The Restriction Activity Investigation of Rv2528c, an Mrr-like Modification-Dependent Restriction Endonuclease from Mycobacterium tuberculosis
Abstract
:1. Introduction
| Protein | Feature | Function |
|---|---|---|
| Rv0058 | PI-MtuHIP | putative intein homing REase |
| Rv1461 | PI-MtuHIIP | putative intein homing REase |
| Rv2024c | MamB | MTase [9] |
| Rv2528c | Mrr | putative Type IV REase |
| Rv2737c | PI-MtuI | intein homing REase [20,21,22] |
| Rv2755c | HsdS.1 | Type I specificity subunit (inactive) [9] |
| Rv2756c | M.MtuHI | Type I MTase [8,9,10,11] |
| Rv2761c | HsdS | Type I specificity subunit [9] |
| Rv2966c | secretory MTase [23,24] | |
| Rv3263 | M.MtuHIII (MamA) | MTase [8] |
2. Materials and Methods
2.1. Strains, Enzymes and Plasmid Constructions
2.2. Sequence Alignment and Tertiary Structure Superposition
2.3. Competent Cell Preparation and Electroporation Transformation
2.4. Growth Curve Measurement
2.5. Western Blot Detection for In Vivo Recombinant Rv2528c Expression
2.6. In Vivo DNA Cleavage Assay (TUNEL Kit) and Flow Cytometry Detection
2.7. In Vitro Modification of pCDFDuet and pRSFDuet
2.8. Protein Expression, Purification, and In Vitro Cleavage Condition Explorations
3. Results
3.1. Protein Structures of Rv2528c Compared with EcoKMrr
3.2. In Vivo Genotoxicity of Rv2528c in E. coli BL21(DE3)pLysS Strain
3.3. In Vitro Cleavage Assay of Rv2528c
3.4. Investigation of DNA Methylation Type Recognized by Rv2528c In Vivo
4. Discussion
4.1. Difficulties for Determination of In Vitro Cleavage Activity of Rv2528c
4.2. The Possible Role of Rv2528c
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Global Tuberculosis Report 2021; World Health Organization: Geneva, Switzerland, 2022. [Google Scholar]
- Cohen, S.B.; Gern, B.H.; Delahaye, J.L.; Adams, K.N.; Plumlee, C.R.; Winkler, J.K.; Sherman, D.R.; Gerner, M.Y.; Urdahl, K.B. Alveolar Macrophages Provide an Early Mycobacterium tuberculosis Niche and Initiate Dissemination. Cell Host Microbe 2018, 24, 439–446.e434. [Google Scholar] [CrossRef] [PubMed]
- Lavalett, L.; Ortega, H.; Barrera, L.F. Human Alveolar and Splenic Macrophage Populations Display a Distinct Transcriptomic Response to Infection with Mycobacterium tuberculosis. Front Immunol. 2020, 11, 630. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—A database for DNA restriction and modification: Enzymes, genes and genomes. Nucleic Acids Res. 2015, 43, D298–D299. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.J.; Belfort, M.; Bestor, T.; Bhagwat, A.S.; Bickle, T.A.; Bitinaite, J.; Blumenthal, R.M.; Degtyarev, S.; Dryden, D.T.; Dybvig, K.; et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003, 31, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Wion, D.; Casadesus, J. N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006, 4, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Casadesus, J.; Low, D. Epigenetic gene regulation in the bacterial world. Microbiol. Mol. Biol. Rev. 2006, 70, 830–856. [Google Scholar] [CrossRef] [PubMed]
- Shell, S.S.; Prestwich, E.G.; Baek, S.H.; Shah, R.R.; Sassetti, C.M.; Dedon, P.C.; Fortune, S.M. DNA methylation impacts gene expression and ensures hypoxic survival of Mycobacterium tuberculosis. PLoS Pathog. 2013, 9, e1003419. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Zhong, J.; Jia, X.; Liu, G.; Kang, Y.; Dong, M.; Zhang, X.; Li, Q.; Yue, L.; Li, C.; et al. Precision methylome characterization of Mycobacterium tuberculosis complex (MTBC) using PacBio single-molecule real-time (SMRT) technology. Nucleic Acids Res. 2016, 44, 730–743. [Google Scholar] [CrossRef]
- Hu, X.; Zhou, X.; Yin, T.; Chen, K.; Hu, Y.; Zhu, B.; Mi, K. The Mycobacterial DNA Methyltransferase HsdM Decreases Intrinsic Isoniazid Susceptibility. Antibiotics 2021, 10, 1323. [Google Scholar] [CrossRef]
- Chu, H.; Hu, Y.; Zhang, B.; Sun, Z.; Zhu, B. DNA Methyltransferase HsdM Induce Drug Resistance on Mycobacterium tuberculosis via Multiple Effects. Antibiotics 2021, 10, 1544. [Google Scholar] [CrossRef]
- Heitman, J.; Model, P. Site-specific methylases induce the SOS DNA repair response in Escherichia coli. J. Bacteriol. 1987, 169, 3243–3250. [Google Scholar] [CrossRef] [PubMed]
- Waite-Rees, P.A.; Keating, C.J.; Moran, L.S.; Slatko, B.E.; Hornstra, L.J.; Benner, J.S. Characterization and expression of the Escherichia coli Mrr restriction system. J. Bacteriol. 1991, 173, 5207–5219. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Rychlewski, L. Identification of a PD-(D/E)XK-like domain with a novel configuration of the endonuclease active site in the methyl-directed restriction enzyme Mrr and its homologs. Gene 2001, 267, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Aertsen, A.; Tesfazgi Mebrhatu, M.; Michiels, C.W. Activation of the Salmonella typhimurium Mrr protein. Biochem. Biophys. Res. Commun. 2008, 367, 435–439. [Google Scholar] [CrossRef]
- Aertsen, A.; Van Houdt, R.; Vanoirbeek, K.; Michiels, C.W. An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 2004, 186, 6133–6141. [Google Scholar] [CrossRef]
- Aertsen, A.; Michiels, C.W. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol. Microbiol. 2005, 58, 1381–1391. [Google Scholar] [CrossRef]
- Bourges, A.C.; Torres Montaguth, O.E.; Ghosh, A.; Tadesse, W.M.; Declerck, N.; Aertsen, A.; Royer, C.A. High pressure activation of the Mrr restriction endonuclease in Escherichia coli involves tetramer dissociation. Nucleic Acids Res. 2017, 45, 5323–5332. [Google Scholar] [CrossRef] [PubMed]
- Bourges, A.C.; Torres Montaguth, O.E.; Tadesse, W.; Labesse, G.; Aertsen, A.; Royer, C.A.; Declerck, N. An oligomeric switch controls the Mrr-induced SOS response in E. coli. DNA Repair 2021, 97, 103009. [Google Scholar] [CrossRef]
- Guhan, N.; Muniyappa, K. Mycobacterium tuberculosis RecA intein possesses a novel ATP-dependent site-specific double-stranded DNA endonuclease activity. J. Biol. Chem. 2002, 277, 16257–16264. [Google Scholar] [CrossRef]
- Guhan, N.; Muniyappa, K. The RecA intein of Mycobacterium tuberculosis promotes cleavage of ectopic DNA sites. Implications for the dispersal of inteins in natural populations. J. Biol. Chem. 2002, 277, 40352–40361. [Google Scholar] [CrossRef]
- Guhan, N.; Muniyappa, K. Mycobacterium tuberculosis RecA intein, a LAGLIDADG homing endonuclease, displays Mn2+ and DNA-dependent ATPase activity. Nucleic Acids Res. 2003, 31, 4184–4191. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Saigal, K.; Malhotra, K.; Sinha, K.M.; Taneja, B. Structural and functional characterization of Rv2966c protein reveals an RsmD-like methyltransferase from Mycobacterium tuberculosis and the role of its N-terminal domain in target recognition. J. Biol. Chem. 2011, 286, 19652–19661. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Upadhyay, S.; Srilalitha, M.; Nandicoori, V.K.; Khosla, S. The interaction of mycobacterial protein Rv2966c with host chromatin is mediated through non-CpG methylation and histone H3/H4 binding. Nucleic Acids Res. 2015, 43, 3922–3937. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, B.S.; Stoddard, B.L. Homing endonucleases: Structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001, 29, 3757–3774. [Google Scholar] [CrossRef] [PubMed]
- Kapopoulou, A.; Lew, J.M.; Cole, S.T. The MycoBrowser portal: A comprehensive and manually annotated resource for mycobacterial genomes. Tuberculosis 2011, 91, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, W.M., Jr.; Malygin, E.G.; Ovechkina, L.G.; Zinoviev, V.V.; Reich, N.O. Functional analysis of BamHI DNA cytosine-N4 methyltransferase. J. Mol. Biol. 2003, 325, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2012. [Google Scholar]
- Gou, L.; Han, T.; Wang, X.; Ge, J.; Liu, W.; Hu, F.; Wang, Z. A Novel TetR Family Transcriptional Regulator, CalR3, Negatively Controls Calcimycin Biosynthesis in Streptomyces chartreusis NRRL 3882. Front Microbiol. 2017, 8, 2371. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Yamada-Mabuchi, M.; Zhao, G.; Li, L.; Liu, G.; Ou, H.Y.; Deng, Z.; Zheng, Y.; He, X. Recognition and cleavage of 5-methylcytosine DNA by bacterial SRA-HNH proteins. Nucleic Acids Res. 2015, 43, 1147–1159. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Loo, D.T. In situ detection of apoptosis by the TUNEL assay: An overview of techniques. Methods Mol. Biol. 2011, 682, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Tolia, N.H.; Joshua-Tor, L. Strategies for protein coexpression in Escherichia coli. Nat. Methods 2006, 3, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Su, T.J.; Tock, M.R.; Egelhaaf, S.U.; Poon, W.C.; Dryden, D.T. DNA bending by M.EcoKI methyltransferase is coupled to nucleotide flipping. Nucleic Acids Res. 2005, 33, 3235–3244. [Google Scholar] [CrossRef] [PubMed]
- Blattner, F.R.; Plunkett, G., 3rd; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Gonchar, D.; Chernukhin, V.; Abdurashitov, M.; Kileva, E.; Dedkov, V.; Mikhnenkova, N.; Lomakovskaya, E.; Udalyeva, S.; Degtyarev, S.K. Cloning and characterization of a new site specific methyl-directed DNA endonuclease EcoBLI recognizing 5′-G (5mC) NGC-3′/3′-CGN (5mC) G-5′. Biotechnol. Indian J. 2016, 12, 175–181. [Google Scholar]
- Fomenkov, A.; Sun, Z.; Dila, D.K.; Anton, B.P.; Roberts, R.J.; Raleigh, E.A. EcoBLMcrX, a classical modification-dependent restriction enzyme in Escherichia coli B: Characterization in vivo and in vitro with a new approach to cleavage site determination. PLoS ONE 2017, 12, e0179853. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.Y.; Zemlyanskaya, E.V.; Gonchar, D.A.; Sun, Z.; Weigele, P.; Fomenkov, A.; Degtyarev, S.K.; Roberts, R.J. Characterization of BisI Homologs. Front Microbiol. 2021, 12, 689929. [Google Scholar] [CrossRef] [PubMed]
- Tesfazgi Mebrhatu, M.; Wywial, E.; Ghosh, A.; Michiels, C.W.; Lindner, A.B.; Taddei, F.; Bujnicki, J.M.; Van Melderen, L.; Aertsen, A. Evidence for an evolutionary antagonism between Mrr and Type III modification systems. Nucleic Acids Res. 2011, 39, 5991–6001. [Google Scholar] [CrossRef] [PubMed]
- Cao, B.; Cheng, Q.; Gu, C.; Yao, F.; DeMott, M.S.; Zheng, X.; Deng, Z.; Dedon, P.C.; You, D. Pathological phenotypes and in vivo DNA cleavage by unrestrained activity of a phosphorothioate-based restriction system in Salmonella. Mol. Microbiol. 2014, 93, 776–785. [Google Scholar] [CrossRef]
- Zheng, Y.; Cohen-Karni, D.; Xu, D.; Chin, H.G.; Wilson, G.; Pradhan, S.; Roberts, R.J. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010, 38, 5527–5534. [Google Scholar] [CrossRef]
- Cohen-Karni, D.; Xu, D.; Apone, L.; Fomenkov, A.; Sun, Z.; Davis, P.J.; Kinney, S.R.; Yamada-Mabuchi, M.; Xu, S.Y.; Davis, T.; et al. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. USA 2011, 108, 11040–11045. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.R.; Nugent, R.L.; Li, A.; Mabuchi, M.Y.; Fomenkov, A.; Cohen-Karni, D.; Griggs, R.M.; Zhang, X.; Wilson, G.G.; Zheng, Y.; et al. Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI. Sci. Rep. 2014, 4, 4246. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, E.; Coe, L.; Raleigh, E.A. McrBC: A multisubunit GTP-dependent restriction endonuclease. J. Mol. Biol. 1992, 225, 327–348. [Google Scholar] [CrossRef]
- Stewart, F.J.; Raleigh, E.A. Dependence of McrBC cleavage on distance between recognition elements. Biol. Chem. 1998, 379, 611–616. [Google Scholar] [PubMed]
- Ishikawa, K.; Handa, N.; Sears, L.; Raleigh, E.A.; Kobayashi, I. Cleavage of a model DNA replication fork by a methyl-specific endonuclease. Nucleic Acids Res. 2011, 39, 5489–5498. [Google Scholar] [CrossRef]
- Randall, S.E.; Martini, M.C.; Zhou, Y.; Joubran, S.R.; Shell, S.S. MamA essentiality in Mycobacterium smegmatis is explained by the presence of an apparent cognate restriction endonuclease. BMC Res. Notes 2020, 13, 462. [Google Scholar] [CrossRef]


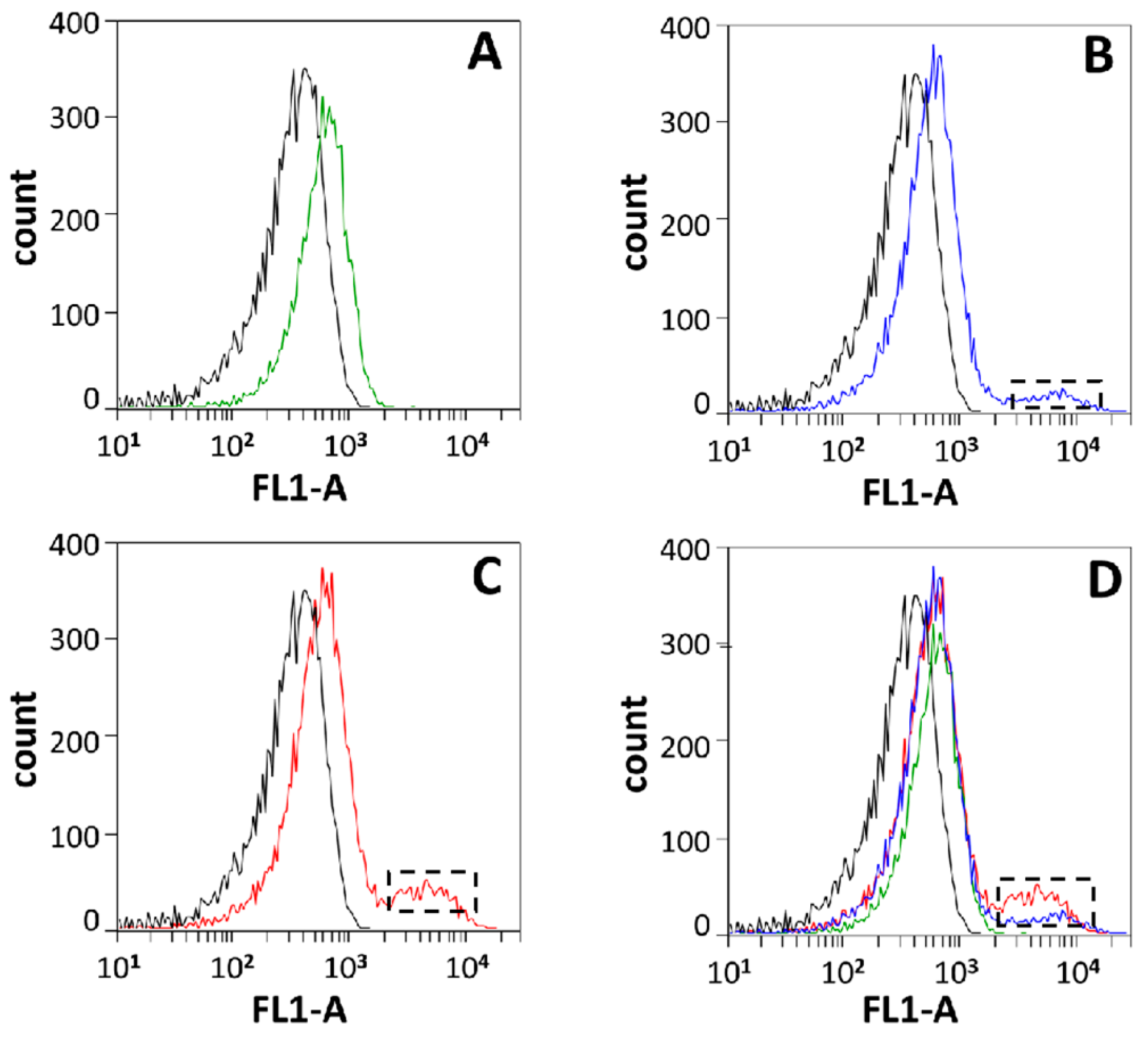
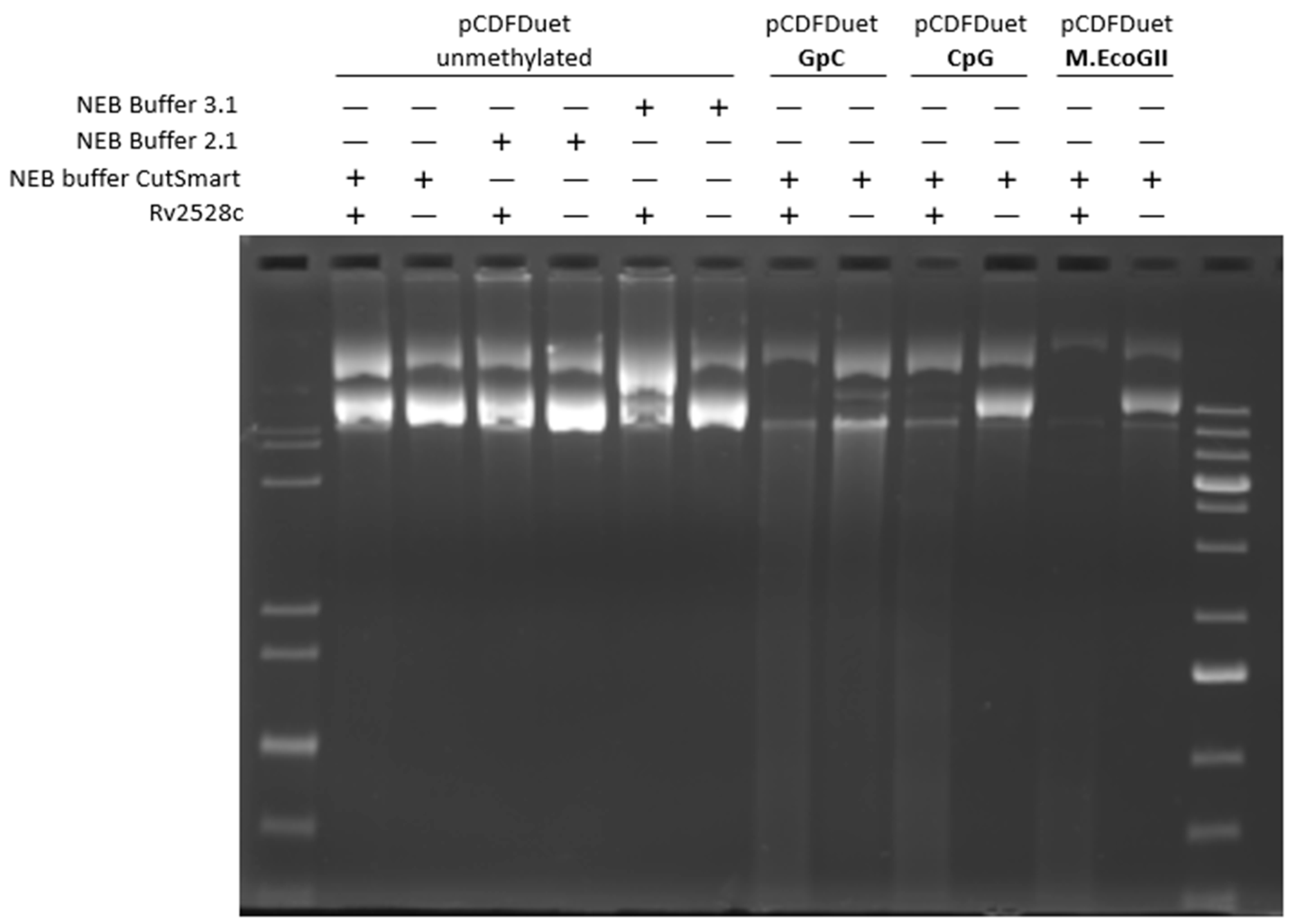
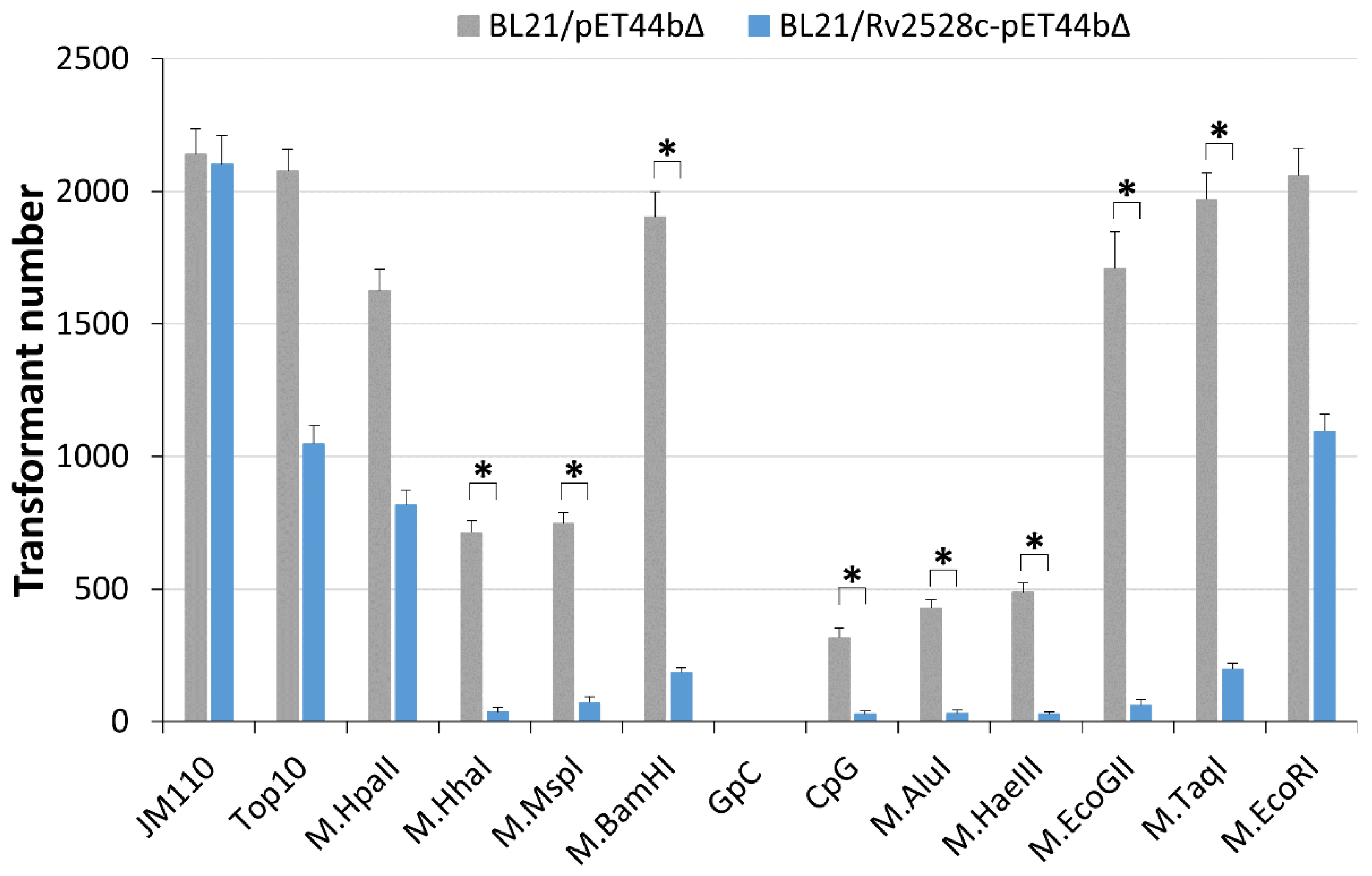
| Strain/MTase | Modification Site | Site Number in pRSFDuet | Sites Overlapping with EcoBLI (GCNGC) |
|---|---|---|---|
| Strains | |||
| JM110 | AAC(N6)GTGC 1,2 | 0 | 0 |
| Top10 | GATC and CCWGG 2 | 10 and 9 | 0 |
| MTases | |||
| M.HpaII | CCGG | 20 | 0 |
| M.HhaI | GCGC | 28 | 0 |
| M.MspI | CCGG | 20 | 0 |
| M.BamHI 3 | GGATCC | 1 | 0 |
| GpC | GC | 288 | 27 |
| CpG | CG | 258 | 12 |
| M.AluI | AGCT | 17 | 3 |
| M.HaeIII | GGCC | 12 | 4 |
| M.EcoGII | A | 979 | 0 |
| M.TaqI | TCGA | 15 | 0 |
| M.EcoRI | GAATTC | 1 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.; Wei, W.; Xu, M.; Ren, Q.; Liu, M.; Pan, X.; Feng, F.; Han, T.; Gou, L. The Restriction Activity Investigation of Rv2528c, an Mrr-like Modification-Dependent Restriction Endonuclease from Mycobacterium tuberculosis. Microorganisms 2024, 12, 1456. https://doi.org/10.3390/microorganisms12071456
Liu T, Wei W, Xu M, Ren Q, Liu M, Pan X, Feng F, Han T, Gou L. The Restriction Activity Investigation of Rv2528c, an Mrr-like Modification-Dependent Restriction Endonuclease from Mycobacterium tuberculosis. Microorganisms. 2024; 12(7):1456. https://doi.org/10.3390/microorganisms12071456
Chicago/Turabian StyleLiu, Tong, Wei Wei, Mingyan Xu, Qi Ren, Meikun Liu, Xuemei Pan, Fumin Feng, Tiesheng Han, and Lixia Gou. 2024. "The Restriction Activity Investigation of Rv2528c, an Mrr-like Modification-Dependent Restriction Endonuclease from Mycobacterium tuberculosis" Microorganisms 12, no. 7: 1456. https://doi.org/10.3390/microorganisms12071456
APA StyleLiu, T., Wei, W., Xu, M., Ren, Q., Liu, M., Pan, X., Feng, F., Han, T., & Gou, L. (2024). The Restriction Activity Investigation of Rv2528c, an Mrr-like Modification-Dependent Restriction Endonuclease from Mycobacterium tuberculosis. Microorganisms, 12(7), 1456. https://doi.org/10.3390/microorganisms12071456





