Different Roles of Dioxin-Catabolic Plasmids in Growth, Biofilm Formation, and Metabolism of Rhodococcus sp. Strain p52
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Bacterial Culture
2.2. Plasmid Curing Experiment
2.3. Cell Growth Rate Analysis
2.4. Scanning Electron Microscope (SEM)
2.5. Flow Cytometric Analysis
2.6. Degradation Experiments
2.7. Biofilm Measurement
2.8. Reverse Transcription Quantitative PCR (RT–qPCR)
2.9. Transcriptome Analysis
2.10. Statistical Analysis
3. Results and Discussion
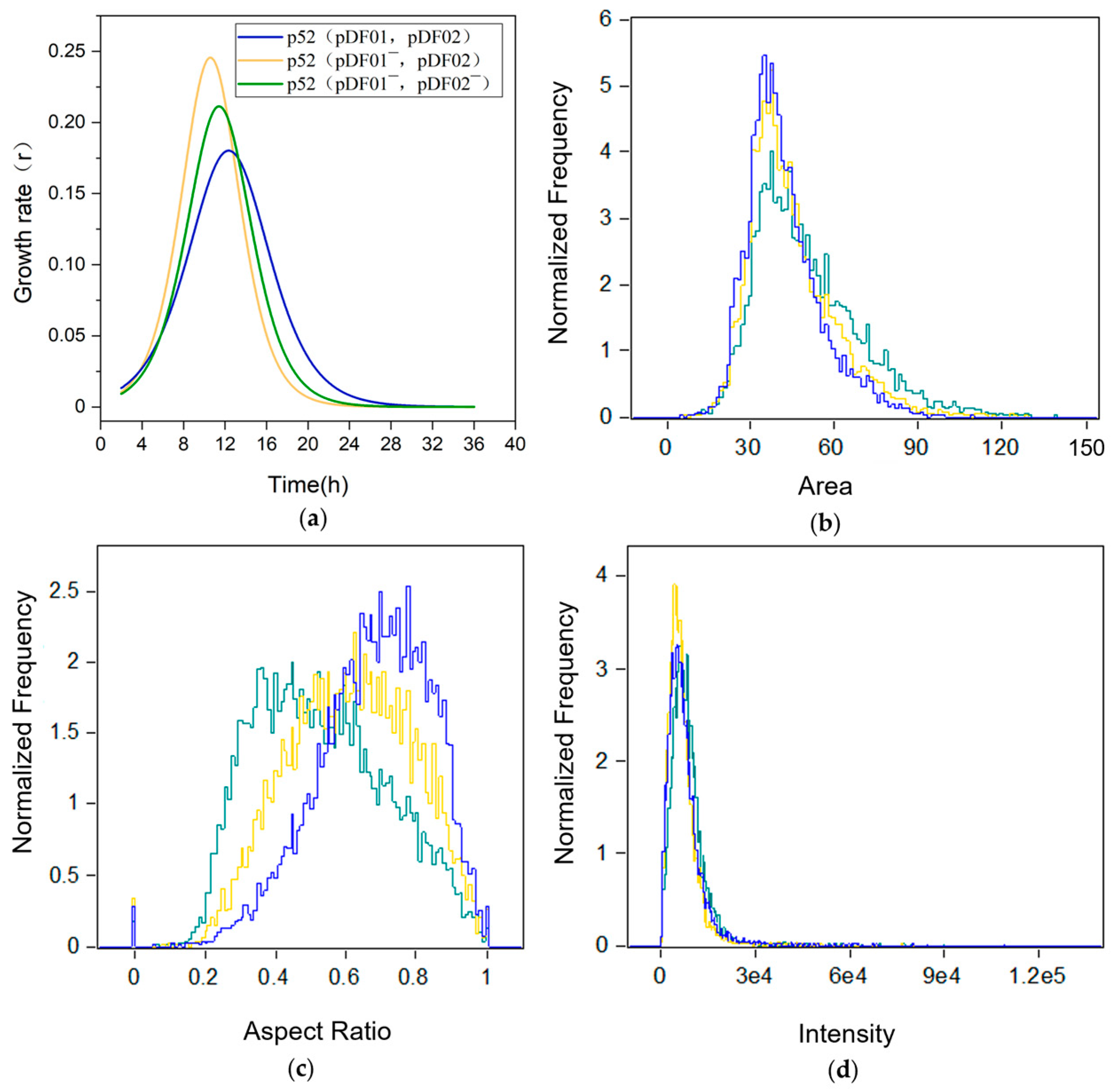
3.1. Effects of pDF01 and pDF02 on Cell Growth
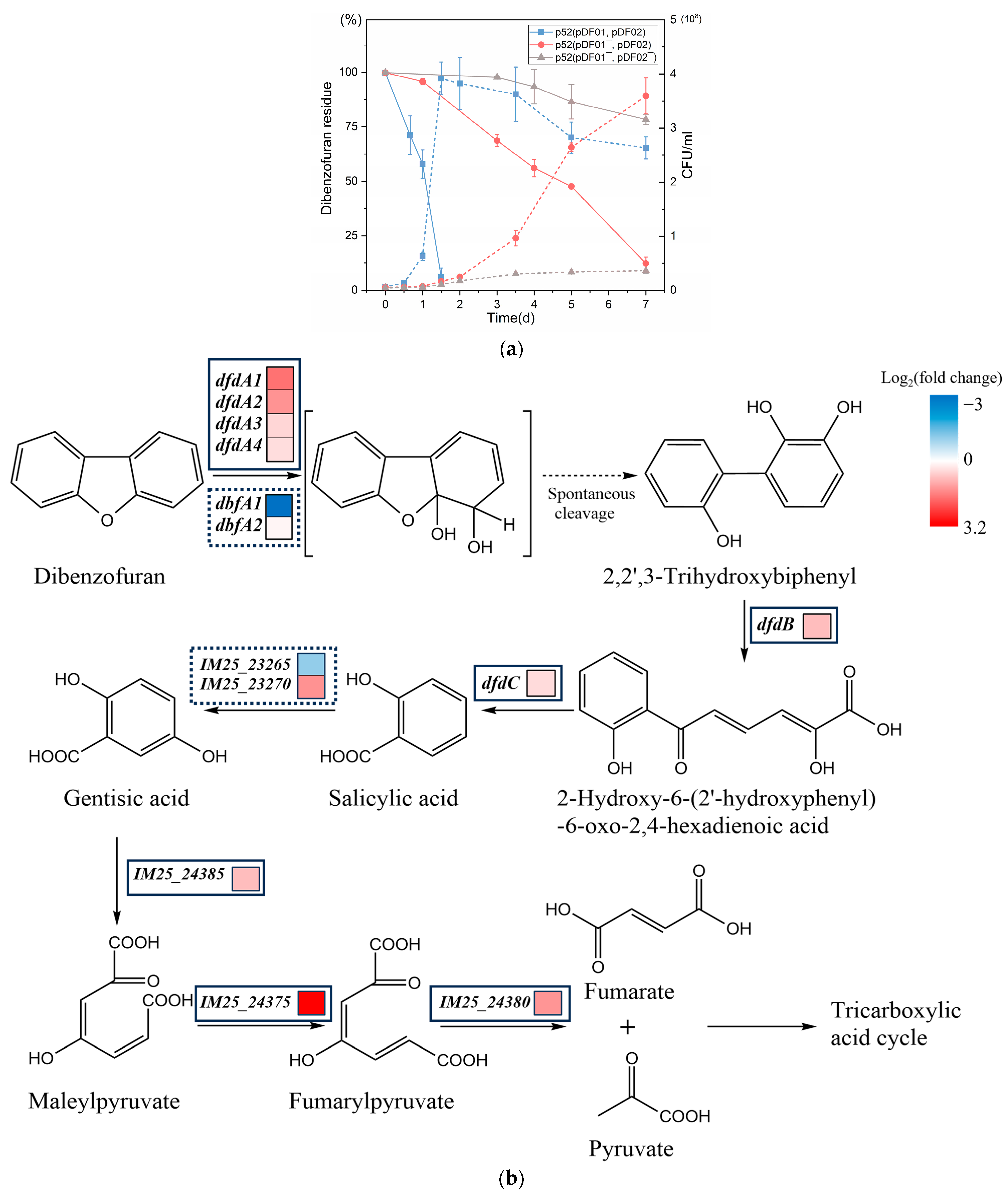
3.2. Distinguishing Roles of pDF01 and pDF02 in Dibenzofuran Degradation
3.3. pDF02 Loss Resulted in Reduced Biofilm Formation
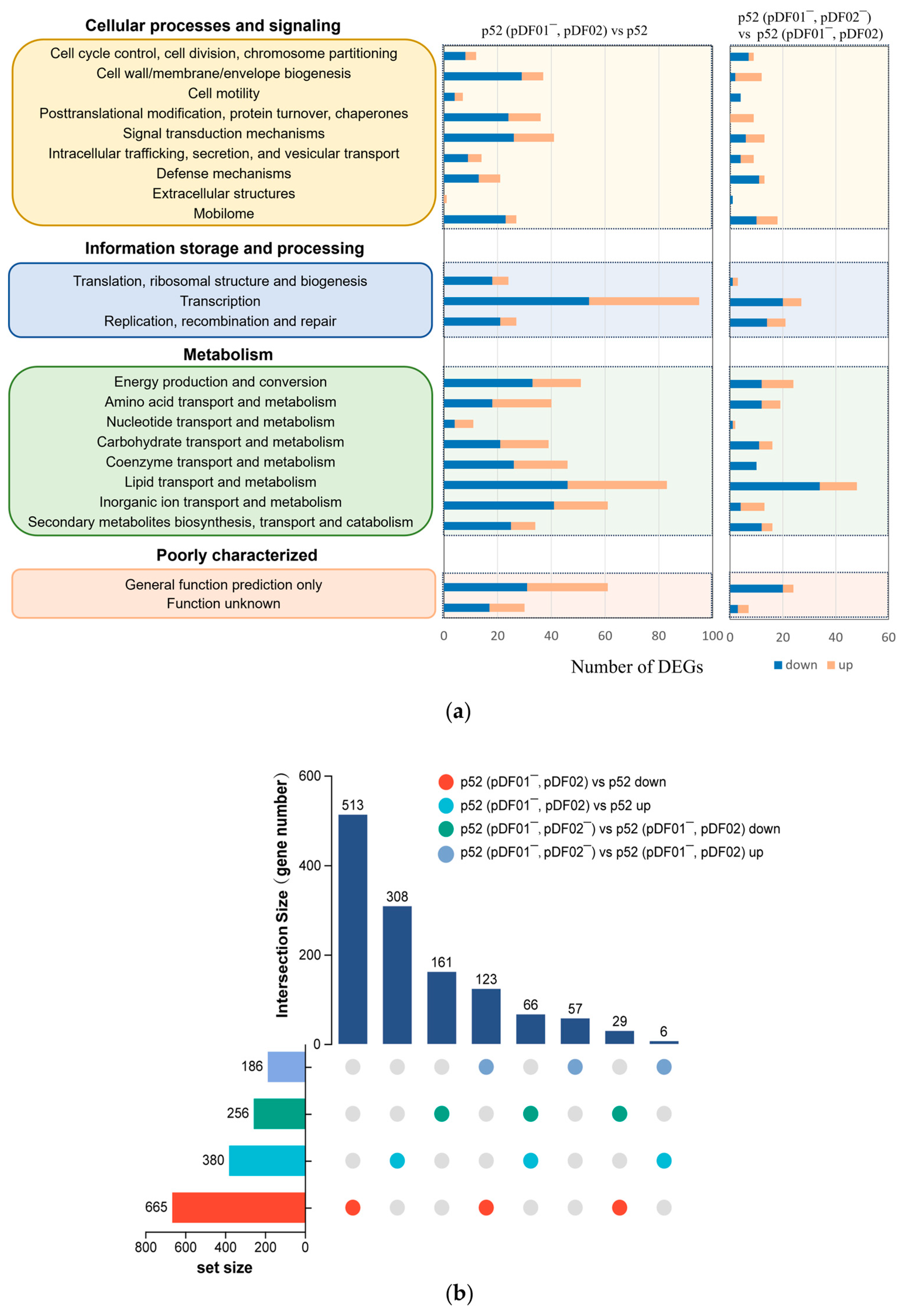
3.4. Metabolic Adaptation in Response to the pDF01 and pDF02 Loss
3.4.1. pDF01 Played Roles in Cellular Metabolism and Transcription
3.4.2. pDF02 Played Roles in Cellular Energy Production and Defense Responses
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wimmerová, S.; van den Berg, M.; Chovancová, J.; Patayová, H.; Jusko, T.A.; van Duursen, M.B.M.; Palkovičová Murínová, Ľ.; Canton, R.F.; van Ede, K.I.; Trnovec, T. Relative effect potency estimates of dioxin-like activity for dioxins, furans, and dioxin-like PCBs in adults based on cytochrome P450 1A1 and 1B1 gene expression in blood. Environ. Int. 2016, 96, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Kirkok, S.K.; Kibet, J.K.; Kinyanjui, T.K.; Okanga, F.I. A review of persistent organic pollutants: Dioxins, furans, and their associated nitrogenated analogues. SN Appl. Sci. 2020, 2, 1729. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, H.; Liu, Q.; Crabbe, M.J.C.; Qu, W. Spatial-temporal distribution, cancer risk, and disease burden attributed to the dietary dioxins exposure of Chinese residents. Sci. Total Environ. 2022, 832, 154851. [Google Scholar] [CrossRef] [PubMed]
- Pajurek, M.; Mikolajczyk, S.; Warenik-Bany, M. Occurrence and dietary intake of dioxins, furans (PCDD/Fs), PCBs, and flame retardants (PBDEs and HBCDDs) in baby food and infant formula. Sci. Total Environ. 2023, 903, 166590. [Google Scholar] [CrossRef] [PubMed]
- Köstlbacher, S.; Collingro, A.; Halter, T.; Domman, D.; Horn, M. Coevolving plasmids drive gene flow and genome plasticity in host-associated intracellular bacteria. Curr. Biol. 2021, 31, 346–357. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, W. Genome analysis of naphthalene-degrading Pseudomonas sp. AS1 harboring the megaplasmid pAS1. J. Microbiol. Biotechnol. 2018, 28, 330–337. [Google Scholar] [CrossRef] [PubMed]
- Dennis, J.J.; Zylstra, G.J. Complete sequence and genetic organization of pDTG1, the 83 kilobase naphthalene degradation plasmid from Pseudomonas putida strain NCIB 9816-4. J. Mol. Biol. 2004, 341, 753–768. [Google Scholar] [CrossRef]
- Shimizu, S.; Kobayashi, H.; Masai, E.; Fukuda, M. Characterization of the 450-kb linear plasmid in a polychlorinated biphenyl degrader, Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol. 2001, 67, 2021–2028. [Google Scholar] [CrossRef]
- Maeda, K.; Nojiri, H.; Shintani, M.; Yoshida, T.; Habe, H.; Omori, T. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J. Mol. Biol. 2003, 326, 21–33. [Google Scholar] [CrossRef]
- Elken, E.; Heinaru, E.; Jõesaar, M.; Heinaru, A. Formation of new PHE plasmids in pseudomonads in a phenol-polluted environment. Plasmid 2020, 110, 102504. [Google Scholar] [CrossRef]
- San Millan, A.; MacLean, R.C. Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 2017, 5, MTBP-0016-2017. [Google Scholar] [CrossRef] [PubMed]
- Vial, L.; Hommais, F. Plasmid-chromosome cross-talks. Environ. Microbiol. 2020, 22, 540–556. [Google Scholar] [CrossRef]
- San Millan, A.; Toll-Riera, M.; Qi, Q.; Betts, A.; Hopkinson, R.J.; McCullagh, J.; MacLean, R.C. Integrative analysis of fitness and metabolic effects of plasmids in Pseudomonas aeruginosa PAO1. ISME J. 2018, 12, 3014–3024. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.M.A.; Hall, J.P.J.; Chandra, G.; Martins, C.; Saalbach, G.; Panturat, S.; Bird, S.M.; Ford, S.; Little, R.H.; Piazza, A.; et al. Plasmids manipulate bacterial behaviour through translational regulatory crosstalk. PLoS Biol. 2023, 21, e3001988. [Google Scholar] [CrossRef] [PubMed]
- Rozkov, A.; Avignone-Rossa, C.A.; Ertl, P.F.; Jones, P.; O'Kennedy, R.D.; Smith, J.J.; Dale, J.W.; Bushell, M.E. Characterization of the metabolic burden on Escherichia coli DH1 cells imposed by the presence of a plasmid containing a gene therapy sequence. Biotechnol. Bioeng. 2004, 88, 909–915. [Google Scholar] [CrossRef]
- Diaz Ricci, J.C.; Hernández, M.E. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Biotechnol. 2000, 20, 79–108. [Google Scholar] [CrossRef]
- Horemans, B.; Raes, B.; Brocatus, H.; T’Syen, J.; Rombouts, C.; Vanhaecke, L.; Hofkens, J.; Springael, D. Genetic (in) stability of 2, 6-dichlorobenzamide catabolism in Aminobacter sp. strain MSH1 biofilms under carbon starvation conditions. Appl. Environ. Microbiol. 2017, 83, e00137-17. [Google Scholar] [CrossRef]
- Takahashi, Y.; Shintani, M.; Li, L.; Yamane, H.; Nojiri, H. Carbazole-degradative IncP-7 plasmid pCAR1.2 is structurally unstable in Pseudomonas fluorescens Pf0-1, which accumulates catechol, the intermediate of the carbazole degradation pathway. Appl. Environ. Microbiol. 2009, 75, 3920–3929. [Google Scholar] [CrossRef]
- Ghigo, J.M. Natural conjugative plasmids induce bacterial biofilm development. Nature 2001, 412, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Yao, S.; Hao, L.; Zhou, R.; Jin, Y.; Huang, J.; Wu, C. Formation of biofilm by Tetragenococcus halophilus benefited stress tolerance and anti-biofilm activity against S. aureus and S. typhimurium. Front. Microbiol. 2022, 13, 819302. [Google Scholar] [CrossRef]
- Howard, S.A.; McCarthy, R.R. Modulating biofilm can potentiate activity of novel plastic-degrading enzymes. NPJ Biofilms Microbiomes 2023, 9, 72. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.J.; Kjellerup, B.V. Applications of biofilms in bioremediation and biotransformation of persistent organic pollutants, pharmaceuticals/personal care products, and heavy metals. Appl. Microbiol. Biotechnol. 2013, 97, 9909–9921. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.P.; Yu, Y.; Huang, S.; Liao, W.; Huang, Q.S.; Du, F.L.; Xiang, T.X.; Wei, D.D.; Wan, L.G.; Zhang, W.; et al. Genetic characterization and passage instability of a novel hybrid virulence plasmid in a ST23 hypervirulent Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 2022, 12, 870779. [Google Scholar] [CrossRef] [PubMed]
- Michael, V.; Frank, O.; Bartling, P.; Scheuner, C.; Göker, M.; Brinkmann, H.; Petersen, J. Biofilm plasmids with a rhamnose operon are widely distributed determinants of the ‘swim-or-stick’ lifestyle in roseobacters. ISME J. 2016, 10, 2498–2513. [Google Scholar] [CrossRef] [PubMed]
- Coulson, G.B.; Miranda-CasoLuengo, A.A.; Miranda-CasoLuengo, R.; Wang, X.; Oliver, J.; Willingham-Lane, J.M.; Meijer, W.G.; Hondalus, M.K. Transcriptome reprogramming by plasmid-encoded transcriptional regulators is required for host niche adaption of a macrophage pathogen. Infect. Immun. 2015, 83, 3137–3145. [Google Scholar] [CrossRef] [PubMed]
- Dineen, R.L.; Bottacini, F.; O’Connell-Motherway, M.; van Sinderen, D. Transcriptional landscape of the pMP7017 megaplasmid and its impact on the Bifidobacterium breve UCC2003 transcriptome. Microb. Biotechnol. 2024, 17, e14405. [Google Scholar] [CrossRef]
- Shintani, M.; Takahashi, Y.; Tokumaru, H.; Kadota, K.; Hara, H.; Miyakoshi, M.; Naito, K.; Yamane, H.; Nishida, H.; Nojiri, H. Response of the Pseudomonas host chromosomal transcriptome to carriage of the IncP-7 plasmid pCAR1. Environ. Microbiol. 2010, 12, 1413–1426. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Yang, H.; Jia, R.; Li, L. Biodegradation of dioxin by a newly isolated Rhodococcus sp. with the involvement of self-transmissible plasmids. Appl. Microbiol. Biotechnol. 2013, 97, 5585–5595. [Google Scholar] [CrossRef]
- Ren, C.; Wang, Y.; Tian, L.; Chen, M.; Sun, J.; Li, L. Genetic bioaugmentation of activated sludge with dioxin-catabolic plasmids harbored by Rhodococcus sp. Strain p52. Environ. Sci. Technol. 2018, 52, 5339–5348. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Zhao, W.; Wang, X.; Yang, Z.; Li, L. Roles of n-hexadecane in the degradation of dibenzofuran by a biosurfactant-producing bacterium Rhodococcus sp. J. Clean. Prod. 2024, 434, 140500. [Google Scholar] [CrossRef]
- Wang, X.; Wu, Y.; Fu, C.; Zhao, W.; Li, L. Metabolic cross-feeding between the competent degrader Rhodococcus sp. strain p52 and an incompetent partner during catabolism of dibenzofuran: Understanding the leading and supporting roles. J. Hazard. Mater. 2024, 471, 134310. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989. [Google Scholar]
- Li, L.; Li, Q.; Li, F.; Shi, Q.; Yu, B.; Liu, F.; Xu, P. Degradation of carbazole and its derivatives by a Pseudomonas sp. Appl. Microbiol. Biotechnol. 2006, 73, 941–948. [Google Scholar] [CrossRef]
- Ghosh, S.; Mahapatra, N.R.; Ramamurthy, T.; Banerjee, P.C. Plasmid curing from an acidophilic bacterium of the genus Acidocella. FEMS Microbiol. Lett. 2000, 183, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Ka, J.O.; Tiedje, J.M. Integration and excision of a 2,4-dichlorophenoxyacetic acid-degradative plasmid in Alcaligenes paradoxus and evidence of its natural intergeneric transfer. J. Bacteriol. 1994, 176, 5284–5289. [Google Scholar] [CrossRef]
- Prieto, G.A.; Snigdha, S.; Baglietto-Vargas, D.; Smith, E.D.; Berchtold, N.C.; Tong, L.; Ajami, D.; LaFerla, F.M.; Rebek, J.; Cotman, C.W. Synapse-specific IL-1 receptor subunit reconfiguration augments vulnerability to IL-1β in the aged hippocampus. Proc. Natl. Acad. Sci. USA 2015, 112, E5078-87. [Google Scholar] [CrossRef]
- Chen, C.H.; Puliafito, A.; Cox, B.D.; Primo, L.; Fang, Y.; Di Talia, S.; Poss, K.D. Multicolor cell barcoding technology for long-term surveillance of epithelial regeneration in zebrafish. Dev. Cell 2016, 36, 668–680. [Google Scholar] [CrossRef]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Lili, L.N.; Britton, N.F.; Feil, E.J. The persistence of parasitic plasmids. Genetics 2007, 177, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, H. Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci. Biotechnol. Biochem. 2012, 76, 1–18. [Google Scholar] [CrossRef]
- Enne, V.I.; Bennett, P.M.; Livermore, D.M.; Hall, L.M. Enhancement of host fitness by the sul2-coding plasmid p9123 in the absence of selective pressure. J. Antimicrob. Chemother. 2004, 53, 958–963. [Google Scholar] [CrossRef]
- Marteyn, B.S.; Karimova, G.; Fenton, A.K.; Gazi, A.D.; West, N.; Touqui, L.; Prevost, M.C.; Betton, J.M.; Poyraz, O.; Ladant, D.; et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 2014, 5, e00022. [Google Scholar] [CrossRef]
- Schubert, K.; Sieger, B.; Meyer, F.; Giacomelli, G.; Böhm, K.; Rieblinger, A.; Lindenthal, L.; Sachs, N.; Wanner, G.; Bramkamp, M. The antituberculosis drug ethambutol selectively blocks apical growth in CMN group bacteria. mBio 2017, 8, e02213-16. [Google Scholar] [CrossRef]
- Taguchi, A.; Welsh, M.A.; Marmont, L.S.; Lee, W.; Sjodt, M.; Kruse, A.C.; Kahne, D.; Bernhardt, T.G.; Walker, S. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 2019, 4, 587–594. [Google Scholar] [CrossRef] [PubMed]
- Aly, H.A.; Huu, N.B.; Wray, V.; Junca, H.; Pieper, D.H. Two angular dioxygenases contribute to the metabolic versatility of dibenzofuran-degrading Rhodococcus sp. strain HA01. Appl. Environ. Microbiol. 2008, 74, 3812–3822. [Google Scholar] [CrossRef]
- Nojiri, H.; Kamakura, M.; Urata, M.; Tanaka, T.; Chung, J.S.; Takemura, T.; Yoshida, T.; Habe, H.; Omori, T. Dioxin catabolic genes are dispersed on the Terrabacter sp. DBF63 genome. Biochem. Biophys. Res. Commun. 2002, 296, 233–240. [Google Scholar] [CrossRef]
- Kasuga, K.; Nitta, A.; Kobayashi, M.; Habe, H.; Nojiri, H.; Yamane, H.; Omori, T.; Kojima, I. Cloning of dfdA genes from Terrabacter sp. strain DBF63 encoding dibenzofuran 4,4a-dioxygenase and heterologous expression in Streptomyces lividans. Appl. Microbiol. Biotechnol. 2013, 97, 4485–4498. [Google Scholar] [CrossRef] [PubMed]
- Carrilero, L.; Kottara, A.; Guymer, D.; Harrison, E.; Hall, J.P.J.; Brockhurst, M.A. Positive selection inhibits plasmid coexistence in bacterial genomes. mBio 2021, 12, e00558-21. [Google Scholar] [CrossRef]
- Hall, J.P.J.; Harrison, E.; Lilley, A.K.; Paterson, S.; Spiers, A.J.; Brockhurst, M.A. Environmentally co-occurring mercury resistance plasmids are genetically and phenotypically diverse and confer variable context-dependent fitness effects. Environ. Microbiol. 2015, 17, 5008–5022. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Ahmed, A.; Prasad, K.N.; Khanduja, S.; Singh, S.K.; Srivastava, J.K.; Gajbhiye, N.S. Antibiofilm and membrane-damaging potential of cuprous oxide nanoparticles against Staphylococcus aureus with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 2015, 59, 6882–6890. [Google Scholar] [CrossRef]
- Moye, Z.D.; Gormley, C.M.; Davey, M.E. Galactose impacts the size and intracellular composition of the asaccharolytic oral pathobiont Porphyromonas gingivalis. Appl. Environ. Microbiol. 2019, 85, e02268-18. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Beauregard, P.B.; Vlamakis, H.; Losick, R.; Kolter, R. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 2012, 3, e00184-12. [Google Scholar] [CrossRef] [PubMed]
- Barreto, M.; Jedlicki, E.; Holmes, D.S. Identification of a gene cluster for the formation of extracellular polysaccharide precursors in the chemolithoautotroph Acidithiobacillus ferrooxidans. Appl. Environ. Microbiol. 2005, 71, 2902–2909. [Google Scholar] [CrossRef]
- Ha, D.G.; O'Toole, G.A. c-di-GMP and its effects on biofilm formation and dispersion: A Pseudomonas aeruginosa review. Microbial Spectr. 2015, 3, MB-0003-2014. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sagaram, U.S.; Kim, J.S.; Wang, N. Requirement of the galU gene for polysaccharide production by and pathogenicity and growth in planta of Xanthomonas citri subsp. citri. Appl. Environ. Microbiol. 2010, 76, 2234–2242. [Google Scholar] [CrossRef] [PubMed]
- Billane, K.; Harrison, E.; Cameron, D.; Brockhurst, M.A. Why do plasmids manipulate the expression of bacterial phenotypes? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2022, 377, 20200461. [Google Scholar] [CrossRef]
- Lang, K.S.; Johnson, T.J. Transcriptome modulations due to A/C2 plasmid acquisition. Plasmid 2015, 80, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Crismaru, C.G.; Salo, O.; Bovenberg, R.A.; Driessen, A.J. Impact of classical strain improvement of Penicillium rubens on amino acid metabolism during β-lactam production. Appl. Environ. Microbiol. 2020, 86, e01561-19. [Google Scholar] [CrossRef]
- Janssen, D.B.; Herst, P.M.; Joosten, H.M.; van der Drift, C. Nitrogen control in Pseudomonas aeruginosa: A role for glutamine in the regulation of the synthesis of NADP-dependent glutamate dehydrogenase, urease and histidase. Arch. Microbiol. 1981, 128, 398–402. [Google Scholar] [CrossRef]
- Khan, Y.A.; White, K.I.; Brunger, A.T. The AAA+ superfamily: A review of the structural and mechanistic principles of these molecular machines. Crit. Rev. Biochem. Mol. Biol. 2022, 57, 156–187. [Google Scholar] [CrossRef]
- Blacker, T.S.; Duchen, M.R.; Bain, A.J. NAD(P)H binding configurations revealed by time-resolved fluorescence and two-photon absorption. Biophys. J. 2023, 122, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- Loenen, W.A.; Dryden, D.T.; Raleigh, E.A.; Wilson, G.G.; Murray, N.E. Highlights of the DNA cutters: A short history of the restriction enzymes. Nucleic Acids Res. 2014, 42, 3–19. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Wu, Y.; Chen, M.; Fu, C.; Xu, H.; Li, L. Different Roles of Dioxin-Catabolic Plasmids in Growth, Biofilm Formation, and Metabolism of Rhodococcus sp. Strain p52. Microorganisms 2024, 12, 1700. https://doi.org/10.3390/microorganisms12081700
Wang X, Wu Y, Chen M, Fu C, Xu H, Li L. Different Roles of Dioxin-Catabolic Plasmids in Growth, Biofilm Formation, and Metabolism of Rhodococcus sp. Strain p52. Microorganisms. 2024; 12(8):1700. https://doi.org/10.3390/microorganisms12081700
Chicago/Turabian StyleWang, Xu, Yanan Wu, Meng Chen, Changai Fu, Hangzhou Xu, and Li Li. 2024. "Different Roles of Dioxin-Catabolic Plasmids in Growth, Biofilm Formation, and Metabolism of Rhodococcus sp. Strain p52" Microorganisms 12, no. 8: 1700. https://doi.org/10.3390/microorganisms12081700




