Occurrence and Persistence of Saccharomyces cerevisiae Population in Spontaneous Fermentation and the Relation with “Winery Effect”
Abstract
1. Introduction
2. Materials and Methods
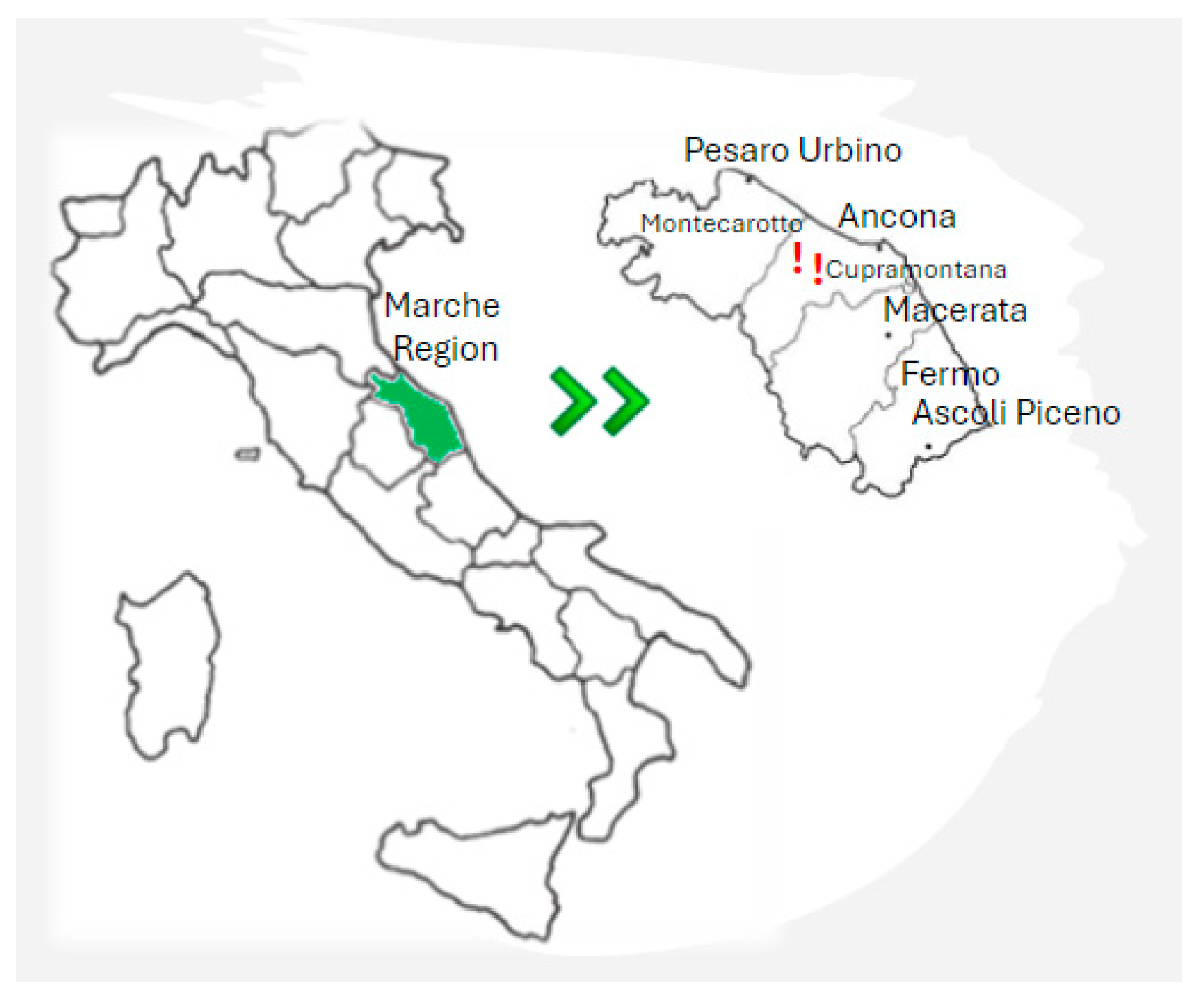
2.1. S. cerevisiae Strains Origin and Sampling
2.2. Genotyping Characterization of S. cerevisiae Strains
2.3. Analyses of Biotypes for H2S Production and Killer Activity
2.4. Fermentation Trials
2.5. Main Analytical Characters and Volatile Compounds of Microfermentation Trials
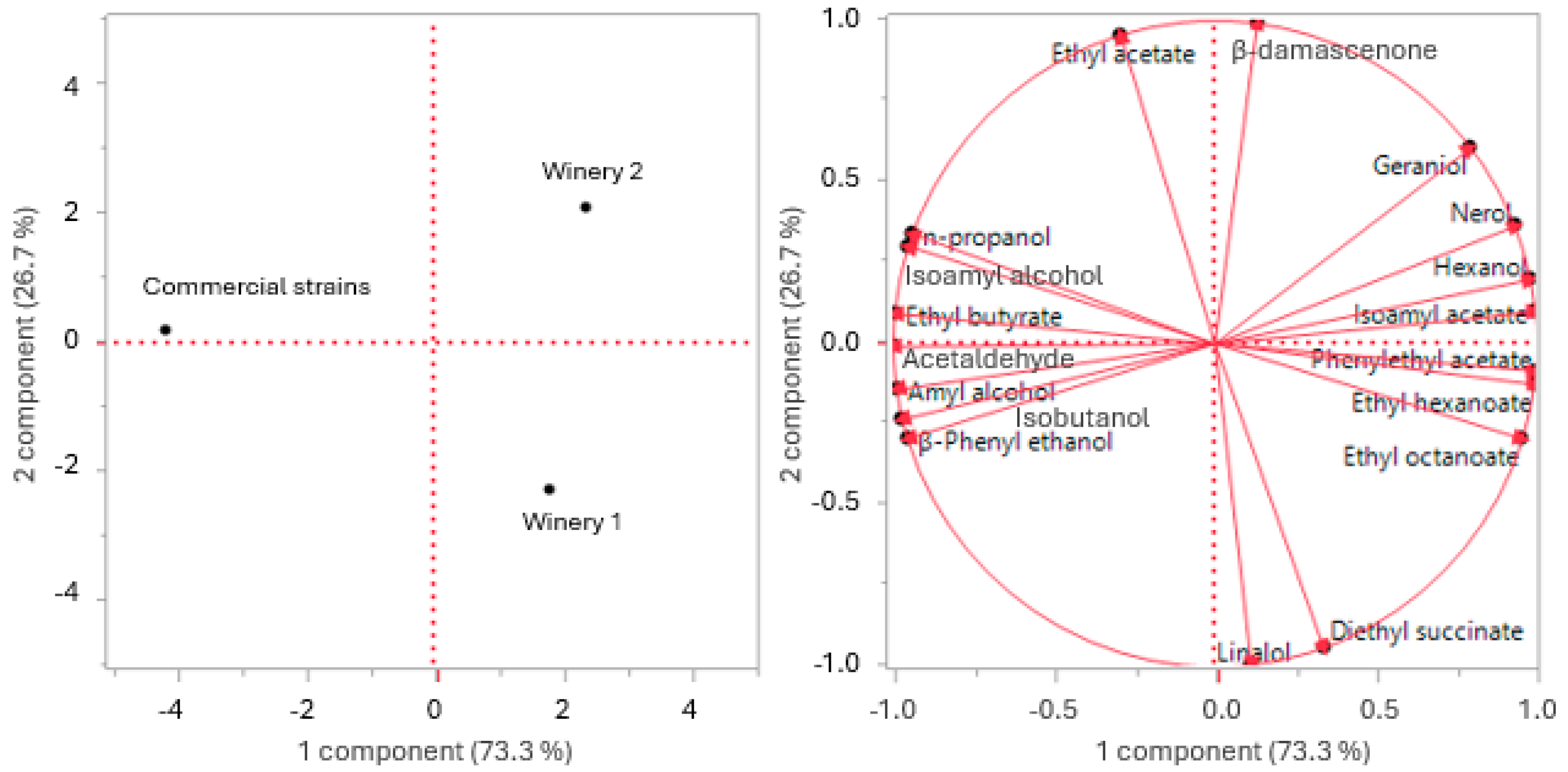
2.6. Data Analyses
3. Results
3.1. Genotyping, Occurrence, H2S Production and Killer Activity of S. cerevisiae Strains
3.2. Oenologically Characterization
3.2.1. Fermentation Rate, Volatile Acidity and Ethanol Production of Biotypes
3.2.2. Main By-Products of Fermentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Longo, E.; Cansado, J.; Agrelo, D.; Villa, T.G. Effect of Climatic Conditions on Yeast Diversity in Grape Musts from Northwest Spain. Am. J. Enol. Vitic. 1991, 42, 141–144. [Google Scholar] [CrossRef]
- Capece, A.; Pietrafesa, R.; Siesto, G.; Romaniello, R.; Condelli, N.; Romano, P. Selected Indigenous Saccharomyces cerevisiae Strains as Profitable Strategy to Preserve Typical Traits of Primitivo Wine. Fermentation 2019, 5, 87. [Google Scholar] [CrossRef]
- Pulcini, L.; Gamalero, E.; Costantini, A.; Vaudano, E.T.; Tsolakis, C.; Garcia-Moruno, E. An Overview on Saccharomyces cerevisiae Indigenous Strains Selection Methods. In Grapes and Wine; Intechopen: London, UK, 2022; p. 165. [Google Scholar]
- Morata, A.; Loira, I.; Del Fresno, J.M.; Escott, C.; Bañuelos, M.A.; Tesfaye, W.; González, C.; Palomero, F.; Suárez-Lepe, J.A. Strategies to Improve the Freshness in Wines from Warm Areas. In Advances in Grape and Wine Biotechnology; Morata, A., Iris, L., Eds.; InTech: London, UK, 2019. [Google Scholar]
- Capece, A.; Romaniello, R.; Siesto, G.; Romano, P. Diversity of Saccharomyces cerevisiae Yeasts Associated to Spontaneously Fermenting Grapes from an Italian “Heroic Vine-Growing Area”. Food Microbiol. 2012, 31, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Torrado, R.; Rantsiou, K.; Perrone, B.; Navarro-Tapia, E.; Querol, A.; Cocolin, L. Ecological Interactions among Saccharomyces cerevisiae Strains: Insight into the Dominance Phenomenon. Sci. Rep. 2017, 7, 43603. [Google Scholar] [CrossRef] [PubMed]
- Granchi, L.; Ganucci, D.; Buscioni, G.; Mangani, S.; Guerrini, S. The Biodiversity of Saccharomyces cerevisiae in Spontaneous Wine Fermentation: The Occurrence and Persistence of Winery-Strains. Fermentation 2019, 5, 86. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Comitini, F.; Ciani, M. Ecological Distribution and Oenological Characterization of Native Saccharomyces cerevisiae in an Organic Winery. Fermentation 2022, 8, 224. [Google Scholar] [CrossRef]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in Alcoholic Fermentation Processes: Role of Physiological Fitness and Microbial Interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef] [PubMed]
- Ganucci, D.; Guerrini, S.; Mangani, S.; Vincenzini, M.; Granchi, L. Quantifying the Effects of Ethanol and Temperature on the Fitness Advantage of Predominant Saccharomyces cerevisiae Strains Occurring in Spontaneous Wine Fermentations. Front. Microbiol. 2018, 9, 1563. [Google Scholar] [CrossRef]
- Börlin, M.; Venet, P.; Claisse, O.; Salin, F.; Legras, J.-L.; Masneuf-Pomarede, I. Cellar-Associated Saccharomyces cerevisiae Population Structure Revealed High-Level Diversity and Perennial Persistence at Sauternes Wine Estates. Appl. Environ. Microbiol. 2016, 82, 2909–2918. [Google Scholar] [CrossRef]
- Lappa, I.K.; Kachrimanidou, V.; Pateraki, C.; Koulougliotis, D.; Eriotou, E.; Kopsahelis, N. Indigenous Yeasts: Emerging Trends and Challenges in Winemaking. Curr. Opin. Food Sci. 2020, 32, 133–143. [Google Scholar] [CrossRef]
- Legras, J.-L.; Karst, F. Optimisation of Interdelta Analysis for Saccharomyces cerevisiae Strain Characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Ciani, M.; Comitini, F. Fitness of Selected Indigenous Saccharomyces cerevisiae Strains for White Piceno DOC Wines Production. Fermentation 2018, 4, 37. [Google Scholar] [CrossRef]
- Agarbati, A.; Canonico, L.; Comitini, F.; Ciani, M. Reduction of Sulfur Compounds through Genetic Improvement of Native Saccharomyces cerevisiae Useful for Organic and Sulfite-Free Wine. Foods 2020, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- Comitini, F.; Agarbati, A.; Canonico, L.; Galli, E.; Ciani, M. Purification and Characterization of WA18, a New Mycocin Produced by Wickerhamomyces anomalus Active in Wine Against Brettanomyces bruxellensis Spoilage Yeasts. Microorganisms 2020, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Volatile Acidity (Type-I)|OIV. Available online: https://www.oiv.int/standards/annex-a-methods-of-analysis-of-wines-and-musts/section-3-chemical-analysis/section-3-1-organic-compounds/section-3-1-3-acids/volatile-acidity-%28type-i%29 (accessed on 5 June 2024).
- Total Acidity (Type-I)|OIV. Available online: https://www.oiv.int/standards/annex-a-methods-of-analysis-of-wines-and-musts/section-3-chemical-analysis/section-3-1-organic-compounds/section-3-1-3-acids/total-acidity-%28type-i%29 (accessed on 5 June 2024).
- Determination of Total Ethanol in Wine by High-Performance Liquid Chromatography (Type-IV)|OIV. Available online: https://www.oiv.int/standards/annex-a-methods-of-analysis-of-wines-and-musts/section-3-chemical-analysis/section-3-1-organic-compounds/section-3-1-3-acids/determination-of-total-ethanol-in-wine-by-high-performance-liquid-chromatography-%28type-iv%29 (accessed on 5 June 2024).
- pH (Type-I)|OIV. Available online: https://www.oiv.int/standards/annex-a-methods-of-analysis-of-wines-and-musts/section-3-chemical-analysis/section-3-1-organic-compounds/section-3-1-3-acids/ph-%28type-i%29 (accessed on 5 June 2024).
- Sulphur Dioxide (Type-IV)|OIV. Available online: https://www.oiv.int/standards/compendium-of-international-methods-of-wine-and-must-analysis/annex-f/annex-f-specific-methods-for-the-analysis-of-grape-sugar/sulphur-dioxide-%28type-iv%29 (accessed on 5 June 2024).
- Dukes, B.C.; Butzke, C.E. Rapid Determination of Primary Amino Acids in Grape Juice Using an O-Phthaldialdehyde/N-Acetyl-L-Cysteine Spectrophotometric Assay. Am. J. Enol. Vitic. 1998, 49, 125–134. [Google Scholar] [CrossRef]
- Canonico, L.; Comitini, F.; Ciani, M. Influence of Vintage and Selected Starter on Torulaspora delbrueckii/Saccharomyces cerevisiae Sequential Fermentation. Eur. Food Res. Technol. 2015, 241, 827–833. [Google Scholar] [CrossRef]
- Canonico, L.; Ciani, E.; Galli, E.; Comitini, F.; Ciani, M. Evolution of Aromatic Profile of Torulaspora delbrueckii Mixed Fermentation at Microbrewery Plant. Fermentation 2020, 6, 7. [Google Scholar] [CrossRef]
- Gao, J.; Wang, M.; Huang, W.; You, Y.; Zhan, J. Indigenous Saccharomyces cerevisiae Could Better Adapt to the Physicochemical Conditions and Natural Microbial Ecology of Prince Grape Must Compared with Commercial Saccharomyces cerevisiae FX10. Molecules 2022, 27, 6892. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, H. Wine Yeast Terroir: Separating the Wheat from the Chaff—For an Open Debate. Microorganisms 2020, 8, 787. [Google Scholar] [CrossRef]
- Pretorius, I.S. Tasting the Terroir of Wine Yeast Innovation. FEMS Yeast Res. 2020, 20, foz084. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial Biogeography of Wine Grapes Is Conditioned by Cultivar, Vintage, and Climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional Microbial Signatures Positively Correlate with Differential Wine Phenotypes: Evidence for a Microbial Aspect to Terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef]
- Vigentini, I.; De Lorenzis, G.; Fabrizio, V.; Valdetara, F.; Faccincani, M.; Panont, C.A.; Picozzi, C.; Imazio, S.; Failla, O.; Foschino, R. The Vintage Effect Overcomes the Terroir Effect: A Three Year Survey on the Wine Yeast Biodiversity in Franciacorta and Oltrepò Pavese, Two Northern Italian Vine-Growing Areas. Microbiology 2015, 161, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Torija, M.J.; Rozes, N.; Poblet, M.; Guillamon, J.M.; Mas, A. Yeast Population Dynamics in Spontaneous Fermentations: Comparison between Two Different Wine-Producing Areas over a Period of Three Years. Antonie Van Leeuwenhoek 2001, 79, 345–352. [Google Scholar] [CrossRef]
- Carrau, F.; Boido, E.; Ramey, D. Yeasts for Low Input Winemaking: Microbial Terroir and Flavor Differentiation. Adv. Appl. Microbiol. 2020, 111, 89–121. [Google Scholar] [CrossRef]
- López-Enríquez, L.; Vila-Crespo, J.; Rodríguez-Nogales, J.M.; Fernández-Fernández, E.; Ruipérez, V. Non-Saccharomyces Yeasts from Organic Vineyards as Spontaneous Fermentation Agents. Foods 2023, 12, 3644. [Google Scholar] [CrossRef]
- Castrillo, D.; Blanco, P. Characterization of Indigenous Non-Saccharomyces Yeast Strains with Potential Use in Winemaking. Front. Biosci.-Elite 2023, 15, 1. [Google Scholar] [CrossRef]
- Rosini, G. Assessment of Dominance of Added Yeast in Wine Fermentation and Origin of Saccharomyces cerevisiae in Wine-Making. J. Gen. Appl. Microbiol. 1984, 30, 249–256. [Google Scholar] [CrossRef]
- Cocolin, L.; Pepe, V.; Comitini, F.; Comi, G.; Ciani, M. Enological and Genetic Traits of Saccharomyces cerevisiae Isolated from Former and Modern Wineries. FEMS Yeast Res. 2004, 5, 237–245. [Google Scholar] [CrossRef][Green Version]
- Sabate, J.; Cano, J.; Esteve-Zarzoso, B.; Guillamón, J.M. Isolation and Identification of Yeasts Associated with Vineyard and Winery by RFLP Analysis of Ribosomal Genes and Mitochondrial DNA. Microbiol. Res. 2002, 157, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Lopes, C.A.; Van Broock, M.; Querol, A.; Caballero, A.C. Saccharomyces cerevisiae Wine Yeast Populations in a Cold Region in Argentinean Patagonia. A Study at Different Fermentation Scales. J. Appl. Microbiol. 2002, 93, 608–615. [Google Scholar] [CrossRef]
- Beltran, G.; Torija, M.J.; Novo, M.; Ferrer, N.; Poblet, M.; Guillamón, J.M.; Rozès, N.; Mas, A. Analysis of Yeast Populations during Alcoholic Fermentation: A Six Year Follow-up Study. Syst. Appl. Microbiol. 2002, 25, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Le Jeune, C.; Erny, C.; Demuyter, C.; Lollier, M. Evolution of the Population of Saccharomyces cerevisiae from Grape to Wine in a Spontaneous Fermentation. Food Microbiol. 2006, 23, 709–716. [Google Scholar] [CrossRef] [PubMed]
- Abdo, H.; Catacchio, C.R.; Ventura, M.; D’Addabbo, P.; Calabrese, F.M.; Laurent, J.; David-Vaizant, V.; Alexandre, H.; Guilloux-Bénatier, M.; Rousseaux, S. Colonization of Wild Saccharomyces cerevisiae Strains in a New Winery. Beverages 2020, 6, 9. [Google Scholar] [CrossRef]
- Genetic Diversity of Wine Saccharomyces cerevisiae Strains in an Experimental Winery from Galicia (NW Spain). Available online: http://ouci.dntb.gov.ua/en/works/9Jg2PNA7/ (accessed on 6 June 2024).
- Gutierrez, A.R.; Santamaria, P.; Epifanio, S.; Garijo, P.; Opez, R.L. Ecology of Spontaneous Fermentation in One Winery during 5 Consecutive Years. Lett. Appl. Microbiol. 1999, 29, 411–415. [Google Scholar] [CrossRef]
- Stefanini, I.; Albanese, D.; Cavazza, A.; Franciosi, E.; De Filippo, C.; Donati, C.; Cavalieri, D. Dynamic Changes in Microbiota and Mycobiota during Spontaneous ‘V Ino S Anto T Rentino’ Fermentation. Microb. Biotechnol. 2016, 9, 195–208. [Google Scholar] [CrossRef]
- Jouhten, P.; Ponomarova, O.; Gonzalez, R.; Patil, K.R. Saccharomyces cerevisiae Metabolism in Ecological Context. FEMS Yeast Res. 2016, 16, fow080. [Google Scholar] [CrossRef]
- Conacher, C.; Luyt, N.; Naidoo-Blassoples, R.; Rossouw, D.; Setati, M.; Bauer, F. The Ecology of Wine Fermentation: A Model for the Study of Complex Microbial Ecosystems. Appl. Microbiol. Biotechnol. 2021, 105, 3027–3043. [Google Scholar] [CrossRef]
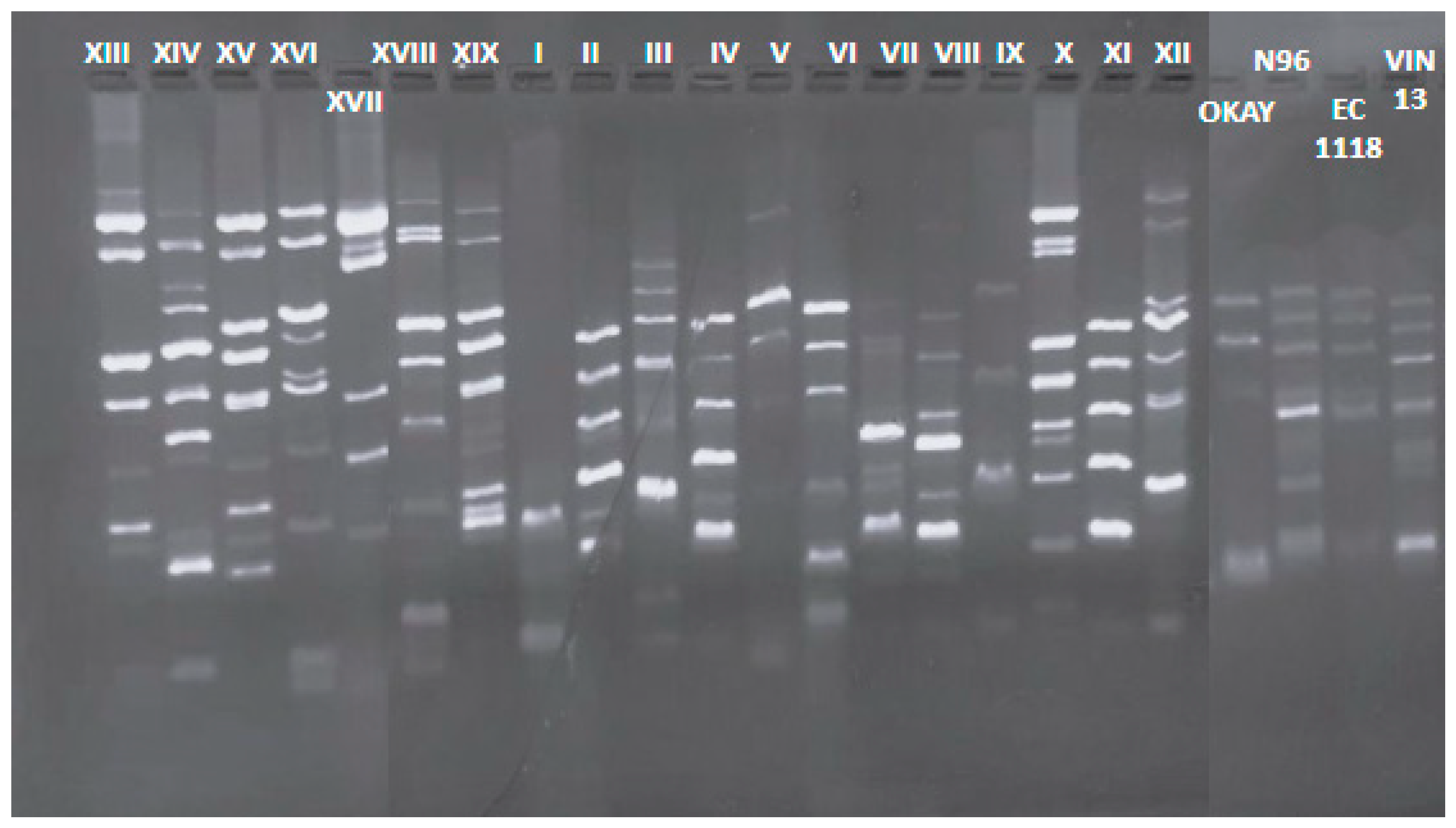
| Winery | Biotype | Frequency (%) | H2S Production | Killer Activity |
|---|---|---|---|---|
| 1 | XIII * | 49.5 | 2.0 | ± |
| XIV * | 43.5 | 0.0 | + | |
| XV | 1.0 | 1.5 | + | |
| XVI * | 2.0 | 0.0 | + | |
| XVII | 1.0 | 0.5 | + | |
| XVIII | 1.0 | 0.5 | ± | |
| XIX | 2.0 | 0.8 | + | |
| 2 | I | 23.4 | 0.5 | + |
| II | 15.0 | 0.0 | − | |
| III | 16.8 | 0.0 | + | |
| IV * | 19.6 | 1.0 | + | |
| V | 2.8 | 0.5 | ± | |
| VI | 7.5 | 0.5 | + | |
| VII | 1.9 | 1.5 | ± | |
| VIII | 1.9 | 0.8 | ± | |
| IX | 3.7 | 0.0 | ± | |
| X | 0.9 | 1.0 | + | |
| XI | 5.6 | 1.0 | + | |
| XII | 0.9 | 1.0 | ± |
| Strains Population | Fermentation Rate (gCO2/day) * | Volatile Acidity (Acetic Acid g/L) | Ethanol (% v/v) |
|---|---|---|---|
| Winery 1 | 1.4 ± 0.2 a | 0.4 ± 0.1 a | 12.8 ± 0.3 a |
| Winery 2 | 1.2 ± 0.1 ab | 0.5 ± 0.1 a | 12.4 ± 0.3 a |
| OKAY® | 0.9 ± 0.0 c | 0.4 ± 0.0 a | 12.4 ± 0.0 a |
| N96 | 1.1 ± 0.0 bc | 0.5 ± 0.0 a | 12.4 ± 0.1 a |
| Fermentation By-Products | Winery 1 | Winery 2 | OKAY® | N96 |
|---|---|---|---|---|
| Alcohols | ||||
| Hexanol (mg/L) | 11.84 ± 0.99 a | 12.66 ± 1.77 a | 10.38 ± 0.61 ab | 9.13 ± 0.74 b |
| β-Phenyl ethanol (mg/L) | 7.48 ± 1.12 a | 7.39 ± 1.11 a | 7.96 ± 0.03 b | 7.32 ± 0.00 b |
| n-propanol (mg/L) | 12.60 ± 8.26 b | 16.88 ± 2.96 b | 42.70 ± 0.09 a | 13.51 ± 0.42 b |
| Amyl alcohol (mg/L) | 7.28 ± 4.44 b | 4.85 ± 2.39 b | 15.42 ± 0.39 a | 15.55 ± 0.38 a |
| Isoamyl alcohol (mg/L) | 36.96 ± 10.7 b | 41.60 ± 12.07 b | 52.01 ± 0.17 ab | 60.78 ± 0.37 a |
| Isobutanol (mg/L) | 5.22 ± 3.94 a | 8.24 ± 1.78 a | 5.29 ± 0.03 a | 7.89 ± 0.04 a |
| Carbonyl compounds | ||||
| Acetaldehyde (mg/L) | 15.59 ± 9.85 b | 12.21 ± 2.06 b | 15.05 ± 0.47 b | 80.97 ± 0.23 a |
| Esters | ||||
| Isoamyl acetate (mg/L) | 0.80 ± 0.39 ab | 0.90 ± 0.20 a | 0.39 ± 0.00 b | 0.35 ± 0.01 b |
| Phenyl ethyl acetate (mg/L) | 0.47 ± 0.16 a | 0.47 ± 0.09 a | 0.21 ± 0.01 b | 0.38 ± 0.02 ab |
| Ethyl hexanoate (mg/L) | 0.65 ± 0.19 a | 0.63 ± 0.22 a | 0.39 ± 0.00 ab | 0.16 ± 0.01 b |
| Ethyl butyrate (mg/L) | 0.03 ± 0.03 a | 0.03 ± 0.05 a | 0.06 ± 0.00 a | 0.05 ± 0.01 a |
| Ethyl octanoate (µg/L) | 3.75 ± 1.04 a | 3.60 ± 1.13 a | 3.45 ± 0.34 a | 2.78 ± 0.05 a |
| Diethyl succinate (mg/L) | 0.04 ± 0.01 b | 0.03 ± 0.01 b | 0.01 ± 0.00 c | 0.06 ± 0.00 a |
| Ethyl acetate (mg/L) | 10.16 ± 3.34 b | 11.61 ± 4.61 ab | 17.62 ± 0.20 a | 5.13 ± 0.03 b |
| Terpenes | ||||
| Linalol (µg/L) | 17.85 ± 8.96 a | 7.12 ± 5.04 a | 8.89 ± 0.53 a | 12.51 ± 1.07 a |
| Nerol (µg/L) | 3.41 ± 1.48 ab | 4.56 ± 1.50 a | 1.95 ± 0.50 b | 1.88 ± 0.39 b |
| Geraniol (µg/L) | 4.71 ± 1.58 a | 5.69 ± 1.53 a | 4.02 ± 0.57 a | 4.49 ± 0.47 a |
| Enones | ||||
| β-Damascenone (µg/L) | 4.56 ± 2.05 a | 4.15 ± 1.41 a | 5.89 ± 0.34 a | 4.98 ± 0.28 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agarbati, A.; Comitini, F.; Ciani, M.; Canonico, L. Occurrence and Persistence of Saccharomyces cerevisiae Population in Spontaneous Fermentation and the Relation with “Winery Effect”. Microorganisms 2024, 12, 1494. https://doi.org/10.3390/microorganisms12071494
Agarbati A, Comitini F, Ciani M, Canonico L. Occurrence and Persistence of Saccharomyces cerevisiae Population in Spontaneous Fermentation and the Relation with “Winery Effect”. Microorganisms. 2024; 12(7):1494. https://doi.org/10.3390/microorganisms12071494
Chicago/Turabian StyleAgarbati, Alice, Francesca Comitini, Maurizio Ciani, and Laura Canonico. 2024. "Occurrence and Persistence of Saccharomyces cerevisiae Population in Spontaneous Fermentation and the Relation with “Winery Effect”" Microorganisms 12, no. 7: 1494. https://doi.org/10.3390/microorganisms12071494
APA StyleAgarbati, A., Comitini, F., Ciani, M., & Canonico, L. (2024). Occurrence and Persistence of Saccharomyces cerevisiae Population in Spontaneous Fermentation and the Relation with “Winery Effect”. Microorganisms, 12(7), 1494. https://doi.org/10.3390/microorganisms12071494








