Efflux Pumps and Porins Enhance Bacterial Tolerance to Phenolic Compounds by Inhibiting Hydroxyl Radical Generation
Abstract
:1. Introduction
2. Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Protein Expression and Gel Electrophoresis Analysis
2.3. Biosynthesis of PG
3. Results
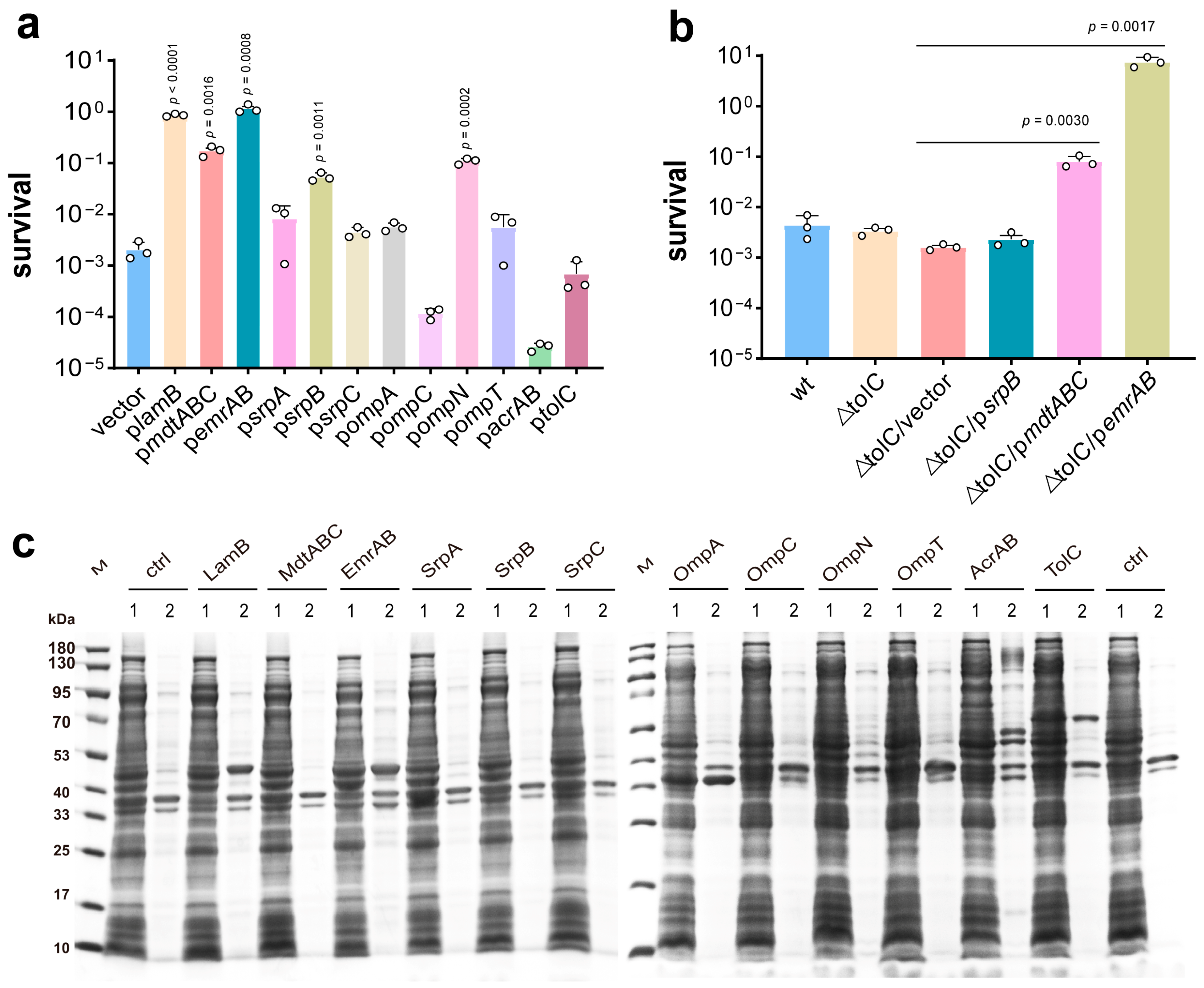
3.1. Identification of Efflux Pumps and Porin Gene Essential to PG Tolerance
3.2. Efflux Pumps and Porins Inhibit the Generation of Hydroxyl Radical
3.3. Comparison of Intracellular Iron Levels
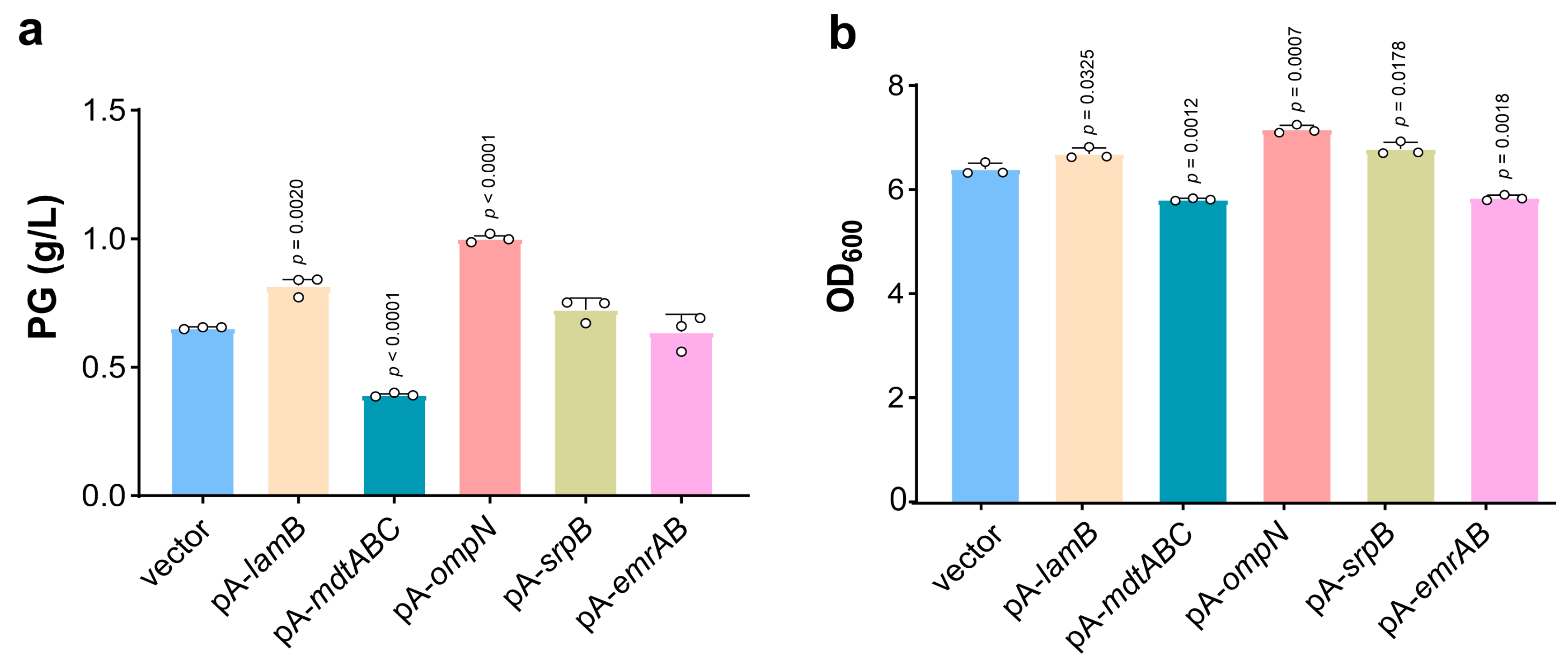
3.4. Porin Overexpression Benefits Biosynthesis of PG
3.5. Efflux Pumps and Porins Improve Bacterial Tolerance to Diverse Phenolic Compounds
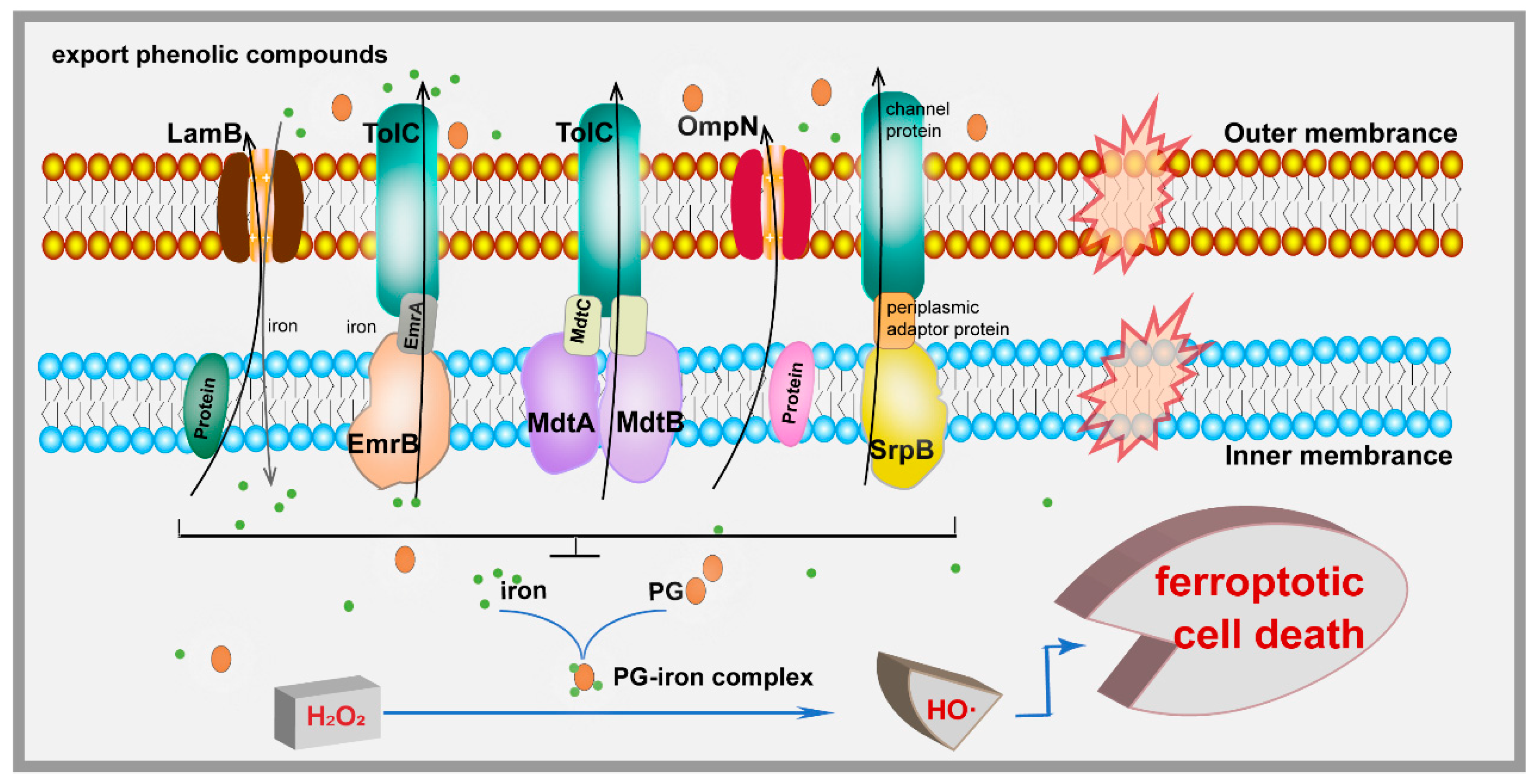
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmidt, R.J. Industrial catalytic processes—Phenol production. Appl. Catal. A Gen. 2005, 280, 89–103. [Google Scholar] [CrossRef]
- Wu, F.; Chen, Y.; Zheng, C.H. Efficacy of phloroglucinol for acceleration of labour: A systematic review and meta-analysis. Arch. Gynecol. Obstet. 2021, 304, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Cao, Y.J. Biosynthesis of phloroglucinol compounds in microorganisms-review. Appl. Microbiol. Biotechnol. 2012, 93, 487–495. [Google Scholar] [CrossRef]
- Krastanov, A.; Alexieva, Z.; Yemendzhiev, H. Microbial degradation of phenol and phenolic derivatives. Eng. Life Sci. 2013, 13, 76–87. [Google Scholar] [CrossRef]
- Bu, Q.; Lei, H.W.; Wang, L.; Wei, Y.; Zhu, L.; Zhang, X.S.; Liu, Y.P.; Yadavalli, G.; Tang, J. Bio-based phenols and fuel production from catalytic microwave pyrolysis of lignin by activated carbons. Bioresour. Technol. 2014, 162, 142–147. [Google Scholar] [CrossRef]
- Thompson, B.; Machas, M.; Nielsen, D.R. Engineering and comparison of non-natural pathways for microbial phenol production. Biotechnol. Bioeng. 2016, 113, 1745–1754. [Google Scholar] [CrossRef] [PubMed]
- Pugh, S.; McKenna, R.; Osman, M.; Thompson, B.; Nielsen, D.R. Rational engineering of a novel pathway for producing the aromatic compounds p-hydroxybenzoate, protocatechuate, and catechol in Escherichia coli. Process Biochem. 2014, 49, 1843–1850. [Google Scholar] [CrossRef]
- Ma, Y.H.; Li, L.J.; Awasthi, M.K.; Tian, H.X.; Li, M.H.; Megharaj, M.; Pan, Y.L.; He, W.X. Time-course transcriptome analysis reveals the mechanisms of Burkholderia sp. adaptation to high phenol concentrations. Appl. Microbiol. Biotechnol. 2020, 104, 5873–5887. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shen, X.L.; Yuan, Q.P.; Yan, Y.J. Microbial synthesis of pyrogallol using genetically engineered Escherichia coli. Metab. Eng. 2018, 45, 134–141. [Google Scholar] [CrossRef]
- Sui, X.Y.; Wang, J.C.; Zhao, Z.Q.; Liu, B.; Liu, M.M.; Liu, M.; Shi, C.; Feng, X.J.; Fu, Y.X.; Shi, D.Y.; et al. Phenolic compounds induce ferroptosis-like death by promoting hydroxyl radical generation in the Fenton reaction. Commun. Biol. 2024, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Wierckx, N.J.; Ballerstedt, H.; de Bont, J.A.; Wery, J. Engineering of solvent-tolerant Pseudomonas putida S12 for bioproduction of phenol from glucose. Appl. Environ. Microbiol. 2005, 71, 8221–8227. [Google Scholar] [CrossRef] [PubMed]
- Keweloh, H.; Weyrauch, G.; Rehm, H.J. Phenol-induced membrane changes in free and immobilized Escherichia coli. Appl. Microbiol. Biotechnol. 1990, 33, 66–71. [Google Scholar] [CrossRef]
- Ibraheem, O.; Ndimba, B.K. Molecular adaptation mechanisms employed by ethanologenic bacteria in response to lignocellulose-derived inhibitory compounds. Int. J. Biol. Sci. 2013, 9, 598–612. [Google Scholar] [CrossRef]
- Lin, R.; Cheng, J.; Ding, L.; Song, W.; Zhou, J.; Cen, K. Inhibitory effects of furan derivatives and phenolic compounds on dark hydrogen fermentation. Bioresour. Technol. 2015, 196, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, E.; Gao, K.; Mercurio, K.; Ali, M.; Baetz, K. Yeast chemogenomic screen identifies distinct metabolic pathways required to tolerate exposure to phenolic fermentation inhibitors ferulic acid, 4-hydroxybenzoic acid and coniferyl aldehyde. Metab. Eng. 2019, 52, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I.; Nikaido, H. Multidrug resistance mechanisms: Drug efflux across two membranes. Mol. Microbiol. 2000, 37, 219–225. [Google Scholar] [CrossRef]
- Nanjan, P.; Bose, V. Efflux-mediated Multidrug Resistance in Critical Gram-negative Bacteria and Natural Efflux Pump Inhibitors. Curr. Drug Res. Rev. 2024, 16, 349–368. [Google Scholar] [CrossRef] [PubMed]
- Kornelsen, V.; Kumar, A. Update on Multidrug Resistance Efflux Pumps in Acinetobacter spp. Antimicrob. Agents Chemother. 2021, 65, e00514-21. [Google Scholar] [CrossRef] [PubMed]
- Lekshmi, M.; Ortiz-Alegria, A.; Kumar, S.; Varela, M.F. Major facilitator superfamily efflux pumps in human pathogens: Role in multidrug resistance and beyond. Curr. Res. Microb. Sci. 2024, 7, 100248. [Google Scholar] [CrossRef] [PubMed]
- Gaurav, A.; Bakht, P.; Saini, M.; Pandey, S.; Pathania, R. Role of bacterial efflux pumps in antibiotic resistance, virulence, and strategies to discover novel efflux pump inhibitors. Microbiology 2023, 169, 001333. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.A.; Liu, Q.; Henderson, P.J.F.; Paulsena, I.T. Homologs of the Acinetobacter baumannii AceI Transporter Represent a New Family of Bacterial Multidrug Efflux Systems. mBio 2015, 6, e01982-14. [Google Scholar] [CrossRef]
- Novelli, M.; Bolla, J.M. RND Efflux Pump Induction: A Crucial Network Unveiling Adaptive Antibiotic Resistance Mechanisms of Gram-Negative Bacteria. Antibiotics 2024, 13, 501. [Google Scholar] [CrossRef] [PubMed]
- Alenazy, R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology 2022, 11, 1328. [Google Scholar] [CrossRef] [PubMed]
- Putman, M.; van Veen, H.W.; Konings, W.N. Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 2000, 64, 672–693. [Google Scholar] [CrossRef] [PubMed]
- Sekar, P.; Mamtora, D.; Bhalekar, P.; Krishnan, P. AcrAB-TolC Efflux Pump Mediated Resistance to Carbapenems Among Clinical Isolates of Enterobacteriaceae. J. Pure Appl. Microbiol. 2022, 16, 1982–1989. [Google Scholar] [CrossRef]
- Horiyama, T.; Nishino, K. AcrB, AcrD, and MdtABC Multidrug Efflux Systems Are Involved in Enterobactin Export in Escherichia coli. PLoS ONE 2014, 9, e108642. [Google Scholar] [CrossRef]
- Bui, L.M.; Lee, J.Y.; Geraldi, A.; Rahman, Z.; Lee, J.H.; Kim, S.C. Improved n-butanol tolerance in Escherichia coli by controlling membrane related functions. J. Biotechnol. 2015, 204, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Lewis, K. Emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 1992, 89, 8938–8942. [Google Scholar] [CrossRef] [PubMed]
- Law, C.J.; Maloney, P.C.; Wang, D.N. Ins and Outs of Major Facilitator Superfamily, Antiporters. Annu. Rev. Microbiol. 2008, 62, 289–305. [Google Scholar] [CrossRef]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynie, L.; Acosta-Gutierrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; van den Berg, B.; Winterhalter, M.; et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Pagès, J.M.; James, C.E.; Winterhalter, M. The porin and the permeating antibiotic: A selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008, 6, 893–903. [Google Scholar] [CrossRef]
- Dupont, H.; Gaillot, O.; Goetgheluck, A.S.; Plassart, C.; Emond, J.P.; Lecuru, M.; Gaillard, N.; Derdouri, S.; Lemaire, B.; Girard de Courtilles, M.; et al. Molecular Characterization of Carbapenem-Nonsusceptible Enterobacterial Isolates Collected During a Prospective Interregional Survey in France and Susceptibility to the Novel Ceftazidime-Avibactam and Aztreonam-Avibactam Combinations. Antimicrob. Agents Chemother. 2016, 60, 215–221. [Google Scholar] [CrossRef] [PubMed]
- García-Sureda, L.; Juan, C.; Doménech-Sánchez, A.; Albertí, S. Role of Klebsiella pneumoniae LamB porin in antimicrobial resistance. Antimicrob. Agents Chemother. 2011, 55, 1803–1805. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Shao, Y.; Zhao, X.; Li, C.; Guo, M.; Lv, Z.; Zhang, W. Indole contributes to tetracycline resistance via the outer membrane protein OmpN in Vibrio splendidus. World J. Microbiol. Biotechnol. 2020, 36, 36. [Google Scholar] [CrossRef] [PubMed]
- Bao, X.; Yang, C.; Li, T.; Wang, Y.; Cui, A.; Meng, X.; Huang, Q.; Li, S. Efflux of TolC protein to different antimicrobials can be replaced by other outer membrane proteins with similar β-barrel structures in extraintestinal pathogenic Escherichia coli. J. Appl. Microbiol. 2024, 135, lxae214. [Google Scholar] [CrossRef]
- Cao, Y.J.; Jiang, X.L.; Zhang, R.B.; Xian, M. Improved phloroglucinol production by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 2011, 91, 1545–1552. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, B.; Duan, C.; Sun, B.; Yang, J.; Yang, S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 2015, 81, 2506–2514. [Google Scholar] [CrossRef]
- Wang, J.; Sui, X.; Ding, Y.; Fu, Y.; Feng, X.; Liu, M.; Zhang, Y.; Xian, M.; Zhao, G. A fast and robust iterative genome-editing method based on a Rock-Paper-Scissors strategy. Nucleic Acids Res. 2021, 49, e12. [Google Scholar] [CrossRef] [PubMed]
- Nagakubo, S.; Nishino, K.; Hirata, T.; Yamaguchi, A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 2002, 184, 4161–4167. [Google Scholar] [CrossRef]
- Zhang, R.B.; Cao, Y.J.; Liu, W.; Xian, M.; Liu, H.Z. Improving phloroglucinol tolerance and production in Escherichia coli by GroESL overexpression. Microb. Cell Factories 2017, 16, 227. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.Y.; Li, Z.H.; Wang, X.N.; Wang, J.; Chala, J.; Lu, Y.H.; Zhang, H.R. De novo phenol bioproduction from glucose using biosensor-assisted microbial coculture engineering. Biotechnol. Bioeng. 2019, 116, 3349–3359. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.L.; Wang, J.; Wang, J.; Chen, Z.Y.; Yuan, Q.P.; Yan, Y.J. High-level de novo biosynthesis of arbutin in engineered Escherichia coli. Metab. Eng. 2017, 42, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Balderas-Hernández, V.E.; Treviño-Quintanilla, L.G.; Hernández-Chávez, G.; Martinez, A.; Bolívar, F.; Gosset, G. Catechol biosynthesis from glucose in Escherichia coli anthranilate-overproducer strains by heterologous expression of anthranilate 1,2-dioxygenase from Pseudomonas aeruginosa PAO1. Microb. Cell Factories 2014, 13, 136. [Google Scholar] [CrossRef]
- Wang, J.; Shen, X.L.; Wang, J.; Yang, Y.P.; Yuan, Q.P.; Yan, Y.J. Exploring the Promiscuity of Phenol Hydroxylase from Pseudomonas stutzeri OX1 for the Biosynthesis of Phenolic Compounds. ACS Synth. Biol. 2018, 7, 1238–1243. [Google Scholar] [CrossRef] [PubMed]
- Zang, S.Y.; Lian, B.; Wang, J.; Yang, Y.J. Biodegradation of 2-naphthol and its metabolites by coupling Aspergillus niger with Bacillus subtilis. J. Environ. Sci. 2010, 22, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Bafna, J.A.; Sans-Serramitjana, E.; Acosta-Gutiérrez, S.; Bodrenko, I.V.; Hörömpöli, D.; Berscheid, A.; Brötz-Oesterhelt, H.; Winterhalter, M.; Ceccarelli, M. Kanamycin Uptake into Escherichia coli Is Facilitated by OmpF and OmpC Porin Channels Located in the Outer Membrane. ACS Infect. Dis. 2020, 6, 1855–1865. [Google Scholar] [CrossRef] [PubMed]
- Lesjak, M.; Simin, N.; Srai, S.K.S. Can Polyphenols Inhibit Ferroptosis? Antioxidants 2022, 11, 150. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Wu, T.Y.; Machado, I.M.P.; Huang, W.C.; Chen, P.Y.; Pellegrini, M.; Liao, J.C. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol. Syst. Biol. 2010, 6, 449. [Google Scholar] [CrossRef] [PubMed]


| Strain or Plasmid | Description | Source |
|---|---|---|
| E. coil DH5α | F− supE44 ΔlacU169 (ϕ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Lab collection |
| E. coil BL21(DE3) | F− ompT gal dcm lon hsdSB (rB− mB−) λ(DE3) | Lab collection |
| E. coil W3110 | F- λ- rph-1 INV (rrnD, rrnE) | Lab collection |
| Q3595 | E. coil BL21(DE3)/pA-phlD/marA/acc | [36] |
| Q3861 | E. coil BL21(DE3)/pTrcHis2B | This study |
| Q5660 | E. coil BL21(DE3)/pTRC-tolC | This study |
| Q5661 | E. coil BL21(DE3)/pTRC-ompA | This study |
| Q5662 | E. coil BL21(DE3)/pTRC-ompN | This study |
| Q5663 | E. coil BL21(DE3)/pTRC-lamB | This study |
| Q5664 | E. coil BL21(DE3)/pTRC-acrAB | This study |
| Q5665 | E. coil BL21(DE3)/pTRC-ompC | This study |
| Q5666 | E. coil BL21(DE3)/pTRC-emrAB | This study |
| Q5667 | E. coil BL21(DE3)/pTRC-mdtABC | This study |
| Q6285 | E. coil BL21(DE3)/pTRC-ompT | This study |
| Q6286 | E. coil BL21(DE3)/pTRC-srpA | This study |
| Q6287 | E. coil BL21(DE3)/pTRC-srpB | This study |
| Q6288 | E. coil BL21(DE3)/pTRC-srpC | This study |
| Q6329 | E. coil BL21(DE3)/pA-lamB | This study |
| Q6330 | E. coil BL21(DE3)/pA-ompN | This study |
| Q6331 | E. coil BL21(DE3)/pA-srpB | This study |
| Q6332 | E. coil BL21(DE3)/pA-mdtABC | This study |
| Q6333 | E. coil BL21(DE3)/pA-emrAB | This study |
| Q6424 | E. coil BL21(DE3) ΔtolC | This study |
| Q6425 | E. coil BL21(DE3) ΔtolC/pTrcHis2B | This study |
| Q6426 | E. coil BL21(DE3) ΔtolC/pTRC-emrAB | This study |
| Q6427 | E. coil BL21(DE3) ΔtolC/pTRC-mdtABC | This study |
| Q6428 | E. coil BL21(DE3) ΔtolC/pTRC-srpB | This study |
| Plasmids | ||
| pTrcHis2B | reppBR322 AmpR lacIq Ptrc | Invitrogen |
| pA-phlD/marA/acc | repp15A CmR lacI PT7-phlD-marA-accADBC | [36] |
| pCas | reppSC101Ts KanR Pcas-cas9 ParaB-Red lacIQ Ptrc-sgRNApMB1 | MolecularCloud: MC0000011 [37] |
| pPaper-ΔtolC | repp15A CmR PlacIQ-sgRNA-TetR PJ23119-sgRNA ΔtolC | This study |
| pTRC-lamB | reppBR322 ApR lacIq Ptrc-lamB | This study |
| pTRC-ompN | reppBR322 ApR lacIq Ptrc-ompN | This study |
| pTRC-tolC | reppBR322 ApR lacIq Ptrc-tolC | This study |
| pTRC-srpA | reppBR322 ApR lacIq Ptrc-srpA | This study |
| pTRC-srpB | reppBR322 ApR lacIq Ptrc-srpB | This study |
| pTRC-srpC | reppBR322 ApR lacIq Ptrc-emrAB | This study |
| pTRC-emrAB | reppBR322 ApR lacIq Ptrc-emrAB | This study |
| pTRC-mdtABC | reppBR322 ApR lacIq Ptrc-mdtABC | This study |
| pTRC-acrAB | reppBR322 ApR lacIq Ptrc-acrAB | This study |
| pTRC-ompA | reppBR322 ApR lacIq Ptrc-ompA | This study |
| pTRC-ompC | reppBR322 ApR lacIq Ptrc-ompC | This study |
| pTRC-ompN | reppBR322 ApR lacIq Ptrc-ompN | This study |
| pTRC-ompT | reppBR322 ApR lacIq Ptrc-ompT | This study |
| pA-lamB | repp15A CmR lacI PT7-phlD-marA-accADBC-Ptrc-lamB | This study |
| pA-ompN | repp15A CmR lacI PT7-phlD-marA-accADBC-Ptrc-ompN | This study |
| pA-srpB | repp15A CmR lacI PT7-phlD-marA-accADBC-Ptrc-srpB | This study |
| pA-mdtABC | repp15A CmR lacI PT7-phlD-marA-accADBC-Ptrc-mdtABC | This study |
| pA-emrAB | repp15A CmR lacI PT7-phlD-marA-accADBC-Ptrc-emrAB | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sui, X.; Guo, L.; Bao, Z.; Xian, M.; Zhao, G. Efflux Pumps and Porins Enhance Bacterial Tolerance to Phenolic Compounds by Inhibiting Hydroxyl Radical Generation. Microorganisms 2025, 13, 202. https://doi.org/10.3390/microorganisms13010202
Sui X, Guo L, Bao Z, Xian M, Zhao G. Efflux Pumps and Porins Enhance Bacterial Tolerance to Phenolic Compounds by Inhibiting Hydroxyl Radical Generation. Microorganisms. 2025; 13(1):202. https://doi.org/10.3390/microorganisms13010202
Chicago/Turabian StyleSui, Xinyue, Likun Guo, Zixian Bao, Mo Xian, and Guang Zhao. 2025. "Efflux Pumps and Porins Enhance Bacterial Tolerance to Phenolic Compounds by Inhibiting Hydroxyl Radical Generation" Microorganisms 13, no. 1: 202. https://doi.org/10.3390/microorganisms13010202
APA StyleSui, X., Guo, L., Bao, Z., Xian, M., & Zhao, G. (2025). Efflux Pumps and Porins Enhance Bacterial Tolerance to Phenolic Compounds by Inhibiting Hydroxyl Radical Generation. Microorganisms, 13(1), 202. https://doi.org/10.3390/microorganisms13010202







