Fungal Biodeterioration Risk in Monastic Libraries without Climate Control
Abstract
1. Introduction
2. Materials and Methods
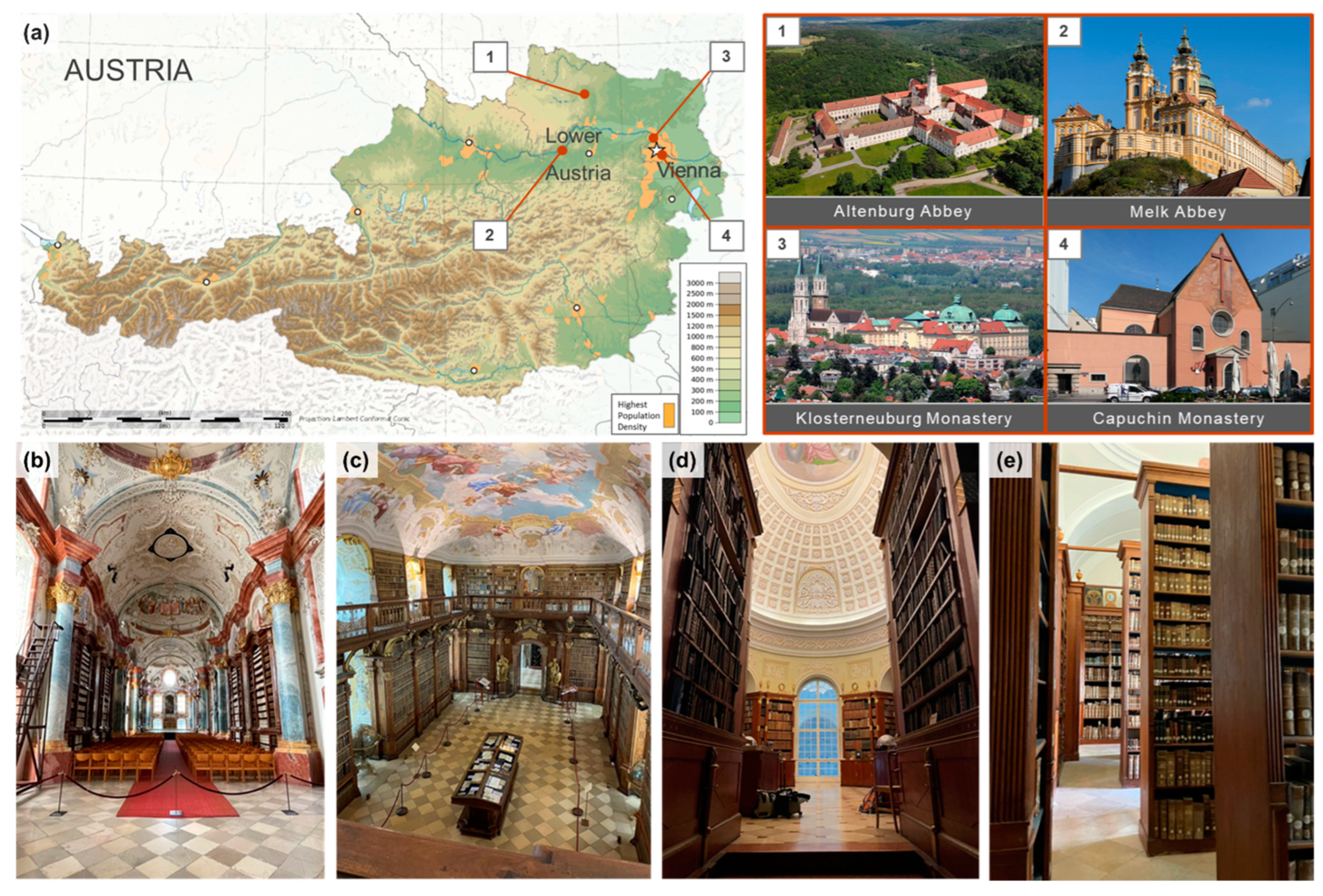
2.1. Description of Sites
2.2. Indoor and Outdoor Climate Data
2.3. Microbiological Data
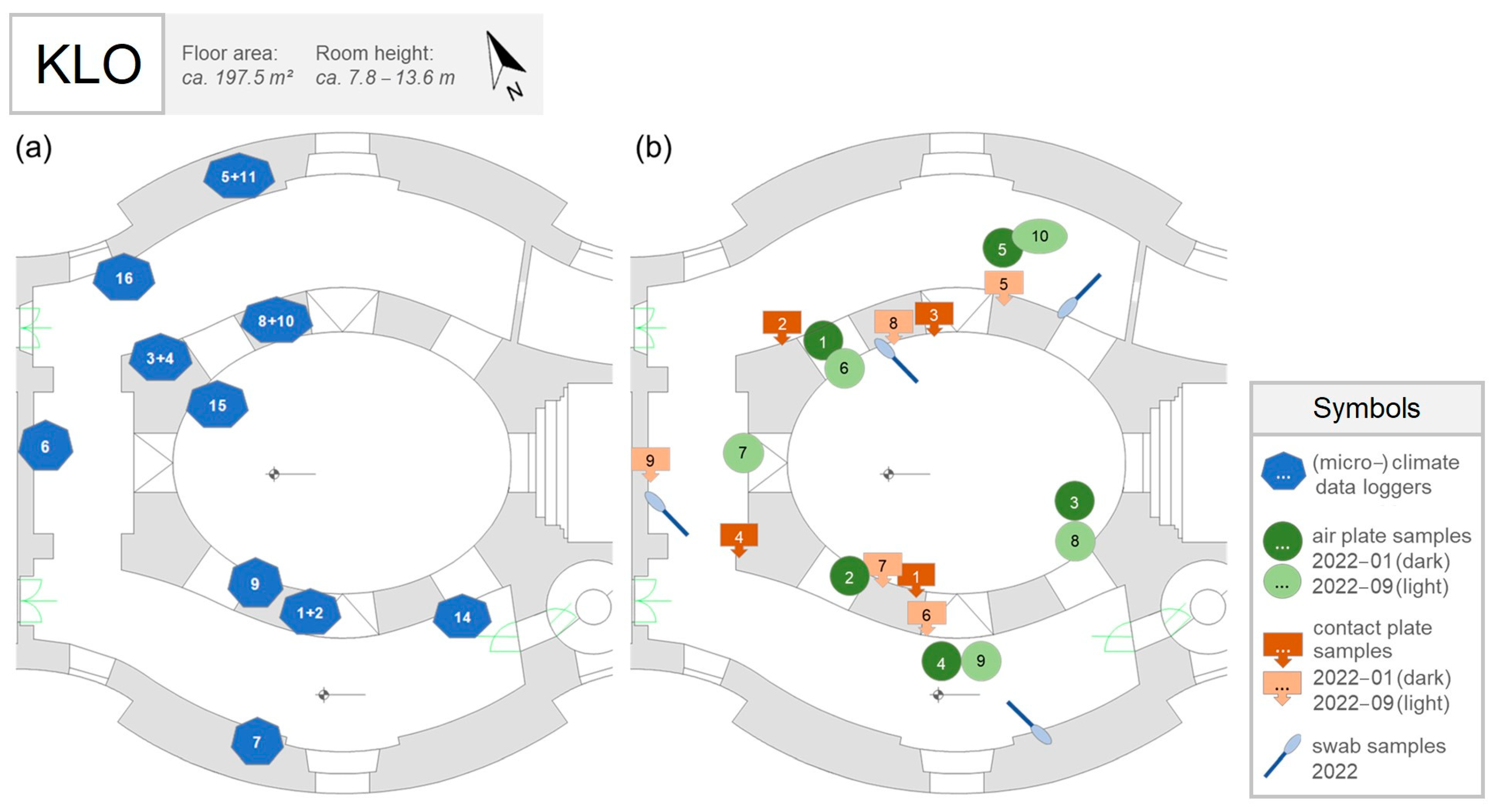
2.3.1. Sampling
2.3.2. Cultivation Plate Analysis
2.3.3. Metagenomic Analysis
2.4. Statistics
3. Results
3.1. Outdoor Climate and Indoor Microclimate
3.1.1. Temperature and Relative Humidity
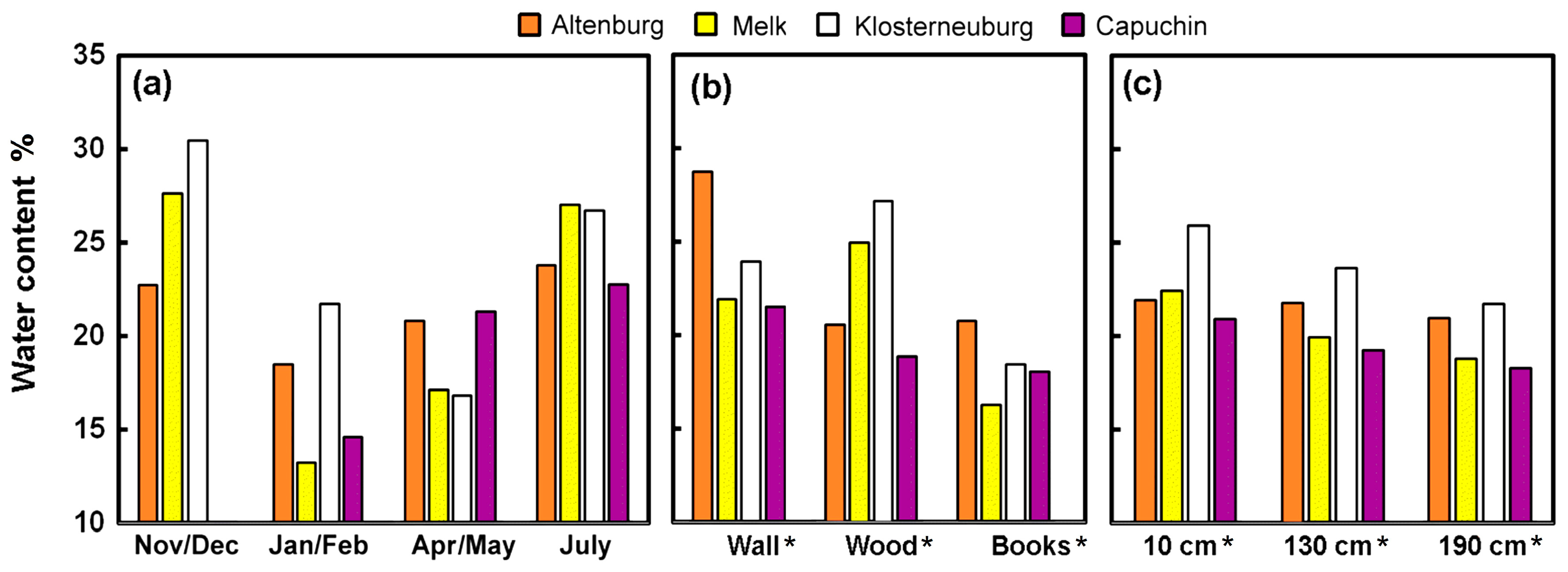
3.1.2. Water Content
3.2. Microbiological Data
3.2.1. Cultivation Data
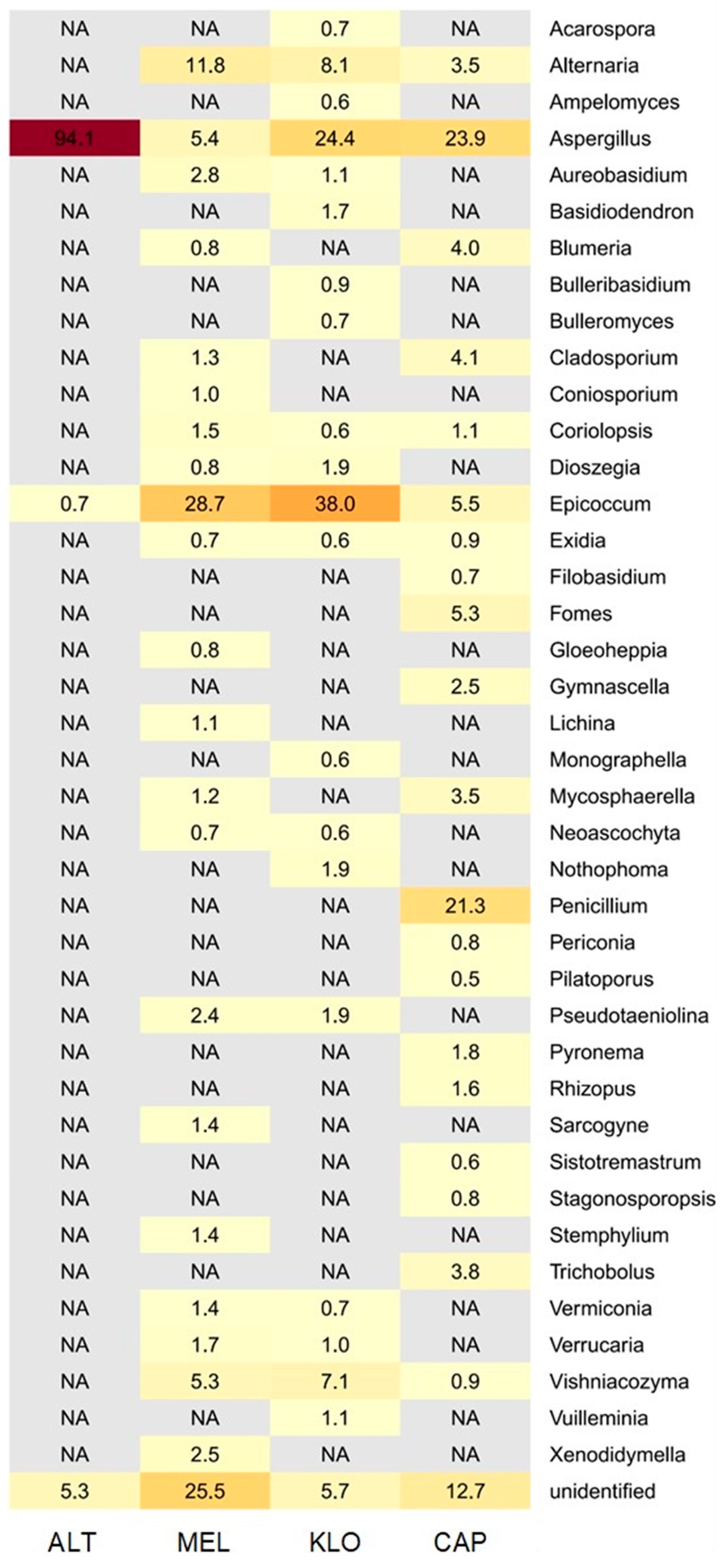
3.2.2. Metagenomic Data
3.2.3. Comparison of Fungal Profiles
4. Discussion
4.1. Historical and Spatial Context
4.2. Humidity Buffering Capacity of Historic Books
4.3. Microbiological Findings
4.4. Microclimate and Fungal Biodeterioration Risk
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A



References
- Gadd, G.; Fomina, M.; Pinzari, F. Fungal biodeterioration and preservation of cultural heritage, artwork, and historical artifacts: Extremophily and adaptation. Microbiol. Mol. Biol. Rev. 2024, 88, e0020022. [Google Scholar] [CrossRef] [PubMed]
- Gallo, F. Aerobiological research and problems in libraries. Aerobiologia 1993, 9, 117–130. [Google Scholar] [CrossRef]
- Sterflinger, K.; Pinzari, F. The revenge of time: Fungal deterioration of cultural heritage with particular reference to books, paper and parchment. Environ. Microbiol. 2012, 14, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, A.C.; Sequeira, S.O.; Macedo, M.F. Fungi in archives, libraries, and museums: A review on paper conservation and human health. Crit. Rev. Microbiol. 2019, 45, 686–700. [Google Scholar] [CrossRef] [PubMed]
- Kosel, J.; Ropret, P. Overview of fungal isolates on heritage collections of photographic materials and their biological potency. J. Cult. Herit. 2021, 48, 277–291. [Google Scholar] [CrossRef]
- Pinzari, F.; Gutarowska, B. Extreme Colonizers and Rapid Profiteers: The Challenging World of Microorganisms That Attack Paper and Parchment. In Microorganisms in the Deterioration and Preservation of Cultural Heritage; Joseph, E., Ed.; Springer (Open Access): Cham, Switzerland, 2021; pp. 79–113. [Google Scholar]
- Michalski, S. The Ideal Climate, Risk Management, the ASHRAE Chapter, Proofed Fluctuations, and Toward a Full Risk Analysis Model. In Contribution to the Experts’ Roundtable on Sustainable Climate Management Strategies, Held in April 2007, in Tenerife, Spain; The Paul Getty Institute: Los Angeles, CA, USA, 2009. [Google Scholar]
- Blades, W. The Enemies of Books; Elliot Stock: London, UK, 1887. [Google Scholar]
- Lankester, P.; Brimblecombe, P. The Impact of Future Climate on Historic Interiors. Sci. Total Environ. 2012, 417–418, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Querner, P.; Sterflinger, K.; Derksen, K.; Leissner, J.; Landsberger, B.; Hammer, A.; Brimblecombe, P. Climate Change and Its Effects on Indoor Pests (Insect and Fungi) in Museums. Climate 2022, 10, 103. [Google Scholar] [CrossRef]
- European Commission; Sabbioni, C.; Brimblecombe, P.; Cassar, M. Chapter 4.2 Climate Induced Decay of Outdoor Wooden Structures by Fungal Growth. In The Atlas of Climate Change Impact on European Cultural Heritage—Scientific Analysis and Management Strategies; Sabbioni, C., Brimblecombe, P., Cassar, M., Eds.; Anthem Press: London, UK, 2010; pp. 44–45. [Google Scholar] [CrossRef]
- Leissner, J.; Kilian, R.; Kotova, L.; Jacob, D.; Mikolajewicz, U.; Broström, T.; Ashley-Smith, J.; Schellen, H.; Martens, M.; van Schijndel, J.; et al. Climate for Culture: Assessing the impact of climate change on the future indoor climate in historic buildings using simulations. Herit. Sci. 2015, 3, 38–52. [Google Scholar] [CrossRef]
- Choidis, P.; Kraniotis, D.; Lehtonen, I.; Hellum, B. A Modelling Approach for the Assessment of Climate Change Impact on the Fungal Colonization of Historic Timber Structures. Forests 2021, 12, 819. [Google Scholar] [CrossRef]
- IPCC. Sections 2–3. In Climate Change 2023: Synthesis Report. Contribution of Working Groups I, II and III to the Sixth Assessment Report. of the Intergovernmental Panel on Climate Change; Core Writing Team, Lee, H., Romero, J., Eds.; IPCC: Geneva, Switzerland, 2023; pp. 42–90. [Google Scholar] [CrossRef]
- Camuffo, D.; Bertolin, C.; Bonazzi, A.; Campana, F.; Merlo, C. Past, present and future effects of climate change on a wooden inlay bookcase cabinet: A new methodology inspired by the novel European Standard EN 1575:2010. J. Cult. Herit. 2014, 15, 26–35. [Google Scholar] [CrossRef]
- Silva, H.E.; Henriques, F.M.A. Preventive conservation of historic buildings in temperate climates. The importance of a risk-based analysis on the decision-making process. Energy Build. 2015, 107, 26–36. [Google Scholar] [CrossRef]
- Frasca, F.; Cornaro, C.; Siani, A. A method based on environmental monitoring and building dynamic simulation to assess indoor climate control strategies in the preventive conservation within historical buildings. Sci. Technol. Built Environ. 2019, 25, 1253–1268. [Google Scholar] [CrossRef]
- Ashley-Smith, J.; Burmester, A.; Eibl, M. Climate for Collections: Standards and Uncertainties. Postprints of the Munich Climate Conference 7 to 9 November 2012; in association with Doerner Institut, Munich, Germany; Archetype Publications: London, UK, 2013. [Google Scholar]
- Ankersmit, B.; Stappers, M.L.H.; Kramer, N. Guideline in Jeopardy: Observations on the Application of the ASHRAE Chapter on Climate Control in Museums. Stud. Conserv. 2018, 63 (Suppl. S1), 1–7. [Google Scholar] [CrossRef]
- Camuffo, D. Microclimate for Cultural Heritage: Measurement, Risk Assessment, Conservation, Restoration, and Maintenance of Indoor and Outdoor Monuments; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Camuffo, D.; Becherini, F.; Della Valle, A. The European Standard EN 15757 Concerning Specifications for Relative Humidity: Suggested Improvements for Its Revision. Atmosphere 2022, 13, 1344. [Google Scholar] [CrossRef]
- Fabbri, K.; Pretelli, M. Heritage buildings and historic microclimate without HVAC technology: Malatestiana Library in Cesena, Italy, UNESCO Memory of the World. Energy Build. 2014, 76, 15–31. [Google Scholar] [CrossRef]
- Moretti, E.; Sciurpi, F.; Stamponi, E.; Fiore, M. Microclimatic monitoring for book heritage preservation in historic buildings: Preliminary investigation on ”Sala del Dottorato” in Palazzo Murena, Perugia, Italy. IOP Conf. Ser. Mater. Sci. Eng. 2020, 949, 012098. [Google Scholar] [CrossRef]
- Verticchio, E.; Frasca, F.; Bertolin, C.; Siani, A.M. Climate-induced risk for the preservation of paper collections: Comparative study among three historic libraries in Italy. Build. Environ. 2021, 206, 108394. [Google Scholar] [CrossRef]
- Boeri, A.; Longo, D.; Fabbri, K.; Pretelli, M.; Bonora, A.; Boulanger, S. Library indoor microclimate monitoring with and without heating system. A bologna university library case study. J. Cult. Herit. 2022, 53, 143–153. [Google Scholar] [CrossRef]
- Schito, E.; Pereira, L.D.; Testi, D.; da Silva, M.G. A procedure for identifying chemical and biological risks for books in historic libraries based on microclimate analysis. J. Cult. Herit. 2019, 37, 155–165. [Google Scholar] [CrossRef]
- Verticchio, E.; Frasca, F.; Cavalieri, P.; Teodonio, L.; Fugaro, D.; Siani, A.M. Conservation risks for paper collections induced by the microclimate in the repository of the Alessandrina Library in Rome (Italy). Herit. Sci. 2022, 10, 80. [Google Scholar] [CrossRef] [PubMed]
- Pasquarella, C.; Balocco, C.; Saccani, E.; Capobianco, E.; Viani, I.; Veronesi, L.; Pavani, F.; Pasquariello, G.; Rotolo, V.; Palla, F.; et al. Biological and microclimatic monitoring for conservation of cultural heritage: A case study at the De Rossi room of the Palatina library in Parma. Aerobiologia 2020, 36, 105–111. [Google Scholar] [CrossRef]
- Ilies, D.C.; Blaga, L.; Hassan, T.H.; Ilies, A.; Caciora, T.; Grama, V.; Herman, G.V.; Dejeu, P.; Zdringa, M.; Marshall, T.; et al. Indoor Microclimate and Microbiological Risks in Heritage Buildings: A Case Study of the Neologic Sinagogue, Oradea, Romania. Buildings 2023, 13, 2277. [Google Scholar] [CrossRef]
- Sabatini, L.; Sisti, M.; Campana, R. Evaluation of fungal community involved in the bioderioration process of wooden artworks and canvases in Montefeltro area (Marche, Italy). Microbiol. Res. 2018, 207, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Awad, A.H.; Saeed, Y.; Shakour, A.A.; Abdellatif, N.M.; Ibrahim, Y.H.; Elghanam, M.; Elwakeel, F. Indoor air fungal pollution of a historical museum, Egypt: A case study. Aerobiologia 2020, 36, 197–209. [Google Scholar] [CrossRef]
- Bastholm, C.J.; Madsen, A.; Andersen, B.; Frisvad, J.; Richter, J. The mysterious mould outbreak—A comprehensive fungal colonisation in a climate-controlled museum repository challenges the environmental guidelines for heritage collections. J. Cult. Herit. 2022, 55, 78–87. [Google Scholar] [CrossRef]
- Zalar, P.; Graf Hriberšek, D.; Gostinčar, C.; Breskvar, M.; Džeroski, S.; Matul, M.; Novak Babič, M.; Čremožnik Zupančič, J.; Kujović, A.; Gunde-Cimerman, N.; et al. Xerophilic fungi contaminating historically valuable easel paintings from Slovenia. Front. Microbiol. 2023, 14, 1258670. [Google Scholar] [CrossRef] [PubMed]
- Micheluz, A.; Manente, S.; Tigini, V.; Prigione, V.P.; Pinzari, F.; Ravagnan, G.; Varese, G.C. The extreme environment of a library: Xerophilic fungi inhabiting indoor niches. Int. Biodeter. Biodegr. 2015, 99, 1–7. [Google Scholar] [CrossRef]
- Sterflinger, K.; Voitl, C.; Lopandic, K.; Piñar, G.; Tafer, H. Big sound and extreme fungi—Xerophilic, halotolerant Aspergilli and Penicillia with low optimal temperature as invaders of historic pipe organs. Life 2018, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Bastholm, C.J.; Madsen, A.M.; Frisvad, J.C.; Richter, J. Xerophilic fungi in museum repositories challenge our perception of healthy buildings and the preservation of cultural heritage. In Healthy Buildings 2021—Europe. Proceedings of the 17th International Healthy Buildings Conference 21–23 June 2021; ISIAQ International Society of Indoor Air Quality: Oslo, Norway, 2021; pp. 260–268. [Google Scholar]
- Montanari, M.; Melloni, V.; Pinzari, F.; Innocenti, G. Fungal biodeterioration of historical library materials stored in Compactus movable shelves. Int. Biodeter. Biodegr. 2012, 75, 83–88. [Google Scholar] [CrossRef]
- Polo, A.; Cappitelli, F.; Villa, F.; Pinzari, F. Biological invasion in the indoor environment: The spread of Eurotium halophilicum on library materials. Int. Biodeter. Biodegr. 2017, 118, 34–44. [Google Scholar] [CrossRef]
- Brimblecombe, P.; Sterflinger, K.; Derksen, K.; Haltrich, M.; Querner, P. Thermohygrometric Climate, Insects and Fungi in the Klosterneuburg Monastic Library. Heritage 2022, 5, 4228–4244. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous Hyphomycetes; Commonwealth Mycological Institute, Kew: Surrey, UK, 1971. [Google Scholar]
- Domsch, K.H.; Gams, W.; Anderson, T.-H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag: Eching, Germany, 2007; ISBN 978-3-930167-69-2. [Google Scholar]
- de Hoog, G.S.; Guarro, J. (Eds.) Atlas of Clinical Fungi; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 1995. [Google Scholar]
- Klich, M.A. Identification of Common Aspergillus Species, 1st ed.; Centraalbureau voor Schimmelcultures: Utrecht, The Netherlands, 2002; ISBN 90-70351-46-3. [Google Scholar]
- Pitt, J.I. A Laboratory Guide to Common Penicillium Species, 3rd ed.; Food Science Australia: North Ryde, Australia, 2000; ISBN 0643048375. [Google Scholar]
- Samson, R.A.; Houbraken, J.; Thrane, U.; Frisvad, J.C.; Andersen, B. Food and Indoor Fungi, 2nd ed.; Westerdijk Laboratory Manual Series: Utrecht, The Netherlands, 2019; ISBN 978-94-91751-18-9. [Google Scholar]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. ISBN 9780123721808. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Fungal Barcoding Consortium. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef] [PubMed]
- Tichy, J.; Waldherr, M.; Ortbauer, M.; Graf, A.; Sipek, B.; Jembrih-Simbuerger, D.; Sterflinger, K.; Piñar, G. Pretty in pink? Complementary strategies for analysing pink biofilms on historical buildings. Sci. Total Environ. 2023, 904, 166737. [Google Scholar] [CrossRef]
- Tietze, H. Altenburg. In Die Denkmale des Politischen Bezirkes Horn (ÖKT); Schroll: Vienna, Austria, 1911; pp. 259–323. [Google Scholar]
- Telesko, W. Bibliothek. Tempel der Weisheit. In Benediktinerstift Altenburg. Mittelalterliches Kloster und barocker Kosmos; Telesko, W., Groiß, A., Eds.; Brandstätter Verlag: Vienna, Austria, 2008; pp. 112–121. [Google Scholar]
- Mittendorfer, K.; Kaiser, A. Bibliothek des Benediktinerstiftes Altenburg, In Handbuch der Historischen Buchbestände in Österreich, Bd. 3; Olms-Weidmann: Hildesheim, Germany; Zürich, Switzerland; New York, NY, USA, 1996; pp. 87–93. [Google Scholar]
- Tietze, H. Melk. In Die Denkmale des Politischen Bezirkes Melk (ÖKT); Schroll: Vienna, Austria, 1909; pp. 178–369. [Google Scholar]
- Glassner, G. Bibliothek des Benediktinerstiftes Melk. In Handbuch der Historischen Buchbestände in Österreich, Bd. 3; Olms-Weidmann: Hildesheim, Germany; Zürich, Switzerland; New York, NY, USA, 1996; pp. 146–157. [Google Scholar]
- Haltrich, M. Die Stiftsbibliothek. In Das Stift Klosterneuburg; Huber, W.C., Ed.; Stejkovic: Wettin-Löbejün, Germany, 2014; pp. 216–229. [Google Scholar]
- Opl, K. Bibliothek des Augustiner-Chorherrenstiftes Klosterneuburg. In Handbuch der Historischen Buchbestände in Österreich, Bd. 3; Olms-Weidmann: Hildesheim, Germany; Zürich, Switzerland; New York, NY, USA, 1996; pp. 130–136. [Google Scholar]
- Buchinger, G. Bau-, Ausstattungs- und Restaurierungsgeschichte des Wiener Kapuzinerklosters und seiner Kirche. In Die Kapuziner in Österreich. Geschichte—Kunst—Spiritualität; Buchinger, G., Ed.; Imhof: Petersberg, Germany, 2021; pp. 105–133. [Google Scholar]
- Pichl, R. Bibliothek des Kapuzinerklosters Innere Stadt (Wien 48). In Handbuch der Historischen Buchbestände in Österreich, Bd. 2/2; Olms-Weidmann: Hildesheim, Germany; Zürich, Switzerland; New York, NY, USA, 1995; pp. 115–119. [Google Scholar]
- Andretta, M.; Coppola, F.; Seccia, L. Investigation on the interaction between the outdoor environment and the indoor microclimate of a historical library. J. Cult. Herit. 2015, 17, 75–86. [Google Scholar] [CrossRef]
- Kupczak, A.; Sadłowska-Sałęga, A.; Krzemień, L.; Sobczyk, J.; Radoń, J.; Kozłowski, R. Impact of paper and wooden collections on humidity stability and energy consumption in museums and libraries. Energy Build. 2018, 158, 77–85. [Google Scholar] [CrossRef]
- Di Bonaventura, M.P.; DeSalle, R.; Eveleigh, D.E.; Baldwin, A.M.; Koestler, R.J. Studies of Fungal Infestations of Tiffany’s Drawings: Limits and Advantages of Classical and Molecular Techniques. In Art, Biology, and Conservation: Biodeterioration of Works of Art; Koestler, R.J., Koestler, V.R., Charola, A.E., Nieto-Fernandez, F.E., Eds.; The Metropolitan Museum of Art, Yale University Press: New York, NY, USA, 2003; pp. 94–109. [Google Scholar]
- Karakasidou, K.; Nikolouli, K.; Amoutzias, G.D.; Pournou, A.; Manassis, C.; Tsiamis, G.; Mossialos, D. Microbial diversity in biodeteriorated Greek historical documents dating back to the 19th and 20th century: A case study. MicrobiologyOpen 2018, 7, e00596. [Google Scholar] [CrossRef] [PubMed]
- Sterflinger, K.; Piñar, G. Molecular-Based Techniques for the Study of Microbial Communities in Artworks. In Microorganisms in the Deterioration and Preservation of Cultural Heritage; Joseph, E., Ed.; Springer (Open Access): Cham, Switzerland, 2021; pp. 59–77. [Google Scholar]
- Rao, C.Y.; Burge, H.A.; Chang, J.C. Review of quantitative standards and guidelines for fungi in indoor air. J. Air Waste Manag. Assoc. 1996, 46, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Afshari, A.; Anderson, H.; Cohen, A.; De Oliveira Fernandes, E.; Douwes, J.; Górny, R.; Hirvonen, M.-R.; Jaakkola, J.; Kirchner, S.; Kurnitski, J.; et al. WHO Guidelines for Indoor Air Quality: Dampness and Mould; WHO Regional Office for Europe: Geneva, Switzerland, 2009; ISBN 7989289041683.
- Krus, M.; Kilian, R.; Sedlbauer, K. Mould growth prediction by computational simulation on historic buildings. In Museum Microclimates; Padfield, T., Borchersen, K., Eds.; National Museum of Denmark: Copenhagen, Denmark, 2007; ISBN 978-87-7602-080-4. [Google Scholar]



| Location | Altenburg Abbey | Melk Abbey | Klosterneuburg Monastery | Capuchin Monastery | |
|---|---|---|---|---|---|
| Average T (°C) | Outdoor | 11.1 ± 7.9 | 12.2 ± 7.8 | 13.6 ± 8.0 | 14.7 ± 8.2 |
| Library | 12.2 ± 7.9 | 15.3 ± 6.2 | 15.9 ± 6.5 | 18.0 ± 5.8 | |
| Average RH (%) | Outdoor | 69.1 ± 13.7 | 71.0 ± 14.0 | 62.4 ± 14.4 | 57.2 ± 13.6 |
| Library | 66.1 ± 6.1 | 59.2 ± 6.4 | 55.4 ± 4.5 | 52.4 ± 1.4 | |
| Total CFUs | Aspergillus | Penicillium | Cladosporium | Alternaria | Fusarium | Epicoccum | Mucoromycota | Others | |
|---|---|---|---|---|---|---|---|---|---|
| Indoor air [CFUs/m3] | |||||||||
| ALT-W | 370.0 | 19.0 | 84.0 | 213.0 | 10.0 | 0 | 0 | 1.0 | 24.0 |
| ALT-S | 152.0 | 3.0 | 14.0 | 27.0 | 2.0 | 0 | 0 | 0 | 69.0 |
| MEL-W | 81.3 | 11.3 | 27.5 | 16.3 | 1.3 | 0 | 0 | 2.5 | 23.8 |
| MEL-S | 630.0 | 15.0 | 42.5 | 253.8 | 45.0 | 2.5 | 15.0 | 3.8 | 222.5 ** |
| KLO-W | 91.0 | 15.0 | 21.0 | 40.0 | 3.0 | 0 | 0 | 0 | 7.0 |
| KLO-S | 373.0 * | 28.0 * | 53.0 | 172.0 | 4.0 | 1.0 | 1.0 | 5.0 | 46.0 |
| CAP-W | 106.3 | 7.5 | 76.3 | 17.5 | 0 | 0 | 0 | 0 | 8.8 |
| CAP-S | 48.8 | 6.3 | 11.3 | 7.5 | 6.3 | 0 | 2.5 | 0 | 13.8 |
| Indoor surfaces [CFUs/m2] | |||||||||
| ALT-W | 27 × 104 | 1813 | 15,063 | 138,125 | 34,438 | 1063 | 375 | 875 | >19,688 |
| ALT-S | 7 × 104 | 750 | 1563 | 50,938 | 1188 | 63 | 1125 | 313 | >6688 |
| MEL-W | 15 × 104 | 1016 | 3594 | 61,641 | 23,047 | 391 | 1016 | 313 | >60,938 |
| MEL-S | 5 × 104 | 781 | 2656 | 26,094 | 2031 | 0 | 547 | 313 | >15,781 |
| KLO-W | 8 × 104 | 3984 | 8906 | 17,422 | 2109 | 625 | 78 | 1797 | >3906 |
| KLO-S | 5 × 104 | 1797 | 78 | 20,547 | 0 | 78 | 0 | 234 | >20,938 |
| CAP-W | 10 × 104 | 1875 | 10,234 | 52,109 | 7891 | 469 | 0 | 469 | >9375 |
| CAP-S | 3 × 104 | 2266 | 2266 | 9219 | 2344 | 78 | 313 | 391 | >8906 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Derksen, K.; Brimblecombe, P.; Piñar, G.; Waldherr, M.; Graf, A.; Haltrich, M.; Querner, P.; Sterflinger, K. Fungal Biodeterioration Risk in Monastic Libraries without Climate Control. Microorganisms 2024, 12, 1450. https://doi.org/10.3390/microorganisms12071450
Derksen K, Brimblecombe P, Piñar G, Waldherr M, Graf A, Haltrich M, Querner P, Sterflinger K. Fungal Biodeterioration Risk in Monastic Libraries without Climate Control. Microorganisms. 2024; 12(7):1450. https://doi.org/10.3390/microorganisms12071450
Chicago/Turabian StyleDerksen, Katharina, Peter Brimblecombe, Guadalupe Piñar, Monika Waldherr, Alexandra Graf, Martin Haltrich, Pascal Querner, and Katja Sterflinger. 2024. "Fungal Biodeterioration Risk in Monastic Libraries without Climate Control" Microorganisms 12, no. 7: 1450. https://doi.org/10.3390/microorganisms12071450
APA StyleDerksen, K., Brimblecombe, P., Piñar, G., Waldherr, M., Graf, A., Haltrich, M., Querner, P., & Sterflinger, K. (2024). Fungal Biodeterioration Risk in Monastic Libraries without Climate Control. Microorganisms, 12(7), 1450. https://doi.org/10.3390/microorganisms12071450







