Genetic Characteristics of Multidrug-Resistant Salmonella Isolated from Poultry Meat in South Korea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Salmonella Isolation and Identification
2.3. Antimicrobial Susceptibility
2.4. WGS Analysis
2.5. Serotyping and Homology Analysis
2.6. In Silico Characterization of WGS
2.7. Nucleotide Sequence Accession Numbers
3. Results
3.1. Prevalence of Salmonella and MDR Salmonella in Poultry Meat Samples
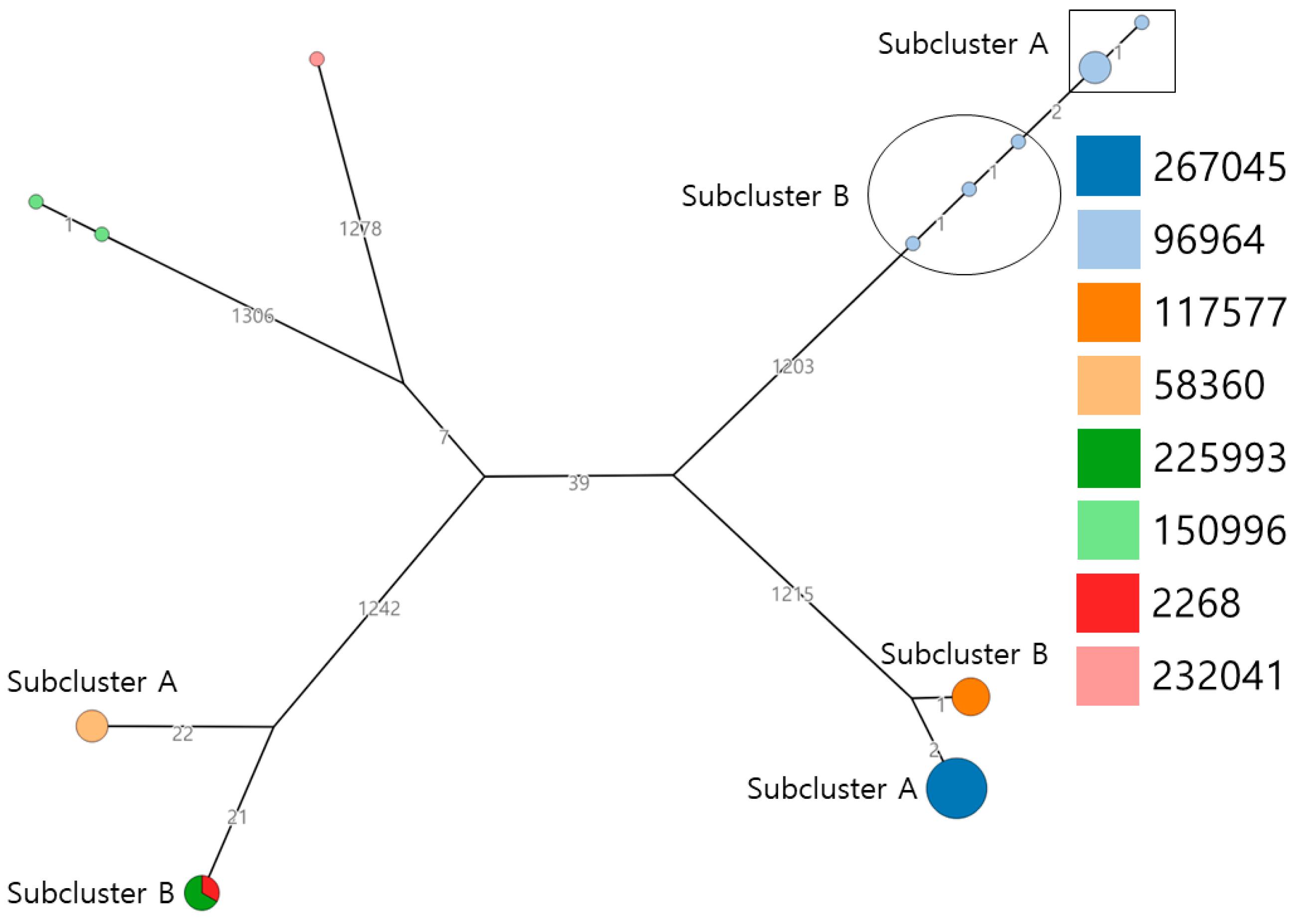
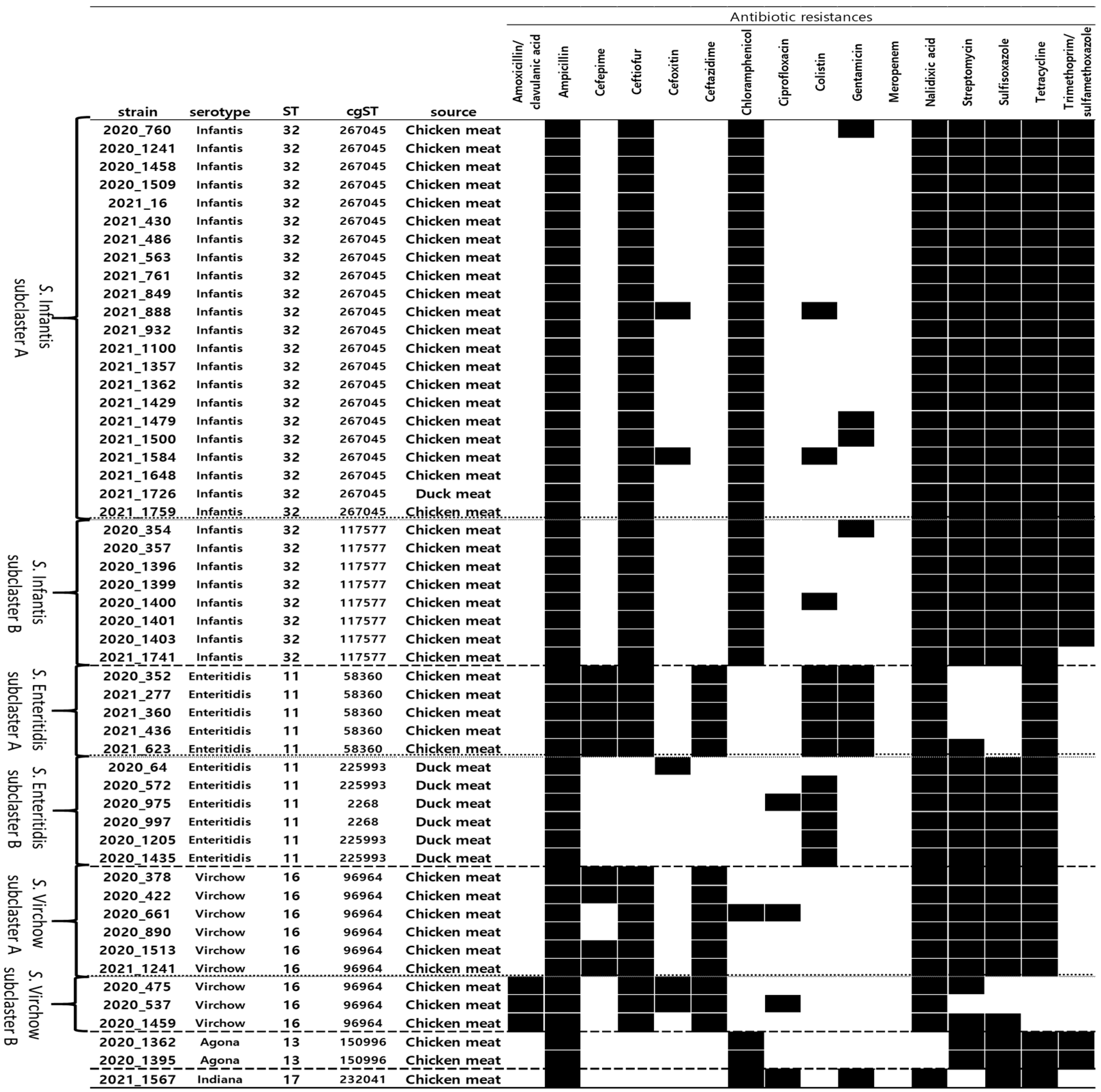
3.2. Serotyping and Phylogenetic Analysis
3.3. Antimicrobial Resistance Patterns of MDR Salmonella
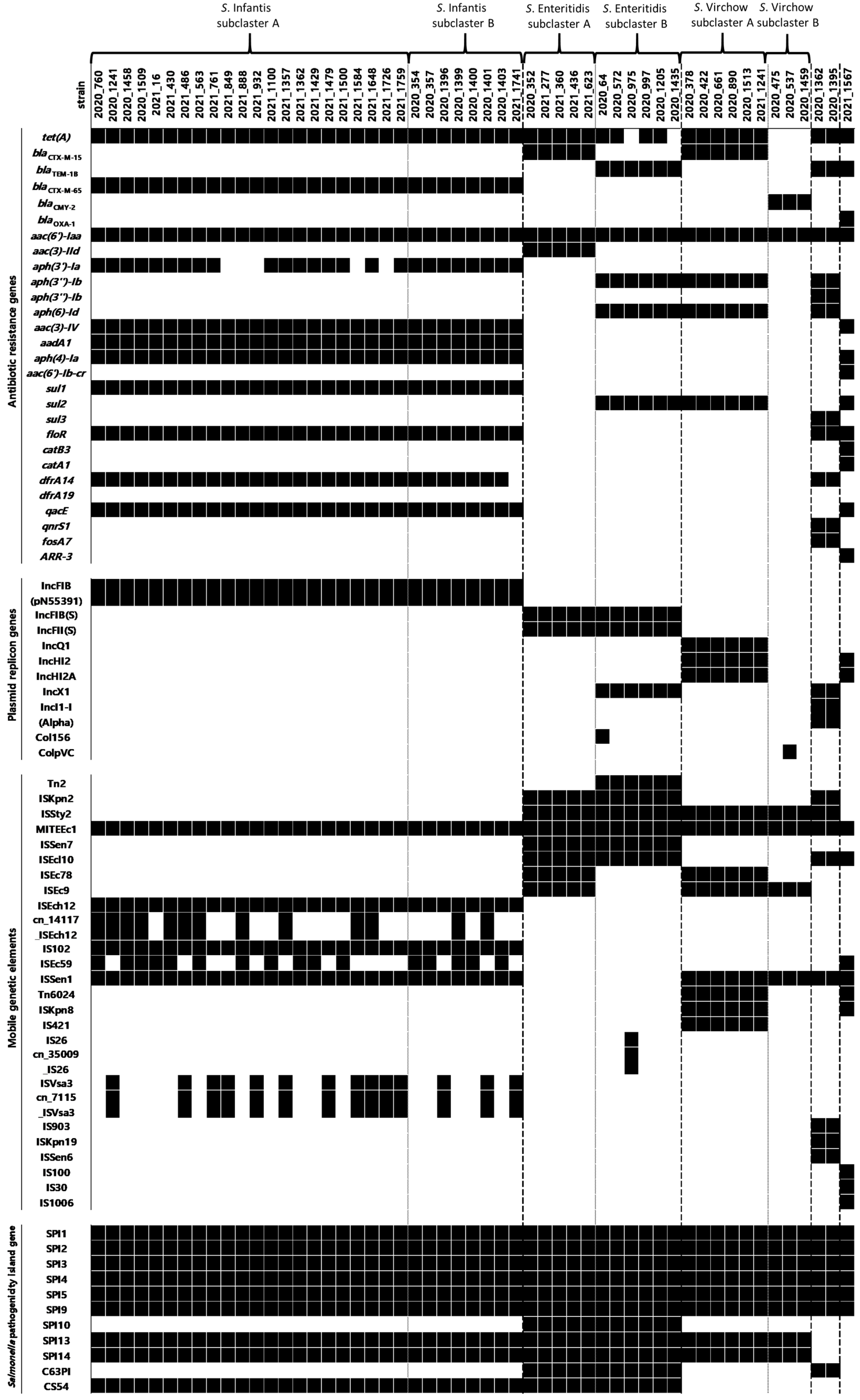
3.4. Detection of Antimicrobial Resistance Genes, Plasmid Genes, Salmonella Pathogenicity Island, and Mobile Genetic Elements
3.5. Detection of Virulence Factor Genes
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Salmonella (Non-Typhoidal). Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 20 February 2018).
- Popa, G.L.; Papa, M.I. Salmonella spp. infection-a continuous threat worldwide. GERMS 2021, 11, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Food and Drug Safety. Food Safety Korea. Available online: https://www.foodsafetykorea.go.kr/portal/healthyfoodlife/foodPoisoningStat.do?menu_no=4425&menu_grp=MENU_NEW02 (accessed on 5 February 2024).
- Tiseo, K.; Huber, L.; Gilbert, M.; Robinson, T.P.; Van Boeckel, T.P. Global trends in antimicrobial use in food animals from 2017 to 2030. Antibiotics 2020, 9, 918. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Feng, J.; Liu, J.; Fu, W.; Li, X.; Li, B. Deciphering of microbial community and antibiotic resistance genes in activated sludge reactors under high selective pressure of different antibiotics. Water Res. 2019, 151, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Bezabih, Y.M.; Sabiiti, W.; Alamneh, E.; Bezabih, A.; Peterson, G.M.; Bezabhe, W.M.; Roujeinikova, A. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J. Antimicrob. Chemother. 2021, 76, 22–29. [Google Scholar] [CrossRef]
- Khoshbakht, R.; Derakhshandeh, A.; Jelviz, L.; Azhdari, F. Tetracycline resistance genes in Salmonella enterica serovars with animal and human origin. Int. J. Enteric Pathog. 2018, 6, 60–64. [Google Scholar] [CrossRef]
- Loharikar, A.; Vawter, S.; Warren, K.; Deasy, M.; Moll, M.; Sandt, C.; Gilhousen, R.; Villamil, E.; Rhorer, A.; Briere, E.; et al. Outbreak of human Salmonella Typhimurium infections linked to contact with baby poultry from a single agricultural feed store chain and mail-order hatchery, 2009. Pediatr. Infect. Dis. J. 2013, 32, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, K.O.; Al-Reshoodi, F.M.; Alshdokhi, E.A.; Alhamed, A.S.; Al Hadlaq, M.A.; Mujallad, M.I.; Mukhtar, L.E.; Alsufyani, A.T.; Alajlan, A.A.; Al Rashidy, M.S.; et al. Antimicrobial resistance and genomic characterization of Salmonella enterica isolates from chicken meat. Front. Microbiol. 2023, 14, 1104164. [Google Scholar] [CrossRef]
- Hyeon, J.Y.; Li, S.; Mann, D.A.; Zhang, S.; Kim, K.J.; Lee, D.H.; Deng, X.; Song, C.S. Whole-genome sequencing analysis of Salmonella Enterica serotype Enteritidis isolated from poultry sources in South Korea, 2010–2017. Pathogens 2021, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- La, T.M.; Kim, T.; Lee, H.J.; Lee, J.B.; Park, S.Y.; Choi, I.S.; Lee, S.W. Whole-genome analysis of multidrug-resistant Salmonella Enteritidis strains isolated from poultry sources in Korea. Pathogens 2021, 10, 1615. [Google Scholar] [CrossRef]
- Ministry of Food and Drug Safety. Food Code. Available online: https://various.foodsafetykorea.go.kr/fsd/#/ (accessed on 5 February 2024).
- CLSI. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twentieth Informational Supplement; CLSI Document m100; CLSI: Wayne, PA, USA, 2021. [Google Scholar]
- NARMS. National Antimicrobial Resistance Monitoring System. NARMS Integrated Report: The National Antimicrobial Resistance Monitoring System: Enteric Bacteria; U.S. Food and Drug Administration: Rockville, MD, USA, 2019.
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Deatherage Kaiser, B.L.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella Serotype Determination Utilizing High-throughput Genome Sequencing Data. J. Clin. Mircobiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef]
- Larsen, M.V.; Cosentino, S.; Rasmussen, S.; Friis, C.; Hasman, H.; Marvig, R.L.; Jelsbak, L.; Sicheritz-Pontén, T.; Ussery, D.W.; Aarestrup, F.M.; et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012, 50, 1355–1361. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Kürekci, C.; Sahin, S.; Iwan, E.; Kwit, R.; Bomba, A.; Wasyl, D. Whole-genome sequence analysis of Salmonella Infantis isolated from raw chicken meat samples and insights into pESI-like megaplasmid. Int. J. Food Microbiol. 2021, 337, 108956. [Google Scholar] [CrossRef]
- McMillan, E.A.; Weinroth, M.D.; Frye, J.G. Increased Prevalence of Salmonella Infantis Isolated from Raw Chicken and Turkey Products in the United States Is Due to a Single Clonal Lineage Carrying the pESI Plasmid. Microorganisms 2022, 10, 1478. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.M.; Montenegro, R.B.; Conejeros, J.; Rivera, D.; Undurraga, E.A.; Switt, A.I.M. A review of the global emergence of multidrug-resistant Salmonella enterica subsp. enterica Serovar Infantis. Int. J. Food Microbiol. 2023, 403, 110297. [Google Scholar] [CrossRef]
- Drauch, V.; Mitra, T.; Liebhart, D.; Hess, M.; Hess, C. Infection dynamics of Salmonella Infantis vary considerably between chicken lines. Avian Pathol. 2022, 51, 561–573. [Google Scholar] [CrossRef]
- Kim, M.B.; Jung, H.R.; Lee, Y.J. Emergence of Salmonella Infantis carrying the pESI megaplasmid in commercial farms of five major integrated broiler operations in Korea. Poult. Sci. 2024, 103, 103516. [Google Scholar] [CrossRef]
- Kim, M.B.; Lee, Y.J. Emergence of Salmonella Infantis carrying the pESI-like plasmid from eggs in egg grading and packing plants in Korea. Food Microbiol. 2024, 122, 104568. [Google Scholar] [CrossRef]
- Jung, H.R.; Lee, Y.J. Prevalence and characterization of non-typhoidal Salmonella in egg from grading and packing plants in Korea. Food Microbiol. 2024, 120, 104464. [Google Scholar] [CrossRef] [PubMed]
- Alba, P.; Leekitcharoenphon, P.; Carfora, V.; Amoruso, R.; Cordaro, G.; Di Matteo, P.; Ianzano, A.; Iurescia, M.; Diaconu, E.L.; ENGAGE-EURL-AR Network Study Group; et al. Molecular epidemiology of Salmonella Infantis in Europe: Insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb. Genom. 2020, 6, e000365. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.; Bae, Y.; Lee, Y.S.; Kang, D.H.; Kim, S.H. Prevalence and characteristics of Salmonella spp. isolated from raw chicken meat in the Republic of Korea. J. Microbiol. Biotechnol. 2022, 32, 1307–1314. [Google Scholar] [CrossRef] [PubMed]
- Mechesso, A.F.; Moon, D.C.; Kim, S.J.; Song, H.J.; Kang, H.Y.; Na, S.H.; Choi, J.H.; Kim, H.Y.; Yoon, S.S.; Lim, S.K. Nationwide surveillance on serotype distribution and antimicrobial resistance profiles of non-typhoidal Salmonella serovars isolated from food-producing animals in South Korea. Int. J. Food Microbiol. 2020, 335, 108893. [Google Scholar] [CrossRef]
- Sin, M.; Yoon, S.; Kim, Y.B.; Noh, E.B.; Seo, K.W.; Lee, Y.J. Molecular characteristics of antimicrobial resistance determinants and integrons in Salmonella isolated from chicken meat in Korea. J. Appl. Poult. Res. 2020, 29, 502–514. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control, European Food Safety Authority. Multi-country outbreak of Salmonella Virchow ST16 infections linked to the consumption of meat products containing chicken meat. EFSA Support. Publ. 2023, 20, 7983E. [Google Scholar]
- Yang, X.; Wu, Q.; Zhang, J.; Huang, J.; Chen, L.; Wu, S.; Zeng, H.; Wang, J.; Chen, M.; Wu, H.; et al. Prevalence, bacterial load, and antimicrobial resistance of Salmonella serovars isolated from retail meat and meat products in China. Front. Microbiol. 2019, 10, 2121. [Google Scholar] [CrossRef] [PubMed]
- García-Soto, S.; Abdel-Glil, M.Y.; Tomaso, H.; Linde, J.; Methner, U. Emergence of multidrug-resistant Salmonella enterica Subspecies enterica serovar Infantis of multilocus sequence Type 2283 in German broiler farms. Front. Microbiol. 2020, 11, 1741. [Google Scholar] [CrossRef]
- Lee, K.Y.; Atwill, E.R.; Pitesky, M.; Huang, A.; Lavelle, K.; Rickard, M.; Shafii, M.; Hung-Fan, M.; Li, X. Antimicrobial resistance profiles of non-typhoidal Salmonella from retail meat products in California, 2018. Front. Microbiol. 2022, 13, 835699. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, H.; Kim, H.; Jeon, J.H.; Kim, S.; Park, Y.; Kim, S.H. Genetic Characteristics of Extended-Spectrum Beta-Lactamase-Producing Salmonella Isolated from Retail Meats in South Korea. J. Microbiol. Biotechnol. 2024, 34, 1101–1108. [Google Scholar] [CrossRef]
- Chu, Y.; Wang, D.; Hao, W.; Sun, R.; Sun, J.; Liu, Y.; Liao, X. Prevalence, antibiotic resistance, virulence genes and molecular characteristics of Salmonella isolated from ducks and wild geese in China. Food Microbiol. 2024, 118, 104423. [Google Scholar] [CrossRef]
- Xiao, Y.; Hu, Y. The major aminoglycoside-modifying enzyme AAC(3)-II found in Escherichia coli determines a significant disparity in its resistance to gentamicin and amikacin in China. Microb. Drug Resist. 2012, 18, 42–46. [Google Scholar] [CrossRef]
- Li, Y.; Kang, X.; Ed-Dra, A.; Zhou, X.; Jia, C.; Müller, A.; Liu, Y.; Kehrenberg, C.; Yue, M. Genome-Based assessment of antimicrobial resistance and virulence potential of isolates of non-pullorum/gallinarum Salmonella serovars recovered from dead poultry in China. Microbiol. Spectr. 2022, 10, e0096522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Z.; Lei, C.W.; Zeng, J.X.; Chen, Y.P.; Kang, Z.Z.; Wang, Y.L.; Ye, X.L.; Zhai, X.W.; Wang, H.N. An IncX1 plasmid isolated from Salmonella enterica subsp. enterica serovar Pullorum carrying blaTEM-1B, sul2, arsenic resistant operons. Plasmid 2018, 100, 14–21. [Google Scholar]
- Fortini, D.; Owczarek, S.; Dionisi, A.M.; Lucarelli, C.; Arena, S.; Carattoli, A.; Enter-Net Italia Colistin Resistance Study Group; Villa, L.; García-Fernández, A. Colistin Resistance Mechanisms in Human Salmonella enterica Strains Isolated by the National Surveillance Enter-Net Italia (2016–2018). Antibiotics 2022, 11, 102. [Google Scholar] [CrossRef]
- Chiu, C.H.; Lee, J.J.; Wang, M.H.; Chu, C. Genetic analysis and plasmid-mediated blaCMY-2 in Salmonella and Shigella and the Ceftriaxone Susceptibility regulated by the ISEcp-1 tnpA-blaCMY-2-blc-sugE. J. Microbiol. Immunol. Infect. 2021, 54, 649–657. [Google Scholar] [CrossRef]
- Shigemura, H.; Maeda, T.; Nakayama, S.; Ohishi, A.; Carle, Y.; Ookuma, E.; Etoh, Y.; Hirai, S.; Matsui, M.; Kimura, H.; et al. Transmission of extended-spectrum cephalosporin-resistant Salmonella harboring a blaCMY-2-carrying IncA/C2 plasmid chromosomally integrated by ISEcp1 or IS26 in layer breeding chains in Japan. J. Vet. Med. Sci. 2021, 83, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Qazi, I.H.; Wang, L.; Zhou, G.; Han, H. Salmonella virulence and immune escape. Microorganisms 2020, 8, 407. [Google Scholar] [CrossRef]
- Shah, D.H.; Lee, M.; Park, J.; Eo, S.; Kwon, J.; Chae, J. Identification of Salmonella gallinarum virulence genes in a chicken infection model using PCR-based signature-tagged mutagenesis. Microbiology 2005, 151, 3957–3968. [Google Scholar] [CrossRef]
- Aviv, G.; Elpers, L.; Mikhlin, S.; Cohen, H.; Vitman Zilber, S.; Grassl, G.A.; Rahav, G.; Hensel, M.; Gal-Mor, O. The plasmid-encoded Ipf and Klf fimbriae display different expression and varying roles in the virulence of Salmonella enterica serovar Infantis in mouse vs. avian hosts. PLoS Pathog. 2017, 13, e1006559. [Google Scholar] [CrossRef]
- Diamant, I.; Adani, B.; Sylman, M.; Rahav, G.; Gal-Mor, O. The transcriptional regulation of the horizontally acquired iron uptake system, yersiniabactin and its contribution to oxidative stress tolerance and pathogenicity of globally emerging salmonella strains. Gut Microbes 2024, 16, 2369339. [Google Scholar] [CrossRef] [PubMed]

| Meat Type | Region | |||||
|---|---|---|---|---|---|---|
| Seoul and Gyeonggi-do | Gyeongsang-do | Chungcheong-do | Jeolla-do | Gangwon-do | Total | |
| Chicken | 197 | 319 | 83 | 104 | 20 | 723 |
| Duck | 151 | 195 | 92 | 62 | 9 | 509 |
| Total | 348 | 514 | 175 | 166 | 29 | 1232 |
| Antimicrobial Subclasses | Antimicrobial Agents | Range Tested |
|---|---|---|
| Aminoglycosides | Gentamicin | 1–64 |
| Streptomycin | 16–128 | |
| Aminopenicillin | Ampicillin | 2–64 |
| β-lactam/β-lactamase inhibitor combinations | Amoxicillin/Clavulanic acid | 2/1–32/16 |
| Cephamycin | Cefoxitin | 1–32 |
| Cephalosporin III | Ceftiofur | 0.5–8 |
| Ceftazidime | 1–16 | |
| Cephalosporin IV | Cefepime | 0.25–16 |
| Carbapenem | Meropenem | 0.25–4 |
| Fluoroquinolone | Ciprofloxacin | 0.12–16 |
| Folate pathway inhibitors | Trimethoprim/Sulfamethoxazole | 0.12/2.38–4/76 |
| Sulfisoxazole | 16–256 | |
| Phenicols | Chloramphenicol | 2–64 |
| Polymyxins | Colistin | 2–16 |
| Quinolone | Nalidixic acid | 2–128 |
| Tetracyclines | Tetracycline | 2–128 |
| VF Class | Virulence Factor | Related Gene |
|---|---|---|
| Fimbrial adherence determinants | Agf/Csg | csgA, csgB, csgC, csgD, csgE, csgF, csgG |
| Bcf | bcfA, bcfB, bcfC, bcfD, bcfE, bcfF, bcfG | |
| Fim | fimA, fimC, fimD, fimF, fimH, fimI, fimZ | |
| Saf | safB, safC | |
| Stb | stbA, stbB, stbC, stbD, stbE | |
| Std | stdA, stdB, stdC | |
| Stf | stfA, stfC, stfD, stfE, stfF, stfG | |
| Sth | sthA, sthB, sthC, sthD, sthE | |
| Macrophage inducible genes | Mig14 | mig14 |
| Magnesium uptake | Mg2+ transport | mgtB, mgtC |
| Non-fimbrial adherence determinants | MisL | misL |
| SinH | sinH | |
| Regulation | PhoPQ | phoP, phoQ |
| Secretion system | TTSS (SPI1 encode) | hilA, hilC, hilD, iacP, iagB, invA, invB, invC, invE, invF, invG, invH, invI, invJ, orgA, orgB, orgC, prgH, prgI, prgJ, prgK, sicA, sicP, sipD, spaO, spaP, spaQ, spaR, spaS, sprB |
| TTSS (SPI2 encode) | ssaC, ssaD, ssaE, ssaG, ssaH, ssaJ, ssaK, ssaL, ssaM, ssaN, ssaO, ssaP, ssaQ, ssaR, ssaT, ssaU, ssaV, sscA, sscB, sseB, sseC, sseD, sseE, ssrA, ssrB | |
| TTSS effectors translocated via both systems | slrP | |
| TTSS1 translocated effectors | sipA, sipB, sipC, sopA, sopB/sigD, sopD, sopE2, sptP | |
| TTSS2 translocated effectors | pipB2, pipB, sifA, sifB, sseF, sseJ, sseL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, H.; Kim, H.; Lee, J.; Jeon, J.H.; Kim, S.; Park, Y.; Joo, I.; Kim, H. Genetic Characteristics of Multidrug-Resistant Salmonella Isolated from Poultry Meat in South Korea. Microorganisms 2024, 12, 1646. https://doi.org/10.3390/microorganisms12081646
Kang H, Kim H, Lee J, Jeon JH, Kim S, Park Y, Joo I, Kim H. Genetic Characteristics of Multidrug-Resistant Salmonella Isolated from Poultry Meat in South Korea. Microorganisms. 2024; 12(8):1646. https://doi.org/10.3390/microorganisms12081646
Chicago/Turabian StyleKang, Haiseong, Hansol Kim, Jonghoon Lee, Ji Hye Jeon, Seokhwan Kim, Yongchjun Park, Insun Joo, and Hyochin Kim. 2024. "Genetic Characteristics of Multidrug-Resistant Salmonella Isolated from Poultry Meat in South Korea" Microorganisms 12, no. 8: 1646. https://doi.org/10.3390/microorganisms12081646





