- CGHFBC. Collaborative Group on Hormonal Factors in Breast Cancer. Type and timing of menopausal hormone therapy and breast cancer risk: Individual participant meta-analysis of the worldwide epidemiological evidence. Lancet 2019, 394, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Piccart-Gebhart, M.J.; Procter, M.; Leyland-Jones, B.; Goldhirsch, A.; Untch, M.; Smith, I.; Gianni, L.; Baselga, J.; Bell, R.; Jackisch, C.; et al. Trastuzumab after Adjuvant Chemotherapy in HER2-Positive Breast Cancer. N. Engl. J. Med. 2005, 353, 1659–1672. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef]
- Pohl-Rescigno, E.; Hauke, J.; Loibl, S.; Möbus, V.; Denkert, C.; Fasching, P.A.; Kayali, M.; Ernst, C.; Weber-Lassalle, N.; Hanusch, C.; et al. Association of Germline Variant Status With Therapy Response in High-risk Early-Stage Breast Cancer: A Secondary Analysis of the GeparOcto Randomized Clinical Trial. JAMA Oncol. 2020, 6, 744–748. [Google Scholar] [CrossRef]
- Luen, S.; Virassamy, B.; Savas, P.; Salgado, R.; Loi, S. The genomic landscape of breast cancer and its interaction with host immunity. Breast 2016, 29, 241–250. [Google Scholar] [CrossRef]
- Ma, T.; Tang, Y.; Wang, T.; Yang, Y.; Zhang, Y.; Wang, R.; Zhang, Y.; Li, Y.; Wu, M.; Tang, M.; et al. Chronic pulmonary bacterial infection facilitates breast cancer lung metastasis by recruiting tumor-promoting MHCIIhi neutrophils. Signal Transduct. Target. Ther. 2023, 8, 296. [Google Scholar] [CrossRef]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef]
- Philley, J.V.; Kannan, A.; Griffith, D.E.; Devine, M.S.; Benwill, J.L.; Wallace, R.J.; Brown-Elliott, B.A.; Thakkar, F.; Taskar, V.S.; Fox, J.G.; et al. Exosome secretome and mediated signaling in breast cancer patients with nontuberculous mycobacterial disease. Oncotarget 2017, 8, 18070–18081. [Google Scholar] [CrossRef] [PubMed]
- Butlin, H.T. Malignant Tumours and Parasitism. Br. Med. J. 1884, 1, 45–46. [Google Scholar] [CrossRef]
- Møller, H.; Heseltine, E.; Vainio, H. Working group report on schistosomes, liver flukes and Helicobacter pylori. Meeting held at IARC, LYON, 7–14 june 1994. Int. J. Cancer 1995, 60, 587–589. [Google Scholar] [CrossRef] [PubMed]
- El Tekle, G.; Garrett, W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 2023, 23, 600–618. [Google Scholar] [CrossRef] [PubMed]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552. [Google Scholar] [CrossRef]
- Laliani, G.; Ghasemian Sorboni, S.; Lari, R.; Yaghoubi, A.; Soleimanpour, S.; Khazaei, M.; Hasanian, S.M.; Avan, A. Bacteria and cancer: Different sides of the same coin. Life Sci. 2020, 246, 117398. [Google Scholar] [CrossRef]
- Song, X.; Wei, C.; Li, X. The Relationship Between Microbial Community and Breast Cancer. Front. Cell. Infect. Microbiol. 2022, 12, 849022. [Google Scholar] [CrossRef] [PubMed]
- Viswanathan, S.; Parida, S.; Lingipilli, B.T.; Krishnan, R.; Podipireddy, D.R.; Muniraj, N. Role of Gut Microbiota in Breast Cancer and Drug Resistance. Pathogens 2023, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Vitorino, M.; Baptista de Almeida, S.; Alpuim Costa, D.; Faria, A.; Calhau, C.; Azambuja Braga, S. Human Microbiota and Immunotherapy in Breast Cancer—A Review of Recent Developments. Front. Oncol. 2022, 11, 815772. [Google Scholar] [CrossRef] [PubMed]
- Parida, S.; Sharma, D. The Microbiome–Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef] [PubMed]
- Alpuim Costa, D.; Nobre, J.G.; Batista, M.V.; Ribeiro, C.; Calle, C.; Cortes, A.; Marhold, M.; Negreiros, I.; Borralho, P.; Brito, M.; et al. Human Microbiota and Breast Cancer—Is There Any Relevant Link?—A Literature Review and New Horizons Toward Personalised Medicine. Front. Microbiol. 2021, 12, 584332. [Google Scholar] [CrossRef] [PubMed]
- Cren, P.-Y.; Bertrand, N.; Le Deley, M.-C.; Génin, M.; Mortier, L.; Odou, P.; Penel, N.; Chazard, E. Is the survival of patients treated with ipilimumab affected by antibiotics? An analysis of 1585 patients from the French National hospital discharge summary database (PMSI). OncoImmunology 2020, 9, 1846914. [Google Scholar] [CrossRef]
- Geller, L.T.; Straussman, R. Intratumoral bacteria may elicit chemoresistance by metabolizing anticancer agents. Mol. Cell. Oncol. 2018, 5, e1405139. [Google Scholar] [CrossRef] [PubMed]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- Zheng, D.-W.; Dong, X.; Pan, P.; Chen, K.-W.; Fan, J.-X.; Cheng, S.-X.; Zhang, X.-Z. Phage-guided modulation of the gut microbiota of mouse models of colorectal cancer augments their responses to chemotherapy. Nat. Biomed. Eng. 2019, 3, 717–728. [Google Scholar] [CrossRef]
- Routy, B.; Lenehan, J.G.; Miller, W.H.; Jamal, R.; Messaoudene, M.; Daisley, B.A.; Hes, C.; Al, K.F.; Martinez-Gili, L.; Punčochář, M.; et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: A phase I trial. Nat. Med. 2023, 29, 2121–2132. [Google Scholar] [CrossRef]
- Mohseni, A.H.; Taghinezhad-S, S.; Casolaro, V.; Lv, Z.; Li, D. Potential links between the microbiota and T cell immunity determine the tumor cell fate. Cell Death Dis. 2023, 14, 154. [Google Scholar] [CrossRef] [PubMed]
- Galván-Peña, S.; Zhu, Y.; Hanna, B.S.; Mathis, D.; Benoist, C. A dynamic atlas of immunocyte migration from the gut. Sci. Immunol. 2024, 9, eadi0672. [Google Scholar] [CrossRef] [PubMed]
- Rescigno, M.; Urbano, M.; Valzasina, B.; Francolini, M.; Rotta, G.; Bonasio, R.; Granucci, F.; Kraehenbuhl, J.-P.; Ricciardi-Castagnoli, P. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2001, 2, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type–specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Schorr, L.; Mathies, M.; Elinav, E.; Puschhof, J. Intracellular bacteria in cancer—Prospects and debates. NPJ Biofilms Microbiomes 2023, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372.e26. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Hu, Y. Microbiome harbored within tumors: A new chance to revisit our understanding of cancer pathogenesis and treatment. Signal Transduct. Target. Ther. 2020, 5, 136. [Google Scholar] [CrossRef]
- Donnet-Hughes, A.; Perez, P.F.; Doré, J.; Leclerc, M.; Levenez, F.; Benyacoub, J.; Serrant, P.; Segura-Roggero, I.; Schiffrin, E.J. Potential role of the intestinal microbiota of the mother in neonatal immune education. Proc. Nutr. Soc. 2010, 69, 407–415. [Google Scholar] [CrossRef]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef]
- Hoskinson, C.; Jiang, R.Y.; Stiemsma, L.T. Elucidating the roles of the mammary and gut microbiomes in breast cancer development. Front. Oncol. 2023, 13, 1198259. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of Human Breast Tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [PubMed]
- Hunt, K.M.; Foster, J.A.; Forney, L.J.; Schütte, U.M.E.; Beck, D.L.; Abdo, Z.; Fox, L.K.; Williams, J.E.; McGuire, M.K.; McGuire, M.A. Characterization of the Diversity and Temporal Stability of Bacterial Communities in Human Milk. PLoS ONE 2011, 6, e21313. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M.; Fernández, L.; Verhasselt, V. The Gut–Breast Axis: Programming Health for Life. Nutrients 2021, 13, 606. [Google Scholar] [CrossRef] [PubMed]
- Jost, T.; Lacroix, C.; Braegger, C.P.; Rochat, F.; Chassard, C. Vertical mother–neonate transfer of maternal gut bacteria via breastfeeding. Environ. Microbiol. 2014, 16, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Shamonki, J.M.; Chung, A.; DiNome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial Dysbiosis Is Associated with Human Breast Cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef]
- Thu, M.S.; Chotirosniramit, K.; Nopsopon, T.; Hirankarn, N.; Pongpirul, K. Human gut, breast, and oral microbiome in breast cancer: A systematic review and meta-analysis. Front. Oncol. 2023, 13, 1144021. [Google Scholar] [CrossRef]
- Samkari, A.A.; Alsulami, M.; Bataweel, L.; Altaifi, R.; Altaifi, A.; Saleem, A.M.; Farsi, A.H.; Iskanderani, O.; Akeel, N.Y.; Malibary, N.H.; et al. Body Microbiota and Its Relationship With Benign and Malignant Breast Tumors: A Systematic Review. Cureus 2022, 14, e25473. [Google Scholar] [CrossRef]
- Bodmer, T.; Miltner, E.; Bermudez, L.E. Mycobacterium avium resists exposure to the acidic conditions of the stomach. FEMS Microbiol. Lett. 2000, 182, 45–49. [Google Scholar] [CrossRef]
- Roth, R.I.; Owen, R.L.; Keren, D.F.; Volberding, P.A. Intestinal infection with Mycobacterium avium in acquired immune deficiency syndrome (AIDS). Dig. Dis. Sci. 1985, 30, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Dam, T.; Danelishvili, L.; Wu, M.; Bermudez, L.E. The fadD2 Gene Is Required for Efficient Mycobacterium avium Invasion of Mucosal Epithelial Cells. J. Infect. Dis. 2006, 193, 1135–1142. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.M.; Caballero, J.D.; van der Schalk, T.E.; De Winter, F.H.R.; Shaw, L.P.; Kapel, N.; Recanatini, C.; Timbermont, L.; Kluytmans, J.; Esser, M.; et al. Gut to lung translocation and antibiotic mediated selection shape the dynamics of Pseudomonas aeruginosa in an ICU patient. Nat. Commun. 2022, 13, 6523. [Google Scholar] [CrossRef] [PubMed]
- Roy, D.; Ehtesham, N.Z.; Hasnain, S.E. Is Mycobacterium tuberculosis carcinogenic to humans? FASEB J. 2021, 35, e21853. [Google Scholar] [CrossRef] [PubMed]
- Fol, M.; Koziński, P.; Kulesza, J.; Białecki, P.; Druszczyńska, M. Dual Nature of Relationship between Mycobacteria and Cancer. Int. J. Mol. Sci. 2021, 22, 8332. [Google Scholar] [CrossRef] [PubMed]
- Malik, A.A.; Sheikh, J.A.; Ehtesham, N.Z.; Hira, S.; Hasnain, S.E. Can Mycobacterium tuberculosis infection lead to cancer? Call for a paradigm shift in understanding TB and cancer. Int. J. Med. Microbiol. 2022, 312, 151558. [Google Scholar] [CrossRef] [PubMed]
- Conic, J.; Lapinel, N.; Ali, J.; Boulmay, B. Association between non-tuberculous mycobacterial infection and aerodigestive cancers: A case series highlighting different features, sequence and association. Respir. Med. Case Rep. 2022, 40, 101751. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.J.; Kim, T.J.; Lee, J.-H.; Park, J.-S. Nontuberculous mycobacterial pulmonary disease mimicking lung cancer: Clinicoradiologic features and diagnostic implications. Medicine 2016, 95, e3978. [Google Scholar] [CrossRef]
- Taira, N.; Kawasaki, H.; Takahara, S.; Chibana, K.; Atsumi, E.; Kawabata, T. The Presence of Coexisting Lung Cancer and Non-Tuberculous Mycobacterium in a Solitary Mass. Am. J. Case Rep. 2018, 19, 748–751. [Google Scholar] [CrossRef]
- Thompson, K.J.; Ingle, J.N.; Tang, X.; Chia, N.; Jeraldo, P.R.; Walther-Antonio, M.R.; Kandimalla, K.K.; Johnson, S.; Yao, J.Z.; Harrington, S.C.; et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS ONE 2017, 12, e0188873. [Google Scholar] [CrossRef]
- Niccolai, E.; Baldi, S.; Nannini, G.; Gensini, F.; Papi, L.; Vezzosi, V.; Bianchi, S.; Orzalesi, L.; Ramazzotti, M.; Amedei, A. Breast cancer: The first comparative evaluation of oncobiome composition between males and females. Biol. Sex Differ. 2023, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Geng, C.; Sang, M.; Gao, W.; Li, S.; Yang, S.; Li, Z. Effect of gastrointestinal microbiome and its diversity on the expression of tumor-infiltrating lymphocytes in breast cancer. Oncol. Lett. 2019, 17, 5050–5056. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Fang, L.T.; Tang, A.Z.; Chen, H.L.; Xu, M.L.; Wei, X.S.; Pang, G.D.; Li, C.Q. Mycobacterium vaccae alleviates allergic airway inflammation and airway hyper-responsiveness in asthmatic mice by altering intestinal microbiota. Immunology 2024, 171, 595–608. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maranha, A.; Alarico, S.; Nunes-Costa, D.; Melo-Marques, I.; Roxo, I.; Castanheira, P.; Caramelo, O.; Empadinhas, N. Drinking Water Microbiota, Entero-Mammary Pathways, and Breast Cancer: Focus on Nontuberculous Mycobacteria. Microorganisms 2024, 12, 1425. https://doi.org/10.3390/microorganisms12071425
Maranha A, Alarico S, Nunes-Costa D, Melo-Marques I, Roxo I, Castanheira P, Caramelo O, Empadinhas N. Drinking Water Microbiota, Entero-Mammary Pathways, and Breast Cancer: Focus on Nontuberculous Mycobacteria. Microorganisms. 2024; 12(7):1425. https://doi.org/10.3390/microorganisms12071425
Chicago/Turabian StyleMaranha, Ana, Susana Alarico, Daniela Nunes-Costa, Inês Melo-Marques, Inês Roxo, Pedro Castanheira, Olga Caramelo, and Nuno Empadinhas. 2024. "Drinking Water Microbiota, Entero-Mammary Pathways, and Breast Cancer: Focus on Nontuberculous Mycobacteria" Microorganisms 12, no. 7: 1425. https://doi.org/10.3390/microorganisms12071425
APA StyleMaranha, A., Alarico, S., Nunes-Costa, D., Melo-Marques, I., Roxo, I., Castanheira, P., Caramelo, O., & Empadinhas, N. (2024). Drinking Water Microbiota, Entero-Mammary Pathways, and Breast Cancer: Focus on Nontuberculous Mycobacteria. Microorganisms, 12(7), 1425. https://doi.org/10.3390/microorganisms12071425






