Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain Collection and Antimicrobial Susceptibility Testing
2.2. Clinical Data Collection and Case–Control Selection
2.3. DNA Extraction, Library Construction, and Whole-Genome Sequencing
2.4. Bioinformatic Analyses
2.5. Statistical Analysis
3. Results
3.1. Study Population and Sociodemographic and Clinical Characteristics
3.2. Antimicrobial Susceptibility Profiles
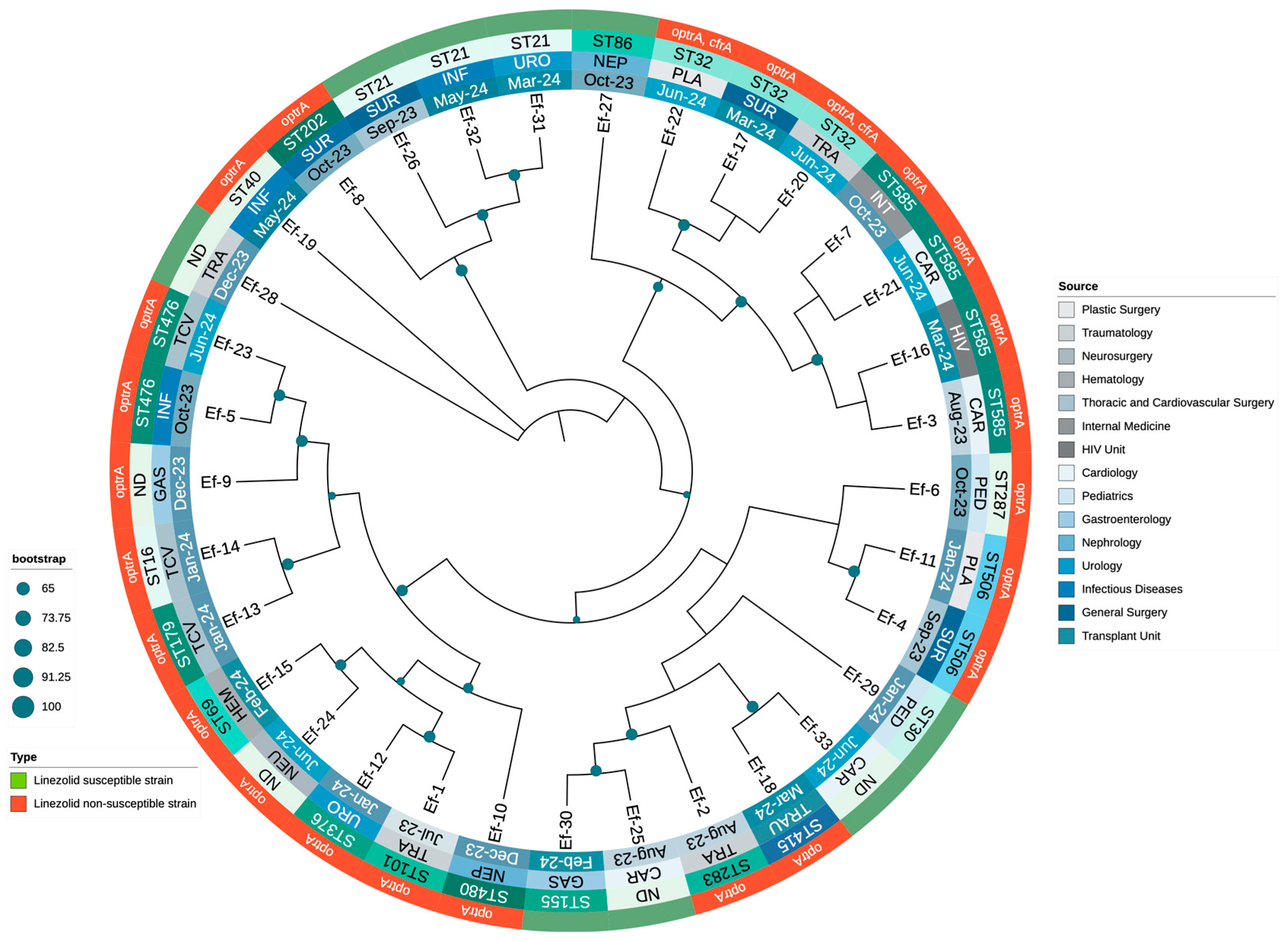
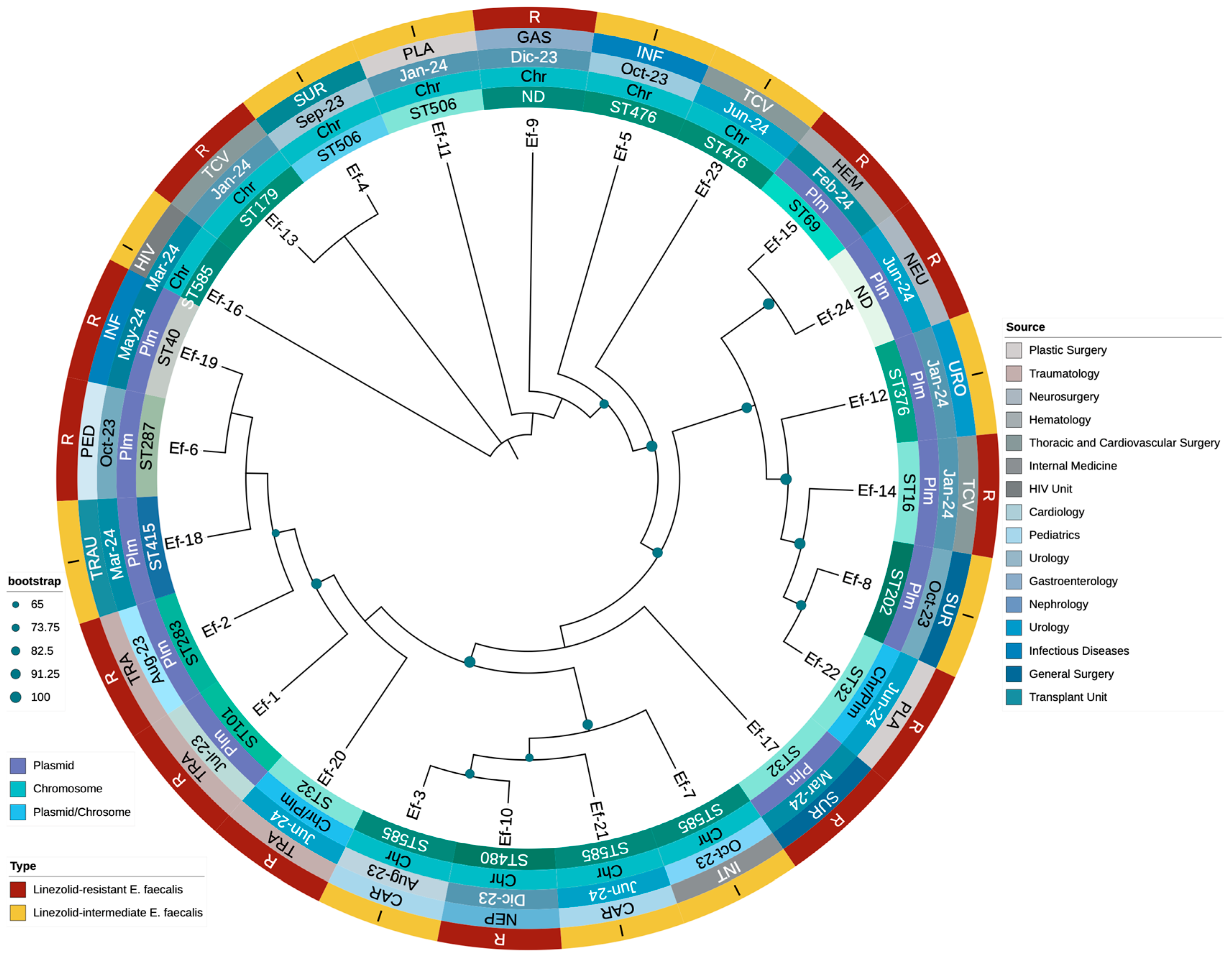
3.3. Molecular Typing and Genotypic Analysis of Linezolid Resistance Mechanisms
3.4. Phylogenetic Analysis of Enterococcus Clinical Isolates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ATP-Binding Cassette |
| AMR | Antimicrobial Resistance |
| AMP | Ampicillin |
| ARGs | Antimicrobial Resistance Genes |
| AST | Antimicrobial Susceptibility Testing |
| ATP | Adenosine Triphosphate |
| CARD | Comprehensive Antibiotic Resistance Database |
| CAR | Cardiology |
| CIP | Ciprofloxacin |
| CLSI | Clinical and Laboratory Standards Institute |
| CSF | Cerebrospinal Fluid |
| DAP | Daptomycin |
| DNA | Deoxyribonucleic Acid |
| FASTQ | Format for Sequence Data |
| GAS | Gastroenterology |
| GEN | Gentamicin |
| HEM | Hematology |
| HIV | Human Immunodeficiency Virus |
| ICU | Intensive Care Unit |
| INF | Infectious Diseases |
| INT | Internal Medicine |
| LEV | Levofloxacin |
| LNZ | Linezolid |
| LNSEf | Linezolid-Non-Susceptible Enterococcus faecalis |
| LRE | Linezolid-Resistant Enterococcus |
| LSEf | Linezolid-Susceptible Enterococcus faecalis |
| MDR-TB | Multidrug-Resistant Tuberculosis |
| MIC | Minimum Inhibitory Concentration |
| ML | Maximum Likelihood |
| MLST | Multilocus Sequence Typing |
| MRSA | Methicillin-Resistant Staphylococcus aureus |
| NEP | Nephrology |
| NEU | Neurosurgery |
| NIT | Nitrofurantoin |
| NR | Non-Redundant Database |
| PCR | Polymerase Chain Reaction |
| PED | Pediatrics |
| PEN | Benzylpenicillin |
| PLA | Plastic Surgery |
| QC | Quality Control |
| rRNA | Ribosomal Ribonucleic Acid |
| RGI | Resistance Gene Identifier |
| SNP | Single Nucleotide Polymorphism |
| ST | Sequence Type |
| STR | Streptomycin |
| SUR | General Surgery |
| TCV | Thoracic and Cardiovascular Surgery |
| TET | Tetracycline |
| TRA | Traumatology |
| TRAU | Transplant Unit |
| URO | Urology |
| VA | Vancomycin |
| WHO | World Health Organization |
| WGS | Whole-Genome Sequencing |
| SUR | General Surgery |
| TCV | Thoracic and Cardiovascular Surgery |
| TET | Tetracycline |
| TRA | Traumatology |
| TRAU | Transplant Unit |
| URO | Urology |
| VA | Vancomycin |
| VFDB | Virulence Factor Database |
| WHO | World Health Organization |
| WGS | Whole-Genome Sequencing |
References
- Pinholt, M.; Ostergaard, C.; Arpi, M.; Bruun, N.E.; Schønheyder, H.C.; Gradel, K.O.; Søgaard, M.; Knudsen, J.D.; Danish Collaborative Bacteraemia Network (DACOBAN). Incidence, Clinical Characteristics and 30-Day Mortality of Enterococcal Bacteraemia in Denmark 2006-2009: A Population-Based Cohort Study. Clin. Microbiol. Infect. 2014, 20, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Kajihara, T.; Nakamura, S.; Iwanaga, N.; Oshima, K.; Takazono, T.; Miyazaki, T.; Izumikawa, K.; Yanagihara, K.; Kohno, N.; Kohno, S. Clinical Characteristics and Risk Factors of Enterococcal Infections in Nagasaki, Japan: A Retrospective Study. BMC Infect. Dis. 2015, 15, 426. [Google Scholar] [CrossRef]
- Hayakawa, K.; Marchaim, D.; Palla, M.; Gudur, U.M.; Pulluru, H.; Bathina, P.; Alshabani, K.; Govindavarjhulla, A.; Mallad, A.; Abbadi, D.R.; et al. Epidemiology of Vancomycin-Resistant Enterococcus Faecalis: A Case-Case-Control Study. Antimicrob. Agents Chemother. 2013, 57, 49–55. [Google Scholar] [CrossRef]
- Dahl, A.; Iversen, K.; Tønder, N.; Hoest, N.; Arpi, M.; Dalsgaard, M.; Chehri, M.; Soerensen, L.L.; Fanoe, S.; Junge, S.; et al. Prevalence of Infective Endocarditis in Enterococcus Faecalis Bacteremia. J. Am. Coll. Cardiol. 2019, 74, 193–201. [Google Scholar] [CrossRef]
- Zerbato, V.; Pol, R.; Sanson, G.; Suru, D.A.; Pin, E.; Tabolli, V.; Monticelli, J.; Busetti, M.; Toc, D.A.; Crocè, L.S.; et al. Risk Factors for 30-Day Mortality in Nosocomial Enterococcal Bloodstream Infections. Antibiotics 2024, 13, 601. [Google Scholar] [CrossRef] [PubMed]
- Heidari, H.; Hasanpour, S.; Ebrahim-Saraie, H.S.; Motamedifar, M. High Incidence of Virulence Factors Among Clinical Enterococcus Faecalis Isolates in Southwestern Iran. Infect. Chemother. 2017, 49, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Balaei Gajan, E.; Shirmohammadi, A.; Aghazadeh, M.; Alizadeh, M.; Sighari Deljavan, A.; Ahmadpour, F. Antibiotic Resistance in Enterococcus Faecalis Isolated from Hospitalized Patients. J. Dent. Res. Dent. Clin. Dent. Prospect. 2013, 7, 102–104. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Salamzade, R.; Selleck, E.; Bryan, N.; Mello, S.S.; Manson, A.L.; Earl, A.M. Genes Contributing to the Unique Biology and Intrinsic Antibiotic Resistance of Enterococcus Faecalis. mBio 2020, 11, e02962-20. [Google Scholar] [CrossRef]
- Borgio, J.F.; AlJindan, R.; Alghourab, L.H.; Alquwaie, R.; Aldahhan, R.; Alhur, N.F.; AlEraky, D.M.; Mahmoud, N.; Almandil, N.B.; AbdulAzeez, S. Genomic Landscape of Multidrug Resistance and Virulence in Enterococcus Faecalis IRMC827A from a Long-Term Patient. Biology 2023, 12, 1296. [Google Scholar] [CrossRef]
- Shobo, C.O.; Amoako, D.G.; Allam, M.; Ismail, A.; Essack, S.Y.; Bester, L.A. A Genomic Snapshot of Antibiotic-ResistantEnterococcus Faecalis within Public Hospital Environments in South Africa. Glob. Health Epidemiol. Genom. 2023, 2023, 6639983. [Google Scholar] [CrossRef]
- Balli, E.P.; Venetis, C.A.; Miyakis, S. Systematic Review and Meta-Analysis of Linezolid versus Daptomycin for Treatment of Vancomycin-Resistant Enterococcal Bacteremia. Antimicrob. Agents Chemother. 2014, 58, 734–739. [Google Scholar] [CrossRef]
- Zahedi Bialvaei, A.; Rahbar, M.; Yousefi, M.; Asgharzadeh, M.; Samadi Kafil, H. Linezolid: A Promising Option in the Treatment of Gram-Positives. J. Antimicrob. Chemother. 2017, 72, 354–364. [Google Scholar] [CrossRef] [PubMed]
- Nahid, P.; Mase, S.R.; Migliori, G.B.; Sotgiu, G.; Bothamley, G.H.; Brozek, J.L.; Cattamanchi, A.; Cegielski, J.P.; Chen, L.; Daley, C.L.; et al. Treatment of Drug-Resistant Tuberculosis. An Official ATS/CDC/ERS/IDSA Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2019, 200, e93–e142. [Google Scholar] [CrossRef]
- Leach, K.L.; Brickner, S.J.; Noe, M.C.; Miller, P.F. Linezolid, the First Oxazolidinone Antibacterial Agent. Ann. N. Y. Acad. Sci. 2011, 1222, 49–54. [Google Scholar] [CrossRef]
- Welshman, I.R.; Sisson, T.A.; Jungbluth, G.L.; Stalker, D.J.; Hopkins, N.K. Linezolid Absolute Bioavailability and the Effect of Food on Oral Bioavailability. Biopharm. Drug Dispos. 2001, 22, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Hashemian, S.M.R.; Farhadi, T.; Ganjparvar, M. Linezolid: A Review of Its Properties, Function, and Use in Critical Care. Drug Des. Devel Ther. 2018, 12, 1759–1767. [Google Scholar] [CrossRef]
- Ippolito, J.A.; Kanyo, Z.F.; Wang, D.; Franceschi, F.J.; Moore, P.B.; Steitz, T.A.; Duffy, E.M. Crystal Structure of the Oxazolidinone Antibiotic Linezolid Bound to the 50S Ribosomal Subunit. J. Med. Chem. 2008, 51, 3353–3356. [Google Scholar] [CrossRef] [PubMed]
- Makarov, G.I.; Makarova, T.M. A Noncanonical Binding Site of Linezolid Revealed via Molecular Dynamics Simulations. J. Comput. Aided Mol. Des. 2020, 34, 281–291. [Google Scholar] [CrossRef]
- Alloush, H.M.; Salisbury, V.; Lewis, R.J.; MacGowan, A.P. Pharmacodynamics of Linezolid in a Clinical Isolate of Streptococcus Pneumoniae Genetically Modified to Express Lux Genes. J. Antimicrob. Chemother. 2003, 52, 511–513. [Google Scholar] [CrossRef]
- Brauers, J.; Kresken, M.; Menke, A.; Orland, A.; Weiher, H.; Morrissey, I. Bactericidal Activity of Daptomycin, Vancomycin, Teicoplanin and Linezolid against Staphylococcus Aureus, Enterococcus Faecalis and Enterococcus Faecium Using Human Peak Free Serum Drug Concentrations. Int. J. Antimicrob. Agents 2007, 29, 322–325. [Google Scholar] [CrossRef]
- Gargis, A.S.; Spicer, L.M.; Kent, A.G.; Zhu, W.; Campbell, D.; McAllister, G.; Ewing, T.O.; Albrecht, V.; Stevens, V.A.; Sheth, M.; et al. Sentinel Surveillance Reveals Emerging Daptomycin-Resistant ST736 Enterococcus Faecium and Multiple Mechanisms of Linezolid Resistance in Enterococci in the United States. Front. Microbiol. 2021, 12, 807398. [Google Scholar] [CrossRef] [PubMed]
- Seyedolmohadesin, M.; Kouhzad, M.; Götz, F.; Ashkani, M.; Aminzadeh, S.; Bostanghadiri, N. Emergence of Lineage ST150 and Linezolid Resistance in Enterococcus Faecalis: A Molecular Epidemiology Study of UTIs in Tehran, Iran. Front. Microbiol. 2024, 15, 1464691. [Google Scholar] [CrossRef]
- Lee, S.-M.; Huh, H.J.; Song, D.J.; Shim, H.J.; Park, K.S.; Kang, C.-I.; Ki, C.-S.; Lee, N.Y. Resistance Mechanisms of Linezolid-Nonsusceptible Enterococci in Korea: Low Rate of 23S rRNA Mutations in Enterococcus Faecium. J. Med. Microbiol. 2017, 66, 1730–1735. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, F.; Bai, B.; Lin, Z.; Xu, G.; Chen, Z.; Sun, X.; Zheng, J.; Deng, Q.; Yu, Z. Linezolid Resistance in Enterococcus Faecalis Associated With Urinary Tract Infections of Patients in a Tertiary Hospitals in China: Resistance Mechanisms, Virulence, and Risk Factors. Front. Public Health 2021, 9, 570650. [Google Scholar] [CrossRef]
- Chen, M.; Pan, H.; Lou, Y.; Wu, Z.; Zhang, J.; Huang, Y.; Yu, W.; Qiu, Y. Epidemiological Characteristics and Genetic Structure of Linezolid-Resistant Enterococcus Faecalis. Infect. Drug Resist. 2018, 11, 2397–2409. [Google Scholar] [CrossRef]
- Hua, R.; Xia, Y.; Wu, W.; Yang, M.; Yan, J. Molecular Epidemiology and Mechanisms of 43 Low-Level Linezolid-Resistant Enterococcus Faecalis Strains in Chongqing, China. Ann. Lab. Med. 2019, 39, 36–42. [Google Scholar] [CrossRef]
- Shen, W.; Hu, Y.; Liu, D.; Wang, Y.; Schwarz, S.; Zhang, R.; Cai, J. Prevalence and Genetic Characterization of Linezolid Resistance Gene Reservoirs in Hospital Sewage from Zhejiang Province, China. Sci. Total Environ. 2024, 955, 177162. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Xiao, S.; Han, L.; Wu, Q. Antimicrobial Resistance, Virulence Gene Profiles, and Molecular Epidemiology of Enterococcal Isolates from Patients with Urinary Tract Infections in Shanghai, China. Microbiol. Spectr. 2024, 13, e01217-24. [Google Scholar] [CrossRef] [PubMed]
- Brenciani, A.; Cinthi, M.; Coccitto, S.N.; Massacci, F.R.; Albini, E.; Cucco, L.; Paniccià, M.; Freitas, A.R.; Schwarz, S.; Giovanetti, E.; et al. Global Spread of the Linezolid-Resistant Enterococcus Faecalis ST476 Clonal Lineage Carrying optrA. J. Antimicrob. Chemother. 2024, 79, 846–850. [Google Scholar] [CrossRef]
- Shan, X.; Li, C.; Zhang, L.; Zou, C.; Yu, R.; Schwarz, S.; Shang, Y.; Li, D.; Brenciani, A.; Du, X.-D. poxtA Amplification and Mutations in 23S rRNA Confer Enhanced Linezolid Resistance in Enterococcus Faecalis. J. Antimicrob. Chemother. 2024, 79, 3199–3203. [Google Scholar] [CrossRef]
- Jiang, L.; Xie, N.; Chen, M.; Liu, Y.; Wang, S.; Mao, J.; Li, J.; Huang, X. Synergistic Combination of Linezolid and Fosfomycin Closing Each Other’s Mutant Selection Window to Prevent Enterococcal Resistance. Front. Microbiol. 2020, 11, 605962. [Google Scholar] [CrossRef]
- Cavaco, L.M.; Bernal, J.F.; Zankari, E.; Léon, M.; Hendriksen, R.S.; Perez-Gutierrez, E.; Aarestrup, F.M.; Donado-Godoy, P. Detection of Linezolid Resistance Due to the optrA Gene in Enterococcus Faecalis from Poultry Meat from the American Continent (Colombia). J. Antimicrob. Chemother. 2017, 72, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Long, K.S.; Vester, B. Resistance to Linezolid Caused by Modifications at Its Binding Site on the Ribosome. Antimicrob. Agents Chemother. 2012, 56, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Long, K.S.; Munck, C.; Andersen, T.M.B.; Schaub, M.A.; Hobbie, S.N.; Böttger, E.C.; Vester, B. Mutations in 23S rRNA at the Peptidyl Transferase Center and Their Relationship to Linezolid Binding and Cross-Resistance. Antimicrob. Agents Chemother. 2010, 54, 4705–4713. [Google Scholar] [CrossRef]
- Manoharan, M.; Sawant, A.R.; Prashanth, K.; Sistla, S. Multiple Mechanisms of Linezolid Resistance in Staphylococcus Haemolyticus Detected by Whole-Genome Sequencing. J. Med. Microbiol. 2023, 72, 001737. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.B.; Morales, G.; Hilgers, M.; Kedar, G.C.; Rahawi, S.; José Picazo, J.; Shaw, K.J.; Stein, J.L. Elevated Linezolid Resistance in Clinical Cfr-Positive Staphylococcus Aureus Isolates Is Associated with Co-Occurring Mutations in Ribosomal Protein L3. Antimicrob. Agents Chemother. 2010, 54, 5352–5355. [Google Scholar] [CrossRef]
- Wang, Y.; Lv, Y.; Cai, J.; Schwarz, S.; Cui, L.; Hu, Z.; Zhang, R.; Li, J.; Zhao, Q.; He, T.; et al. A Novel Gene, optrA, That Confers Transferable Resistance to Oxazolidinones and Phenicols and Its Presence in Enterococcus Faecalis and Enterococcus Faecium of Human and Animal Origin. J. Antimicrob. Chemother. 2015, 70, 2182–2190. [Google Scholar] [CrossRef]
- Fu, Y.; Deng, Z.; Shen, Y.; Wei, W.; Xiang, Q.; Liu, Z.; Hanf, K.; Huang, S.; Lv, Z.; Cao, T.; et al. High Prevalence and Plasmidome Diversity of optrA-Positive Enterococci in a Shenzhen Community, China. Front. Microbiol. 2024, 15, 1505107. [Google Scholar] [CrossRef]
- Cai, J.; Schwarz, S.; Chi, D.; Wang, Z.; Zhang, R.; Wang, Y. Faecal Carriage of optrA-Positive Enterococci in Asymptomatic Healthy Humans in Hangzhou, China. Clin. Microbiol. Infect. 2019, 25, e1–e630. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, K.; Liu, Z.; Li, Y.; Xiao, X.; Du, X.-D.; Li, R.; Wang, Z. Genomic Insights into Linezolid-Resistant Enterococci Revealed Its Evolutionary Diversity and poxtA Copy Number Heterogeneity. Int. J. Antimicrob. Agents 2023, 62, 106929. [Google Scholar] [CrossRef]
- Sun, W.; Liu, H.; Liu, J.; Jiang, Q.; Pan, Y.; Yang, Y.; Zhu, X.; Ge, J. Detection of optrA and poxtA Genes in Linezolid-Resistant Enterococcus Isolates from Fur Animals in China. Lett. Appl. Microbiol. 2022, 75, 1590–1595. [Google Scholar] [CrossRef]
- Hou, J.; Xu, Q.; Zhou, L.; Chai, J.; Lin, L.; Ma, C.; Zhu, Y.; Zhang, W. Identification of an Enterococcus Faecium Strain Isolated from Raw Bovine Milk Co-Harbouring the Oxazolidinone Resistance Genes optrA and poxtA in China. Vet. Microbiol. 2024, 293, 110103. [Google Scholar] [CrossRef]
- LaMarre, J.; Mendes, R.E.; Szal, T.; Schwarz, S.; Jones, R.N.; Mankin, A.S. The Genetic Environment of the Cfr Gene and the Presence of Other Mechanisms Account for the Very High Linezolid Resistance of Staphylococcus Epidermidis Isolate 426-3147L. Antimicrob. Agents Chemother. 2013, 57, 1173–1179. [Google Scholar] [CrossRef]
- Shore, A.C.; Lazaris, A.; Kinnevey, P.M.; Brennan, O.M.; Brennan, G.I.; O’Connell, B.; Feßler, A.T.; Schwarz, S.; Coleman, D.C. First Report of Cfr-Carrying Plasmids in the Pandemic Sequence Type 22 Methicillin-Resistant Staphylococcus Aureus Staphylococcal Cassette Chromosome Mec Type IV Clone. Antimicrob. Agents Chemother. 2016, 60, 3007–3015. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Zhang, W.; Du, X.-D.; Krüger, H.; Feßler, A.T.; Ma, S.; Zhu, Y.; Wu, C.; Shen, J.; Wang, Y. Mobile Oxazolidinone Resistance Genes in Gram-Positive and Gram-Negative Bacteria. Clin. Microbiol. Rev. 2021, 34, e0018820. [Google Scholar] [CrossRef] [PubMed]
- Clinical & Laboratory Standards Institute: CLSI Guidelines. Available online: https://clsi.org/ (accessed on 10 March 2025).
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 10 March 2025).
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast Metagenomic Sequence Classification Using Exact Alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An Algorithm for Hybrid Assembly of Short and Long Reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Mikheenko, A.; Prjibelski, A.; Saveliev, V.; Antipov, D.; Gurevich, A. Versatile Genome Assembly Evaluation with QUAST-LG. Bioinformatics 2018, 34, i142–i150. [Google Scholar] [CrossRef]
- Hunt, M.; Mather, A.E.; Sánchez-Busó, L.; Page, A.J.; Parkhill, J.; Keane, J.A.; Harris, S.R. ARIBA: Rapid Antimicrobial Resistance Genotyping Directly from Sequencing Reads. Microb. Genom. 2017, 3, e000131. [Google Scholar] [CrossRef] [PubMed]
- Hasman, H.; Clausen, P.T.L.C.; Kaya, H.; Hansen, F.; Knudsen, J.D.; Wang, M.; Holzknecht, B.J.; Samulioniené, J.; Røder, B.L.; Frimodt-Møller, N.; et al. LRE-Finder, a Web Tool for Detection of the 23S rRNA Mutations and the optrA, Cfr, Cfr(B) and poxtA Genes Encoding Linezolid Resistance in Enterococci from Whole-Genome Sequences. J. Antimicrob. Chemother. 2019, 74, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- Li, H. A Statistical Framework for SNP Calling, Mutation Discovery, Association Mapping and Population Genetical Parameter Estimation from Sequencing Data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Carattoli, A.; Hasman, H. PlasmidFinder and In Silico pMLST: Identification and Typing of Plasmid Replicons in Whole-Genome Sequencing (WGS). Methods Mol. Biol. 2020, 2075, 285–294. [Google Scholar] [CrossRef]
- Seemann, T. ABRicate: Mass Screening of Contigs for Antiobiotic Resistance Genes 2016. Available online: https://github.com/tseemann/abricate (accessed on 10 March 2025).
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest Suite for Rapid Core-Genome Alignment and Visualization of Thousands of Intraspecific Microbial Genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef]
- Ortiz, E.M. Vcf2phylip v2.0: Convert a VCF Matrix into Several Matrix Formats for Phylogenetic Analysis. 2019. Available online: https://zenodo.org/records/2540861 (accessed on 16 March 2025).
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Seemann, T. Snippy: Rapid Haploid Variant Calling and Core SNP Phylogeny. Available online: https://github.com/tseemann/snippy (accessed on 7 January 2025).
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Interactive Tree of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation|Nucleic Acids Research|Oxford Academic. Available online: https://academic.oup.com/nar/article/49/W1/W293/6246398 (accessed on 10 March 2025).
- Kent, A.G.; Spicer, L.M.; Campbell, D.; Breaker, E.; McAllister, G.A.; Ewing, T.O.; Longo, C.; Balbuena, R.; Burroughs, M.; Burgin, A.; et al. Sentinel Surveillance Reveals Phylogenetic Diversity and Detection of Linear Plasmids Harboring vanA and optrA among Enterococci Collected in the United States. Antimicrob. Agents Chemother. 2024, 68, e00591-24. [Google Scholar] [CrossRef]
- Yang, W.; Li, X.; Chen, J.; Zhang, G.; Li, J.; Zhang, J.; Wang, T.; Kang, W.; Gao, H.; Zhang, Z.; et al. Multicentre Evaluation of in Vitro Activity of Contezolid against Drug-Resistant Staphylococcus and Enterococcus. J. Antimicrob. Chemother. 2024, 79, 3132–3141. [Google Scholar] [CrossRef] [PubMed]
- Dadashi, M.; Sharifian, P.; Bostanshirin, N.; Hajikhani, B.; Bostanghadiri, N.; Khosravi-Dehaghi, N.; van Belkum, A.; Darban-Sarokhalil, D. The Global Prevalence of Daptomycin, Tigecycline, and Linezolid-Resistant Enterococcus Faecalis and Enterococcus Faecium Strains From Human Clinical Samples: A Systematic Review and Meta-Analysis. Front. Med. 2021, 8, 720647. [Google Scholar] [CrossRef] [PubMed]
- Quiles-Melero, I.; Gómez-Gil, R.; Romero-Gómez, M.P.; Sánchez-Díaz, A.M.; de Pablos, M.; García-Rodriguez, J.; Gutiérrez, A.; Mingorance, J. Mechanisms of Linezolid Resistance among Staphylococci in a Tertiary Hospital. J. Clin. Microbiol. 2013, 51, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Pfaller, M.A.; Mendes, R.E.; Streit, J.M.; Hogan, P.A.; Flamm, R.K. Five-Year Summary of In Vitro Activity and Resistance Mechanisms of Linezolid against Clinically Important Gram-Positive Cocci in the United States from the LEADER Surveillance Program (2011 to 2015). Antimicrob. Agents Chemother. 2017, 61, e00609-17. [Google Scholar] [CrossRef]
- Besier, S.; Ludwig, A.; Zander, J.; Brade, V.; Wichelhaus, T.A. Linezolid Resistance in Staphylococcus Aureus: Gene Dosage Effect, Stability, Fitness Costs, and Cross-Resistances. Antimicrob. Agents Chemother. 2008, 52, 1570–1572. [Google Scholar] [CrossRef]
- Alonso, M.; Marín, M.; Iglesias, C.; Cercenado, E.; Bouza, E.; García de Viedma, D. Rapid Identification of Linezolid Resistance in Enterococcus spp. Based on High-Resolution Melting Analysis. J. Microbiol. Methods 2014, 98, 41–43. [Google Scholar] [CrossRef]
- Gawryszewska, I.; Żabicka, D.; Hryniewicz, W.; Sadowy, E. Linezolid-Resistant Enterococci in Polish Hospitals: Species, Clonality and Determinants of Linezolid Resistance. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1279–1286. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, G.; Li, J.; Chen, L.; Liu, H.; Bi, W.; Lu, H.; Zhou, T. A High Incidence and Coexistence of Multiresistance Genes Cfr and optrA among Linezolid-Resistant Enterococci Isolated from a Teaching Hospital in Wenzhou, China. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1441–1448. [Google Scholar] [CrossRef]
- Rodríguez-Lucas, C.; Fernández, J.; Vázquez, X.; de Toro, M.; Ladero, V.; Fuster, C.; Rodicio, R.; Rodicio, M.R. Detection of the optrA Gene Among Polyclonal Linezolid-Susceptible Isolates of Enterococcus Faecalis Recovered from Community Patients. Microb. Drug Resist. 2022, 28, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Egan, S.A.; Shore, A.C.; O’Connell, B.; Brennan, G.I.; Coleman, D.C. Linezolid Resistance in Enterococcus Faecium and Enterococcus Faecalis from Hospitalized Patients in Ireland: High Prevalence of the MDR Genes optrA and poxtA in Isolates with Diverse Genetic Backgrounds. J. Antimicrob. Chemother. 2020, 75, 1704–1711. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Shen, Y.; Schwarz, S.; Cai, J.; Lv, Y.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J.; et al. Genetic Environment of the Transferable Oxazolidinone/Phenicol Resistance Gene optrA in Enterococcus Faecalis Isolates of Human and Animal Origin. J. Antimicrob. Chemother. 2016, 71, 1466–1473. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Y.; Schwarz, S.; Cai, J.; Fan, R.; Li, J.; Feßler, A.T.; Zhang, R.; Wu, C.; Shen, J. Co-Location of the Oxazolidinone Resistance Genes optrA and Cfr on a Multiresistance Plasmid from Staphylococcus Sciuri. J. Antimicrob. Chemother. 2016, 71, 1474–1478. [Google Scholar] [CrossRef]
- Yoon, S.; Son, S.H.; Kim, Y.B.; Seo, K.W.; Lee, Y.J. Molecular Characteristics of optrA-Carrying Enterococcus Faecalis from Chicken Meat in South Korea. Poult. Sci. 2020, 99, 6990–6996. [Google Scholar] [CrossRef]
- Eucast: Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 10 March 2025).

| Variable | Total (n = 24) | Women (%) (n = 7) | Men (%) (n = 17) | p Value |
|---|---|---|---|---|
| Age—median (IQR) | 48 (31.0–57.0) | 56 (53.5–62.25) | 43 (27.0–55.0) | 0.092 |
| Previous antibiotic use in 90 days—n (%) | 11 (45.8) | 3 (27.3) | 8 (72.7) | 1 |
| Previous antibiotic in 14 days—n (%) | 9 (37.5) | 3 (33.3) | 6 (66.7) | 0.643 |
| Previous hospitalization in 90 days—n (%) | 13 (54.2) | 4 (30.8) | 9 (69.2) | 0.66 |
| Comorbidities—n (%) | 16 (66.7) | 6 (37.5) | 10 (62.5) | 0.1243 |
| Charlson comorbidity index—median (IQR) | 1 (0–3) | 3 (1.5–3.75) | 0 (0–2) | 0.059 |
| Diabetes mellitus—n (%) | 6 (25.0) | 3 (50) | 3 (50) | 0.2786 |
| Hypertension—n (%) | 5 (20.8) | 3 (60) | 2 (40) | 0.0886 |
| Cardiovascular disease—n (%) | 0 (0) | 0 (0) | 0 (0) | 1 |
| Obesity—n (%) | 1 (4.2) | 1 (100) | 0 (0) | 0.260 |
| Immunosuppression—n (%) | 3 (12.5) | 1 (33.3) | 2 (66.7) | 1 |
| Chronic kidney disease—n (%) | 2 (8.3) | 0 (0) | 2 (100) | 1 |
| Surgical Intervention—n (%) | 16 (66.7) | 2 (12.5) | 14 (87.5) | 0.045 |
| Central line insertion—n (%) | 9 (37.5) | 3 (33.3) | 6 (66.7) | 0.643 |
| Urinary catheter—n (%) | 16 (66.7) | 5 (31.2) | 11 (68.8) | 0.621 |
| Need for vasopressors—n (%) | 7 (29.2) | 3 (42.9) | 4 (57.1) | 0.318 |
| ICU admission—n (%) | 3 (12.5) | 2 (66.7) | 1 (33.3) | 0.155 |
| Hospital stay—median (IQR) | 14 (11.3–20.3) | 14 (10.3–14.8) | 15 (12.0–23.0) | 0.343 |
| In-hospital mortality—n (%) | 4 (16.7) | 3 (75.0) | 1 (25.0) | 0.040 |
| Variable | Total (n = 72) | LSEf (n = 48) | LNSEf (n = 24) | p Value |
|---|---|---|---|---|
| Age—median (IQR) | 48 (31.25–57) | 48 (31.25–57.25) | 48 (31–57) | 0.9096 |
| Male sex—n (%) | 51 (70.8) | 34 (66.7) | 17 (70.8) | 0.928 |
| Previous antibiotic use in 90 days—n (%) | 34 (47.2) | 23 (47.9) | 11 (45.8) | 1 |
| Previous Antibiotic in 14 days—n (%) | 29 (40.3) | 20 (41.7) | 9 (37.5) | 0.932 |
| Previous hospitalization in 90 days—n (%) | 28 (38.9) | 15 (31.2) | 13 (54.2) | 0.104 |
| Comorbidities—n (%) | 40 (55.6) | 23 (47.9) | 17 (70.8) | 0.111 |
| Charlson comorbidity index—median (IQR) | 1 (0–3) | 1.5 (0–3) | 1 (0–3) | 0.905 |
| Diabetes mellitus—n (%) | 15 (20.8) | 9 (19.6) | 6 (25.0) | 0.826 |
| Hypertension—n (%) | 15 (20.8) | 10 (20.8) | 5 (20.8) | 1 |
| Cardiovascular disease—n (%) | 0 (0) | 0 (0) | 0 (0) | 1 |
| Obesity—n (%) | 2 (2.8) | 1 (2.1) | 1 (4.2) | 1 |
| Immunosuppression—n (%) | 3 (4.2) | 0 (0) | 3 (12.5) | 0.033 |
| Chronic kidney disease—n (%) | 10 (13.9) | 8 (16.7) | 2 (8.3) | 0.478 |
| Surgical intervention—n (%) | 27 (37.5) | 11 (22.9) | 16 (66.7) | <0.001 |
| Central line insertion—n (%) | 25 (34.8) | 15 (31.2) | 10 (41.7) | 0.540 |
| Urinary catheter—n (%) | 40 (55.6) | 23 (47.9) | 17 (70.8) | 0.111 |
| Need for vasopressors—n (%) | 14 (19.4) | 7 (14.6) | 7 (29.2) | 0.246 |
| ICU admission—n (%) | 11 (15.3) | 8 (16.7) | 3 (13.0) | 1 |
| Hospital stay—median (IQR) | 12 (4–17.25) | 9.5 (3.75–15.0) | 14 (11.25–20.25) | 0.02 |
| In-hospital mortality—n (%) | 12 (16.7) | 8 (16.7) | 4 (16.7) | 1 |
| Label | Date | Source | AMP | CIP | DAP | NIT | HLG | HLS | LEV | LNZ | PEN | TET | VA | MLST | Genes Associated with Linezolid Resistance | Locations of Linezolid Resistance Genes | Other AMR Genes | Other Identified Plasmids |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ef-1 | 23 July | Traumatology | ≤2 | 1 | 4 | ≤16 | SYN-S | SYN-S | 1 | ≥8 | 2 | ≥16 | 1 | ST101 | optrA (T10G, T35A, C54T, A91G, A107G, T626G, C949T, A1966G) | Plasmid | erm(B), fexA, lsa(A), NarA, NarB, tet(L) | repUS43 |
| Ef-2 | 23 August | Traumatology | ≤2 | 1 | 1 | ≤16 | SYN-S | SYN-R | 1 | ≥8 | 2 | ≥16 | ≤0.5 | ST283 | optrA (T626G, A1541C) | Plasmid | ant(6)-Ia, aph(2″)-Ic, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lsa(A), tet(L), tet(M) | rep2, rep6, rep9b |
| Ef-3 | 23 August | Cardiology | ≤2 | ≥8 | 2 | ≤16 | SYN-R | SYN-R | ≥8 | 4 | 2 | ≥16 | 1 | ST585 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(6)-Ia, ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E),, parC (S80I), str, tet(L) | rep7a, repUS43 |
| Ef-4 | 23 September | General Surgery | ≤2 | 1 | 4 | ≤16 | SYN-S | SYN-S | 1 | 4 | 1 | ≥16 | 1 | ST506 | optrA (G1879A, C1933T) | Chromosome | ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lsa(A), tet(L) | - |
| Ef-5 | 23 October | Infectious Diseases | ≤2 | ≥8 | 2 | ≤16 | SYN-R | SYN-S | ≥8 | 4 | 2 | ≤1 | 1 | ST476 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, gyrA (S83I), lsa(A), parC (S80I | - |
| Ef-6 | 23 October | Pediatrics | ≤2 | 4 | 2 | ≤16 | SYN-R | SYN-R | 4 | ≥8 | 2 | ≥16 | 1 | ST287 | optrA (G1879A, C1933T) | Plasmid | aac(6′)-aph(2″), ant(6)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lsa(A), NarA, NarB, tet(L) | rep1, rep9b |
| Ef-7 | 23 October | Internal Medicine | ≤2 | ≥8 | 1 | ≤16 | SYN-R | SYN-S | ≥8 | 4 | 2 | ≥16 | 1 | ST585 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, dfrG, erm(A), erm(B), fexA, lsa(A), parC (S80I), tet(L) | repUS43 |
| Ef-8 | 23 October | General Surgery | ≤2 | 4 | 2 | ≤16 | SYN-S | SYN-S | 4 | 4 | 2 | ≥16 | 1 | ST202 | optrA (T10G, T35A, C54T, A91G, A107G, T626G, C949T, A1966G) | Plasmid | ant(9)-Ia, dfrG, erm(B), erm(B), fexA, fosB, lsa(A), NarA, NarB, tet(L) | - |
| Ef-9 | 23 December | Gastroenterology | ≤2 | ≥8 | 2 | ≤16 | SYN-R | SYN-S | ≥8 | ≥8 | 1 | ≥16 | 1 | ND | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, dfrG, erm(A), erm(B), fexA, fosB, gyrA (S83I), lsa(A), NarA, NarB, parC (S80I), tet(L) | - |
| Ef-10 | 23 December | Nephrology | ≤2 | ≥8 | 2 | ≤16 | SYN-S | SYN-S | ≥8 | ≥8 | 2 | ≥16 | 1 | ST480 | optrA ((G1879A, C1933T) | Chromosome | ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lsa(A), NarA, NarB, tet(L) | repUS43 |
| Ef-11 | 24 January | Plastic Surgery | ≤2 | 1 | 2 | ≤16 | SYN-R | SYN-R | 1 | 4 | 1 | ≥16 | ≤0.5 | ST506 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, dfrG, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E), tet(L) | repUS43 |
| Ef-12 | 24 January | Urology | ≤2 | 4 | 2 | ≤16 | SYN-R | SYN-R | 2 | 4 | 8 | ≥16 | 1 | ST376 | optrA (T411G, T626G, G866A) | Plasmid | aac(6′)-aph(2″), ant(6)-Ia, ant(6)-Ia, aph(3′)-III, cat, dfrG, erm(A), fexA, lnu(B), lsa(A), lsa(E), tet(L) | repUS43 |
| Ef-13 | 24 January | Thoracic and Cardiovascular Surgery | ≤2 | ≤0.5 | 4 | ≤16 | SYN-R | SYN-R | 1 | ≥8 | 2 | ≥16 | 1 | ST179 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E), tet(M) | repUS43 |
| Ef-14 | 24 January | Thoracic and Cardiovascular Surgery | ≤2 | ≤0.5 | 2 | ≤16 | SYN-R | SYN-R | 1 | ≥8 | 2 | ≥16 | 1 | ST16 | optrA (T626G, A1541C) | Plasmid | aac(6′)-aph(2″), aph(3′)-III, dfrG, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E), tet(M)c | rep9b, repUS43 |
| Ef-15 | 24 February | Hematology | ≤2 | ≥8 | 2 | ≤16 | SYN-S | SYN-S | ≥8 | ≥8 | 2 | ≥16 | ≤0.5 | ST69 | optrA (T411G, T626G, G866A) | Plasmid | cat, dfrG, erm(B), fexA, lsa(A), parC | rep22, repUS43 |
| Ef-16 | 24 March | HIV Unit | ≤2 | ≥8 | 1 | ≤16 | SYN-R | SYN-R | ≥8 | 4 | 2 | ≥16 | ≤0.5 | ST585 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, dfrG, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E), parC (S80I), str, tet(L) | rep7a, repUS43 |
| Ef-17 | 24 March | General Surgery | ≤2 | ≥8 | 4 | ≤16 | SYN-S | SYN-S | ≥8 | ≥8 | 2 | ≥16 | ≤0.5 | ST32 | optrA (T411G, T626G, G866A) | Plasmid | ant(6)-Ia, aph(3′)-III, erm(A), erm(B), fexA, gyrA (S83I), lsa(A), parC (S80I), tet(L) | repUS43 |
| Ef-18 | 24 March | Transplant Unit | ≤2 | 4 | 2 | ≤16 | SYN-S | SYN-S | 4 | 4 | 2 | ≥16 | 2 | ST415 | optrA (T411G, T626G, G866A) | Plasmid | cat, dfrG, erm(A), erm(B), fexA, fosB3, lsa(A), NarA, NarB, tet(L) | rep1, rep9c |
| Ef-19 | 24 May | Infectious Diseases | ≤2 | ≤0.5 | 2 | ≤16 | SYN-R | SYN-R | 0.5 | ≥8 | 2 | ≥16 | 1 | ST40 | optrA (T411G, T626G, G866A) | Plasmid | aac(6′)-aph(2″), ant(6)-Ia, ant(6)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lnu(B), lsa(A), lsa(E), tet(L) | rep18b, rep9b, repUS43 |
| Ef-20 | 24 June | Traumatology | ≤2 | 4 | 2 | ≤16 | SYN-R | SYN-R | ≥8 | ≥8 | 2 | ≥16 | 1 | ST32 | optrA (T10G, T35A, C54T, A91G, A107G, A134T, T626G, C949T, G1278A, A1331G, A1541C, C1933T), cfrA | optrA, chromosome; cfrA, plasmid | aadD, ant(9)-Ia, aph(2″)-Ic, aph(3′)-III, bleO, cat, dfrG, erm(A), erm(B), fexA, fosB3, lnu(A), lsa(A), tet(L) | repUS43 |
| Ef-21 | 24 June | Cardiology | ≤2 | ≥8 | 1 | ≤16 | SYN-R | SYN-S | ≥8 | 4 | 2 | ≥16 | 1 | ST585 | optrA (G1879A, C1933T) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, cat, dfrG, erm(A), fexA, lnu(B), lsa(A), lsa(E), parC (S80I), str, tet(L) | rep7a, repUS43 |
| Ef-22 | 24 June | Plastic Surgery | ≤2 | 4 | 2 | ≤16 | SYN-R | SYN-R | ≥8 | ≥8 | 2 | ≥16 | 1 | ST32 | optrA (T10G, T35A, C54T, A91G, A107G, A134T, T626G, C949T, G1278A, A1331G, A1541C, C1933T), cfrA | optrA, chromosome; cfrA, plasmid | aadD, ant(9)-Ia, aph(2″)-Ic, aph(3′)-III, bleO, cat, dfrG, erm(A), erm(B), fexA, fosB3, lnu(A), lsa(A), tet(L) | repUS43 |
| Ef-23 | 24 June | Thoracic and Cardiovascular Surgery | ≤2 | ≥8 | 2 | ≤16 | SYN-R | SYN-S | ≥8 | 4 | 2 | ≤1 | 1 | ST476 | optrA (T411G, T626G, G866A) | Chromosome | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, gyrA (S83I), lnu(B), lsa(A), lsa(E), parC (S80I) | rep9b, rep9c |
| Ef-24 | 24 June | Neurosurgery | ≤2 | ≤0.5 | 2 | ≤16 | SYN-R | SYN-R | 0.5 | ≥8 | 2 | ≥16 | 1 | ND | optrA (T411G, T626G, G866A) | Plasmid | aac(6′)-aph(2″), ant(6)-Ia, ant(9)-Ia, aph(3′)-III, dfrG, erm(A), erm(B), fexA, lsa(A), parC (S80I), tet(L) | repUS43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Ayala, P.; Perales-Guerrero, L.; Gómez-Quiroz, A.; Avila-Cardenas, B.B.; Gómez-Portilla, K.; Rea-Márquez, E.A.; Vera-Cuevas, V.C.; Gómez-Quiroz, C.A.; Briseno-Ramírez, J.; De Arcos-Jiménez, J.C. Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital. Microorganisms 2025, 13, 684. https://doi.org/10.3390/microorganisms13030684
Martínez-Ayala P, Perales-Guerrero L, Gómez-Quiroz A, Avila-Cardenas BB, Gómez-Portilla K, Rea-Márquez EA, Vera-Cuevas VC, Gómez-Quiroz CA, Briseno-Ramírez J, De Arcos-Jiménez JC. Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital. Microorganisms. 2025; 13(3):684. https://doi.org/10.3390/microorganisms13030684
Chicago/Turabian StyleMartínez-Ayala, Pedro, Leonardo Perales-Guerrero, Adolfo Gómez-Quiroz, Brenda Berenice Avila-Cardenas, Karen Gómez-Portilla, Edson Alberto Rea-Márquez, Violeta Cassandra Vera-Cuevas, Crisoforo Alejandro Gómez-Quiroz, Jaime Briseno-Ramírez, and Judith Carolina De Arcos-Jiménez. 2025. "Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital" Microorganisms 13, no. 3: 684. https://doi.org/10.3390/microorganisms13030684
APA StyleMartínez-Ayala, P., Perales-Guerrero, L., Gómez-Quiroz, A., Avila-Cardenas, B. B., Gómez-Portilla, K., Rea-Márquez, E. A., Vera-Cuevas, V. C., Gómez-Quiroz, C. A., Briseno-Ramírez, J., & De Arcos-Jiménez, J. C. (2025). Whole-Genome Sequencing of Linezolid-Resistant and Linezolid-Intermediate-Susceptibility Enterococcus faecalis Clinical Isolates in a Mexican Tertiary Care University Hospital. Microorganisms, 13(3), 684. https://doi.org/10.3390/microorganisms13030684







