The Microbiome of an Outpatient Sports Medicine Clinic During a Global Pandemic: Effects of Implementation of a Microbiome-Specific Cleaning Program
Abstract
1. Introduction
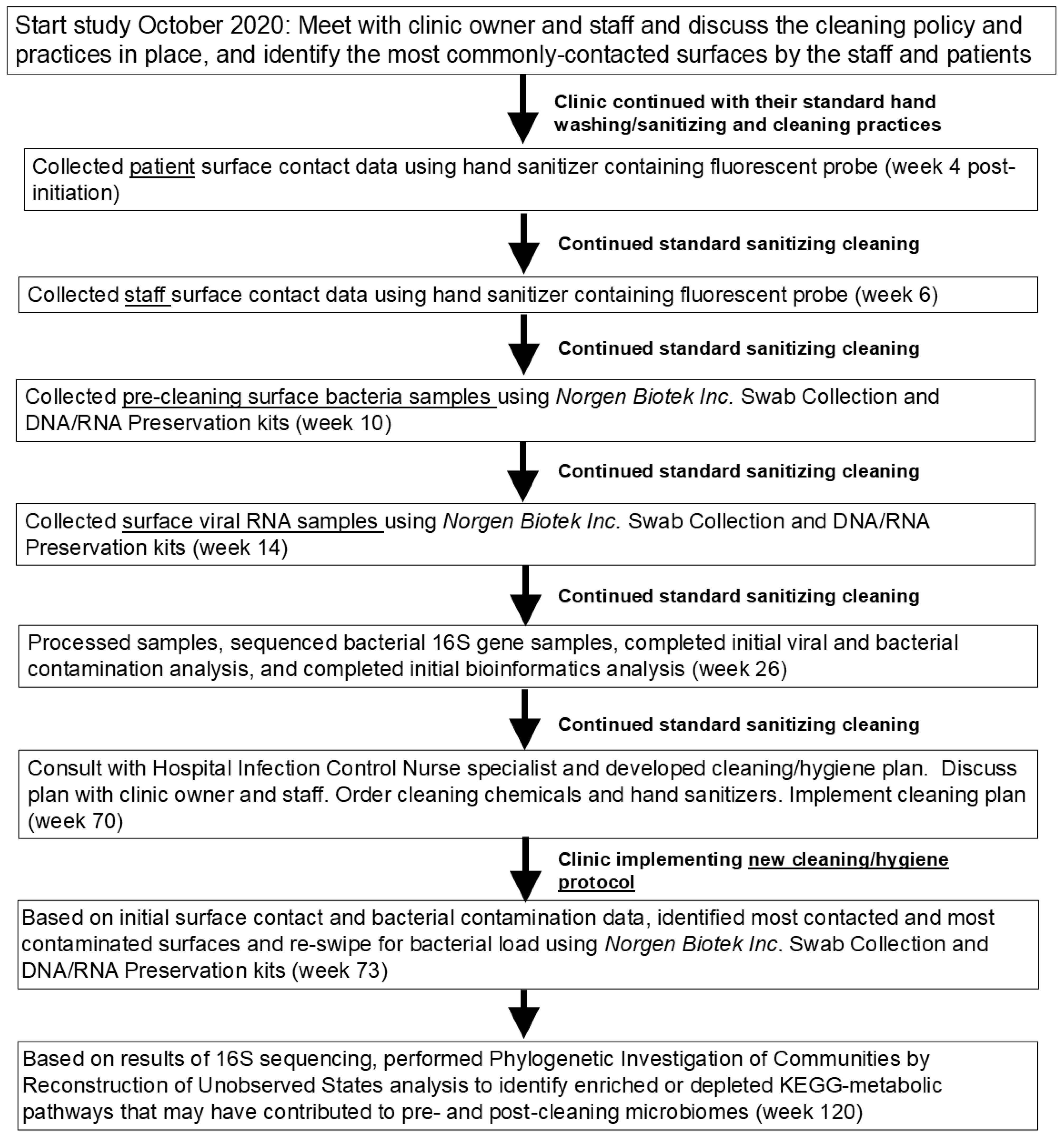
2. Materials and Methods
2.1. Experimental Design, Setting, and Subjects
2.2. Determining Degree of Contact
2.3. Bacterial DNA Sample Collection, Quantification, and Sequencing
2.4. Viral RNA Collection and Detection of SARS-CoV-2
2.5. Bioinformatics and Statistical Analysis
2.6. Clinic Cleaning and Re-Sampling for Bacterial Analysis
3. Results
3.1. Clinic Samples and Cleaning
3.2. Degree of Contact as Measured by Florescent Probe
3.3. Presence of SARS-CoV-2
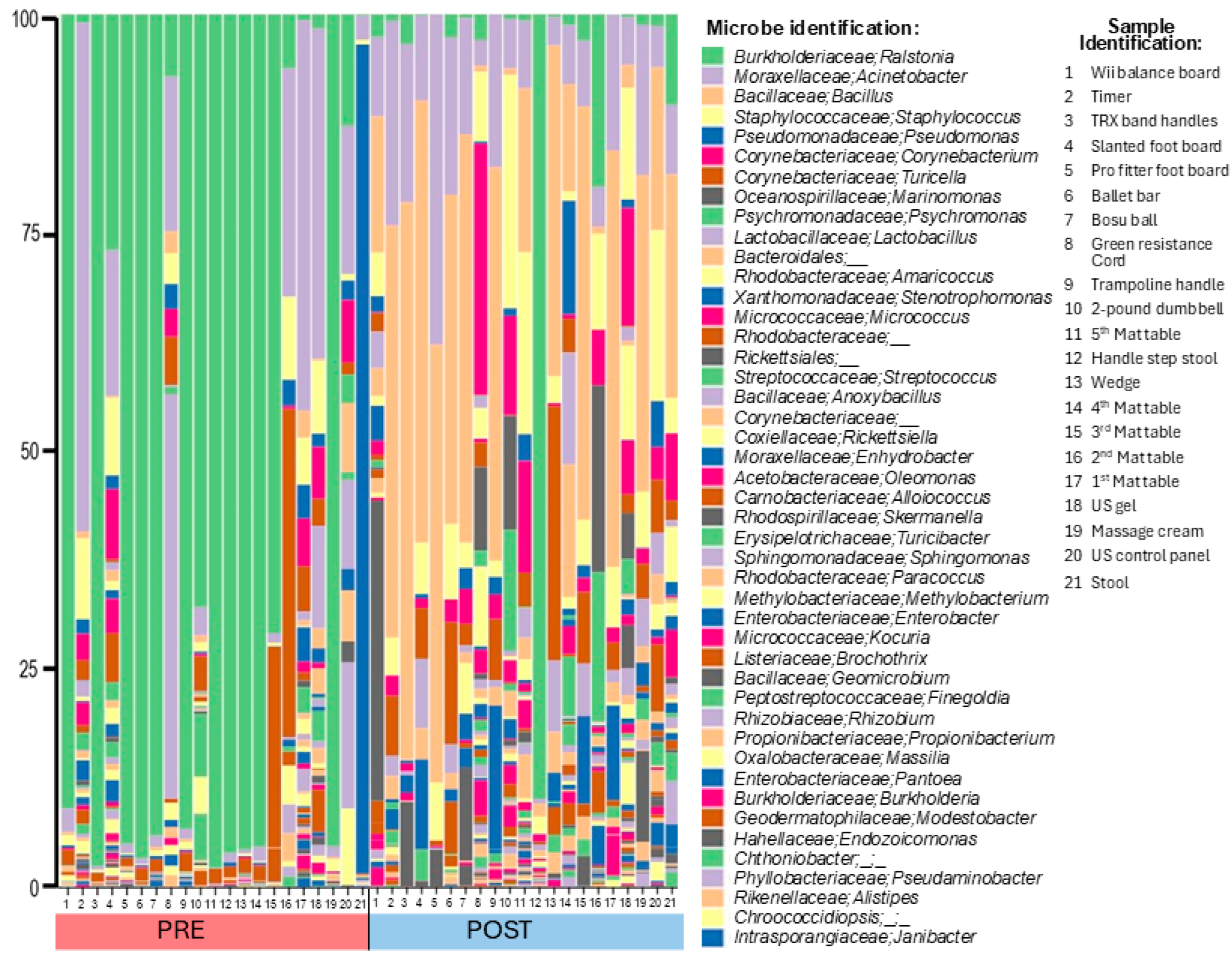
3.4. Bacterial Abundance on Surfaces
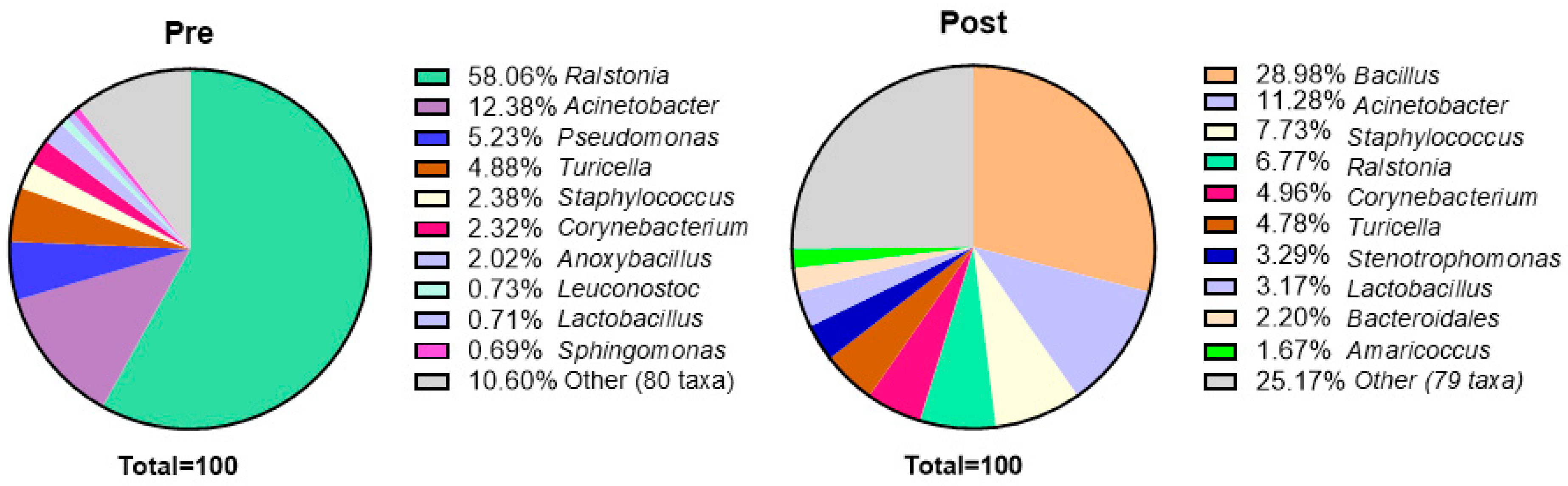
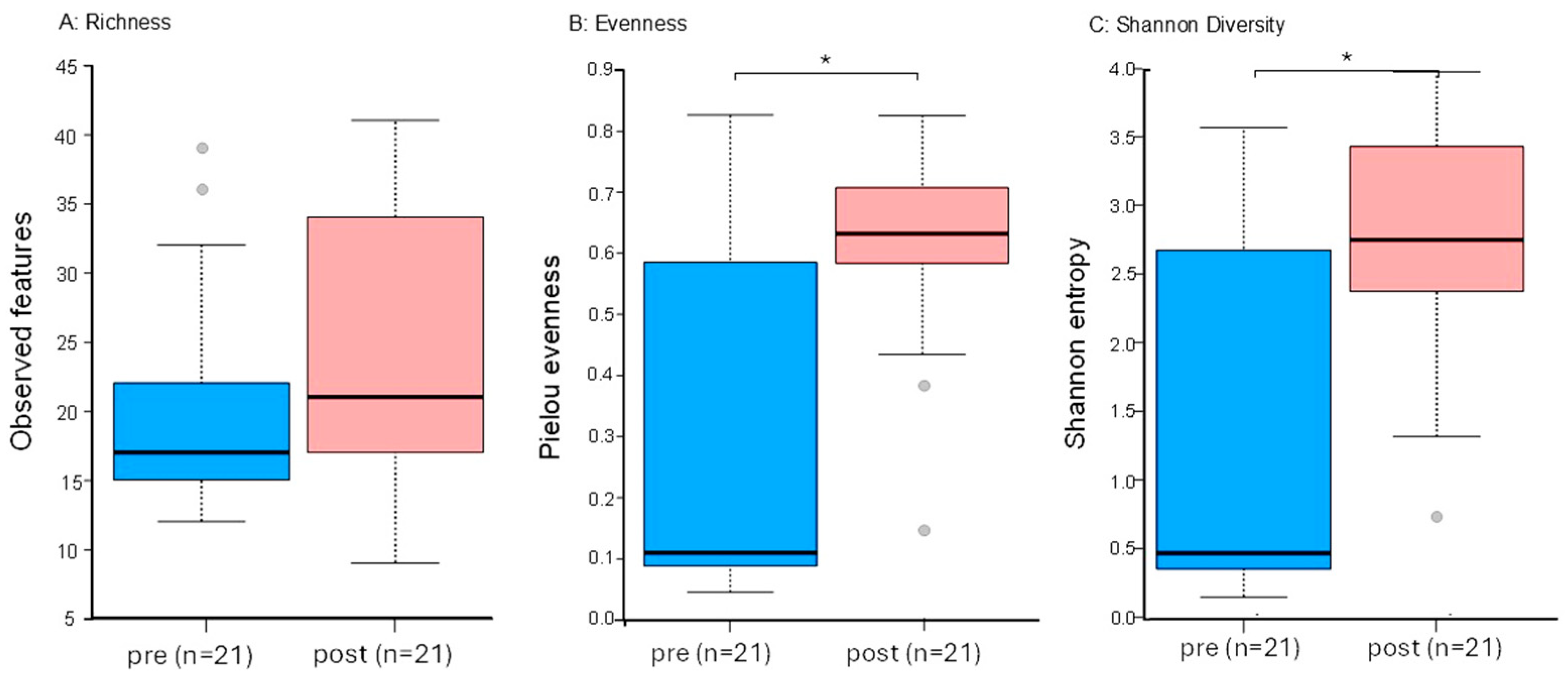
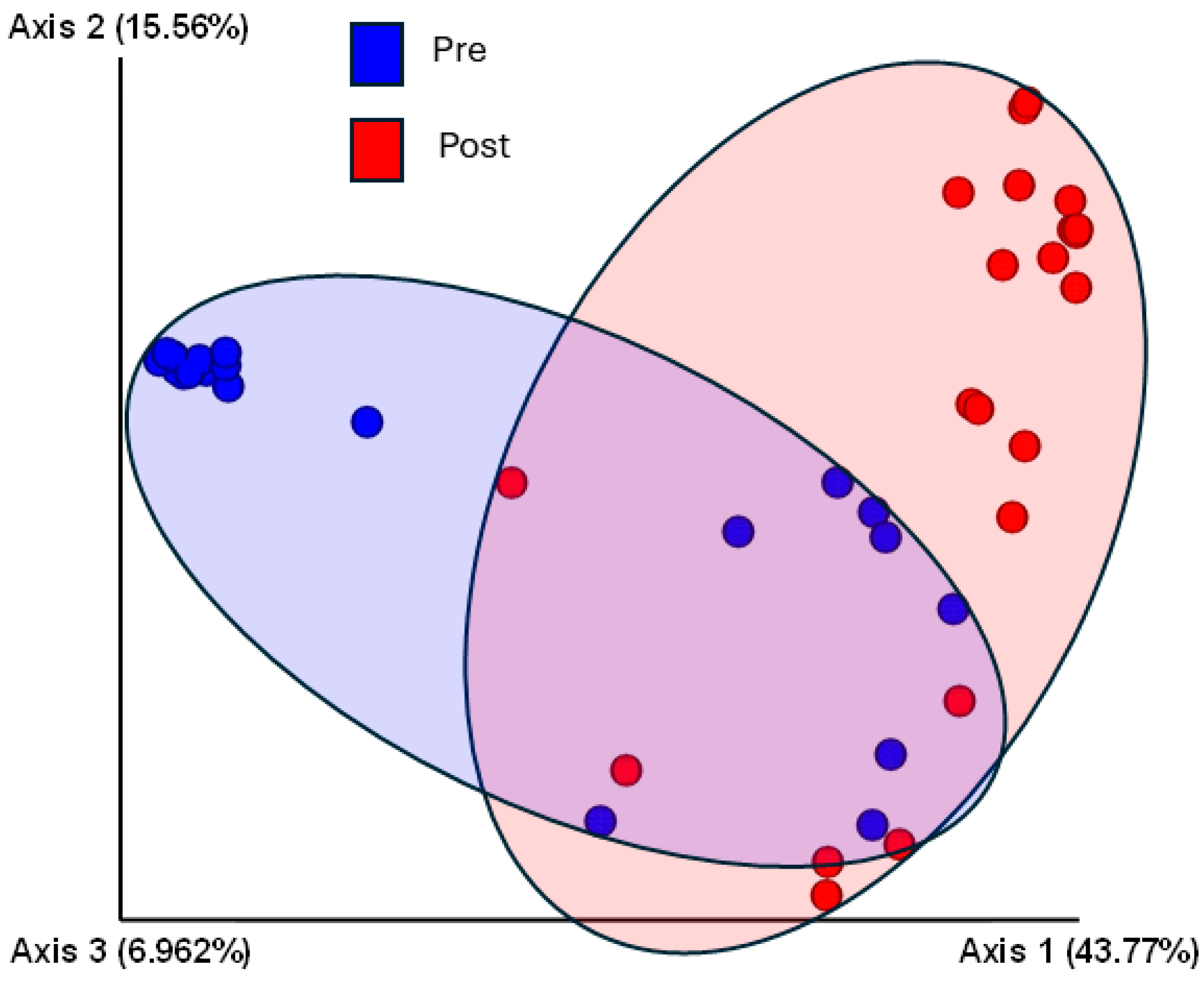
3.5. Sequencing Analysis
3.6. Associated Metabolic Pathways
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McDermott, K.W.; Elixhauser, A.; Sun, R. Trends in hospital inpatient stays in the United States, 2005–2014. Healthc. Cost Util. Proj. 2017, 225, 1–18. [Google Scholar]
- D’Accolti, M.; Soffritti, I.; Mazzacane, S.; Caselli, E. Fighting AMR in the healthcare environment: Microbiome-based sanitation approaches and monitoring tools. Int. J. Mol. Sci. 2019, 20, 1535. [Google Scholar] [CrossRef] [PubMed]
- Galloway, R.V.; Karmarkar, A.M.; Graham, J.E.; Tan, A.; Raji, M.; Granger, C.V.; Ottenbacher, K.J. Hospital Readmission Following Discharge From Inpatient Rehabilitation for Older Adults With Debility. Phys. Ther. 2016, 96, 241–251. [Google Scholar] [CrossRef]
- Gidey, K.; Gidey, M.T.; Hailu, B.Y.; Gebreamlak, Z.B.; Niriayo, Y.L. Clinical and economic burden of healthcare-associated infections: A prospective cohort study. PLoS ONE 2023, 18, e0282141. [Google Scholar]
- Cox, J.; Christensen, B.; Burton, N.; Dunn, K.H.; Finnegan, M.; Ruess, A.; Estill, C. Transmission of SARS-CoV-2 in the workplace: Key findings from a rapid review of the literature. Aerosol Sci. Technol. 2023, 57, 233–254. [Google Scholar]
- Geng, Y.; Wang, Y. Stability and transmissibility of SARS-CoV-2 in the environment. J. Med. Virol. 2023, 95, e28103. [Google Scholar] [PubMed]
- Smith, D.R.; Shirreff, G.; Temime, L.; Opatowski, L. Collateral impacts of pandemic COVID-19 drive the nosocomial spread of antibiotic resistance: A modelling study. PLoS Med. 2023, 20, e1004240. [Google Scholar] [CrossRef]
- Langford, B.J.; Soucy, J.P.; Leung, V.; So, M.; Kwan, A.T.; Portnoff, J.S.; Bertagnolio, S.; Raybardhan, S.; MacFadden, D.R.; Daneman, N. Antibiotic resistance associated with the COVID-19 pandemic: A systematic review and meta-analysis. Clin. Microbiol. Infect. 2023, 29, 302–309. [Google Scholar]
- Chang, H.C.; Chang, C.H.; Tien, K.L.; Tai, C.H.; Lin, L.M.; Lee, T.F.; Ku, S.C.; Fang, C.T.; Chen, Y.C.; Sheng, W.H. Impact of coronavirus disease 2019 (COVID-19) on antimicrobial resistance among major pathogens causing healthcare-associated infection. J. Formos. Med. Assoc. 2024, 123, 123–132. [Google Scholar] [CrossRef]
- Aitken, C.; Jeffries, D.J. Nosocomial spread of viral disease. Clin. Microbiol. Rev. 2001, 14, 528–546. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gontjes, K.J.; Gibson, K.E.; Lansing, B.; Cassone, M.; Mody, L. Contamination of Common Area and Rehabilitation Gym Environment with Multidrug-Resistant Organisms. J. Am. Geriatr. Soc. 2020, 68, 478–485. [Google Scholar] [PubMed]
- Badinski, T.; Seiffert, S.N.; Grässli, F.; Babouee Flury, B.; Besold, U.; Betschon, E.; Biggel, M.; Brucher, A.; Cusini, A.; Dörr, T.; et al. Colonization with resistant bacteria in hospital employees: An epidemiological surveillance and typing study. Antimicrob. Agents Chemother. 2024, 68, e00985-24. [Google Scholar] [PubMed]
- Visalachy, S.; Palraj, K.K.; Kopula, S.S.; Sekar, U. Carriage of Multidrug Resistant Bacteria on Frequently Contacted Surfaces and Hands of Health Care Workers. J. Clin. Diagn. Res. 2016, 10, Dc18. [Google Scholar]
- World Health Organization. Infection Prevention and Control During Health Care When Coronavirus Disease (COVID-19) Is Suspected Or Confirmed: Interim Guidance, 12 July 2021; No. WHO/2019-nCoV/IPC/2021.1; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Olsen, R.J.; Lynch, P.; Coyle, M.B.; Cummings, J.; Bokete, T.; Stamm, W.E. Examination gloves as barriers to hand contamination in clinical practice. JAMA 1993, 270, 350–353. [Google Scholar] [PubMed]
- Raoofi, S.; Pashazadeh Kan, F.; Rafiei, S.; Hosseinipalangi, Z.; Noorani Mejareh, Z.; Khani, S.; Abdollahi, B.; Seyghalani Talab, F.; Sanaei, M.; Zarabi, F.; et al. Global prevalence of nosocomial infection: A systematic review and meta-analysis. PLoS ONE 2023, 18, e0274248. [Google Scholar]
- Dalman, M.; Bhatta, S.; Nagajothi, N.; Thapaliya, D.; Olson, H.; Naimi, H.M.; Smith, T.C. Characterizing the molecular epidemiology of Staphylococcus aureus across and within fitness facility types. BMC Infect. Dis. 2019, 19, 69. [Google Scholar]
- Drew, J.L.; Turner, J.; Mugele, J.; Hasty, G.; Duncan, T.; Zaiser, R.; Cooper, D. Beating the Spread. Simul. Healthc. 2016, 11, 100–105. [Google Scholar]
- Brigando, G.; Sutton, C.; Uebelhor, O.; Pitsoulakis, N.; Pytynia, M.; Dillon, T.; Elliott-Burke, T.; Hubert, N.; Martinez-Guryn, K.; Bolch, C.; et al. The microbiome of an outpatient rehabilitation clinic and predictors of contamination: A pilot study. PLoS ONE 2023, 18, e0281299. [Google Scholar]
- Ugo, C.H.; Cardona, C.J.; Chowanadisai, W.; Lucas, E.A.; Montgomery, M.R. Isolation of gDNA From Fecal Samples of Healthy Subjects for Assessing the Influence of Dietary Pulse Consumption on Tumor Suppressor Gene Expression and Methylation Status. Curr. Dev. Nutr. 2024, 8, 102576. [Google Scholar]
- Poteres, E.; Hubert, N.; Poludasu, S.; Brigando, G.; Moore, J.; Keeler, K.; Isabelli, A.; Ibay, I.C.; Alt, L.; Pytynia, M.; et al. Selective regional alteration of the gut microbiota by diet and antibiotics. Front. Physiol. 2020, 11, 797. [Google Scholar]
- Eren, A.M.; Maignien, L.; Sul, W.J.; Murphy, L.G.; Grim, S.L.; Morrison, H.G.; Sogin, M.L. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 2013, 4, 1111–1119. [Google Scholar] [PubMed]
- Kudo, E.; Israelow, B.; Vogels, C.B.; Lu, P.; Wyllie, A.L.; Tokuyama, M.; Venkataraman, A.; Brackney, D.E.; Ott, I.M.; Petrone, M.E.; et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 2020, 18, e3000867. [Google Scholar] [PubMed]
- Eren, A.M.; Morrison, H.G.; Lescault, P.J.; Reveillaud, J.; Vineis, J.H.; Sogin, M.L. Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015, 9, 968–979. [Google Scholar] [CrossRef]
- Huse, S.M.; Dethlefsen, L.; Huber, J.A.; Welch, D.M.; Relman, D.A.; Sogin, M.L. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008, 4, e1000255. [Google Scholar]
- Fasolo, A.; Deb, S.; Stevanato, P.; Concheri, G.; Squartini, A. ASV vs OTUs clustering: Effects on alpha, beta, and gamma diversities in microbiome metabarcoding studies. PLoS ONE 2024, 19, e0309065. [Google Scholar]
- Leray, M.; Knowlton, N. Visualizing patterns of marine eukaryotic diversity from metabarcoding data using QIIME. In Marine Genomics. Methods in Molecular Biology; Humana Press: New York, NY, USA, 2016; pp. 219–235. [Google Scholar]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar]
- Yang, C.; Mai, J.; Cao, X.; Burberry, A.; Cominelli, F.; Zhang, L. ggpicrust2: An R package for PICRUSt2 predicted functional profile analysis and visualization. Bioinformatics 2023, 39, btad470. [Google Scholar] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar]
- Lampropoulos, P.; Gkentzi, D.; Tzifas, S.; Kapnisi, G.; Karatza, A.; Kolonitsiou, F.; Dimitriou, G. Ralstonia mannitolilytica, an unusual pathogen in the neonatal intensive care unit: A case of neonatal sepsis and literature review. Infect. Disord.-Drug TargetsDisorders) 2021, 21, 168–172. [Google Scholar]
- Viana-Cárdenas, E.; Triana, A.; Cárdenas-Álvarez, J.; Carvajal-Diaz, E.; Mendoza, H.; Viasus, D. A large multicenter Ralstonia pickettii outbreak in critically ill patients during the COVID-19 pandemic: Epidemiological and clinical characteristics of 66 cases. J. Infect. Prev. 2024, 25, 85–88. [Google Scholar]
- Selim, N.A.; Saeed, A.M.; Ibrahim, M.K. Monitoring and controlling bacteria in pharmaceutical industries water system. J. Appl. Microbiol. 2020, 129, 1079–1090. [Google Scholar] [PubMed]
- Cogen, A.L.; Nizet, V.; Gallo, R.L. Skin microbiota: A source of disease or defence? Br. J. Dermatol. 2008, 158, 442–455. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nasir, N.; Sayeed, M.A.; Jamil, B. Ralstonia pickettii bacteremia: An emerging infection in a tertiary care hospital setting. Cureus 2019, 11, e5084. [Google Scholar]
- Gilbert, J.A.; Stephens, B. Microbiology of the built environment. Nat. Rev. Microbiol. 2018, 16, 661–670. [Google Scholar]
- Gilbert, J.A.; Hartmann, E.M. The indoors microbiome and human health. Nat. Rev. Microbiol. 2024, 19, 742–755. [Google Scholar]
- Bosch, T.C.; Wigley, M.; Colomina, B.; Bohannan, B.; Meggers, F.; Amato, K.R.; Azad, M.B.; Blaser, M.J.; Brown, K.; Dominguez-Bello, M.G.; et al. The potential importance of the built-environment microbiome and its impact on human health. Proc. Natl. Acad. Sci. USA 2024, 121, e2313971121. [Google Scholar] [CrossRef]
- Ryan, M.P.; Adley, C.C. Ralstonia spp.: Emerging global opportunistic pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 291–304. [Google Scholar]
- Labarca, J.A.; Trick, W.E.; Peterson, C.L.; Carson, L.A.; Holt, S.C.; Arduino, M.J.; Meylan, M.; Mascola, L.; Jarvis, W.R. Multistate nosocomial outbreak of Ralstonia pickettii colonization associated with an intrinsically contaminated respiratory care solution. Clin. Infect. Dis. 1999, 29, 1281–1286. [Google Scholar]
- Tüzemen, N.Ü.; Önal, U.; Kazak, E.; Tezgeç, N.; Eren, H.; Şimşek, H.; Bakkaloğlu, Z.; Ünaldı, Ö.; Çelebi, S.; Yılmaz, E.; et al. An outbreak of Ralstonia insidiosa bloodstream infections caused by contaminated heparinized syringes. J. Infect. Chemother. 2022, 28, 1387–1392. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; An, T.; Li, X.; Zou, J.; Lin, Z.; Gu, J.; Hu, R.; Fang, Z. Genomic analysis of Ralstonia pickettii reveals the genetic features for potential pathogenicity and adaptive evolution in drinking water. Front. Microbiol. 2024, 14, 1272636. [Google Scholar] [CrossRef] [PubMed]
- Fiore, F.; Cacciatore, S.; Tupputi, S.; Agostino, C.; Montenero, R.; Spaziani, G.; Elmi, D.; Medei, M.; Antocicco, M.; Mammarella, F.; et al. A case of Ralstonia pickettii bloodstream infection and the growing problem of healthcare associated infections in frail older adults. Ann. Geriatr. Med. Res. 2022, 26, 363. [Google Scholar] [CrossRef]
- Adley, C.C.; Ryan, M.P.; Pembroke, J.T.; Saieb, F.M. Ralstonia pickettii: Biofilm formation in high-purity water. In Biofilms: Persistence and Ubiquity; Biofilm Club: Manchester, UK, 2005; pp. 261–271. [Google Scholar]
- Brindle, C.T.; Porter, S.; Bijlani, K.; Arumugam, S.; Matias, R.; Najafi, R.; Fisher, J. Preliminary results of the use of a stabilized hypochlorous acid solution in the management of Ralstonia pickettii biofilm on silicone breast implants. Aesthetic Surg. J. 2018, 38 (Suppl. S2), S52–S61. [Google Scholar] [CrossRef] [PubMed]
- Lompo, P.; Heroes, A.S.; Agbobli, E.; Kühne, V.; Tinto, H.; Affolabi, D.; Jacobs, J. Bacterial Contamination of Antiseptics, Disinfectants and Hand Hygiene Products in Healthcare Facilities in High-Income Countries: A Scoping Review. Hygiene 2023, 3, 136–175. [Google Scholar] [CrossRef]
- Kampf, G. Didecyldimethylammonium Chloride. In Antiseptic Stewardship; Springer: Cham, Switzerland, 2024. [Google Scholar] [CrossRef]
- Spratt, H.G., Jr.; Levine, D.; Bage, J.; Giles, D.K.; Collier, A.G. Topical lotions utilized in outpatient rehabilitation clinics as a potential source of bacterial contamination. Physiother. Theory Pract. 2019, 35, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Spratt, H.G., Jr.; Levine, D.; Tillman, L. Physical therapy clinic therapeutic ultrasound equipment as a source for bacterial contamination. Physiother. Theory Pract. 2014, 30, 507–511. [Google Scholar] [CrossRef]
- Jan, I.; Chen, K.; Sayan, M.; Uprety, P.; Laumbach, R.J.; Ennis, R.D.; Haffty, B.G. Prevalence of Surface Contamination With SARS-CoV-2 in a Radiation Oncology Clinic. JAMA Oncol. 2020, 6, 1632–1634. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]



| Site # | Surface Description | Type of Contact | High Contact | Patients Contact (mg/mL) | Staff Contact (mg/mL) | PRE Total Bacteria (ng/mL) | POST Total Bacteria (ng/mL) | Ralstonia (>0.01% OTUs) | SARS-CoV-2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| PRE | POST | |||||||||
| 1 | Treadmill control panel | hand | 0.0011 | 0.0003 | 0.0004 | NT | + | NT | NA | |
| 2 | NuStep side handles | hand | 0.0113 | 0.0020 | 0.0002 | NT | + | NT | ||
| 3 | NuStep arm rest & seat | hand | 0.0040 | 0.0011 | 0.0003 | NT | + | NT | ||
| 4 | Stationary bike-handle bars | hand | 0.0039 | 0.0004 | 0.0009 | NT | + | NT | ||
| 5 | Stationary bike-control panel | hand | 0.0006 | 0.0002 | 0.0007 | NT | + | NT | NA | |
| 6 | Stair stepper side handles | hand | 0.0011 | 0.0003 | 0.0004 | NT | − | NT | ||
| 7 | Stair stepper control panel | hand | 0.0006 | 0.0002 | 0.0005 | NT | − | NT | ||
| 8 | Pro fitter foot board | foot | * | 0.0039 | 0.0021 | 0.0009 | 0.0014 | − | + | |
| 9 | Wii balance board | foot | * | 0.0012 | 0.0015 | 0.0012 | 0.0001 | + | − | |
| 10 | PT table stand, top surface | hand | 0.0004 | 0.0004 | 0.0005 | NT | − | NT | ||
| 11 | Timer | hand | * | 0.0004 | 0.0001 | 0.0004 | 0.0001 | + | + | |
| 12 | Pulley handles | hand | 0.0005 | 0.0001 | 0.0007 | NT | − | NT | ||
| 14 | Hydrocollator tongs | hand | 0.0004 | 0.0003 | 0.0002 | NT | + | NT | ||
| 15 | TRX band handles | hand | * | 0.0050 | 0.0008 | 0.0090 | 0.0013 | + | + | NA |
| 16 | Slanted foot board | foot | * | 0.0016 | 0.0057 | 0.0011 | 0.0002 | + | − | |
| 17 | FF balance board | foot | 0.0027 | 0.0095 | 0.0002 | NT | + | NT | ||
| 18 | Ballet bar | hand | * | 0.0061 | 0.0020 | 0.0006 | 0.0169 | − | + | |
| 19 | Weight bench | other | 0.0010 | 0.0124 | 0.0181 | + | − | NA | ||
| 20 | Bosu ball | hand | 0.0028 | 0.0070 | 0.0064 | 0.0002 | + | + | ||
| 21 | Purple exercise mat | other | 0.0034 | 0.0011 | 0.0002 | NT | + | NT | ||
| 22 | Green resistance cord | hand | 0.0091 | 0.0005 | 0.0067 | 0.0012 | + | + | NA | |
| 23 | Trampoline handles | hand | 0.0020 | 0.0005 | 0.0040 | 0.0004 | + | + | ||
| 24 | 1 lb dumbbell | hand | 0.0037 | 0.0006 | 0.0004 | NT | + | NT | ||
| 25 | 2 lb dumbbell | hand | 0.0029 | 0.0004 | 0.0076 | 0.0003 | + | + | NA | |
| 26 | 3 lb dumbbell | hand | 0.0019 | 0.0004 | 0.0003 | NT | + | NT | ||
| 27 | 1st mat table | other | 0.0020 | 0.0007 | 0.0005 | 0.0004 | + | + | NA | |
| 28 | 2nd mat table | other | 0.0011 | 0.0008 | 0.0002 | 0.0002 | + | + | NA | |
| 29 | 3rd mat table | other | 0.0022 | 0.0009 | 0.0083 | 0.0003 | + | − | NA | |
| 30 | 4th mat table | other | 0.0043 | 0.0009 | 0.0001 | 0.0232 | + | + | ||
| 31 | 5th mat table | foot | 0.0020 | 0.0007 | 0.0004 | 0.0001 | − | − | ||
| 32 | 6th mat table | foot | 0.0068 | 0.0010 | 0.0056 | NT | + | NT | ||
| 33 | Handle for step stool | hand | 0.0024 | 0.0004 | 0.0058 | 0.0309 | + | + | NA | |
| 34 | Wedge | other | 0.0017 | 0.0006 | 0.0048 | 0.0002 | + | + | ||
| 35 | Ultrasound gel | other | 0.0002 | 0.0003 | 0.0022 | 0.0202 | + | + | ||
| 36 | Massage cream | other | * | 0.0006 | 0.0006 | 0.0090 | 0.0001 | + | − | NA |
| 37 | Massage Gun | hand | 0.0013 | 0.0006 | 0.0002 | NT | + | NT | ||
| 38 | US control panel & head | hand | 0.0011 | 0.0004 | 0.0007 | 0.0001 | + | + | ||
| 39 | Stool | hand | 0.0005 | 0.0003 | 0.0037 | 0.0001 | − | + | ||
| Total hand, foot and other contacts | Median= | 0.00180 | 0.00060 | 0.00295 | 0.0003 | |||||
| 23 Hand, 6 Foot, 8 Other | 25%/75%= | 0.0006/0.0039 | 0.0003/0.0011 | 0.00057/0.00692 | 0.0001/0.00527 | |||||
| Cleaning Strategy | New Products Implemented |
|---|---|
| Switch from a sodium hydroxide-based weekly floor cleaner (Mr. Clean® All Purpose Cleaner) to multiple hospital-based cleaners. Switch to disinfectant cleaners that did not need to be diluted with tap water. | Ecolab® Broad Spectrum Quaternary Disinfectant Cleaning Solution. |
| Switch from CaviWipes (Metrix®) and Lysol® Antibacterial Cleaner to a Chlorine-based cleaner for other clinic surfaces. | Clorox™ Bleach Germicidal Cleaner * |
| Recommend frequent rotation of cleaning agents to ensure microbes do not develop resistance to any one specific agent or compound. | |
| Switch from reliance primarily on alcohol gel-based hand sanitizers (Purell®) to cleaning hands with soap and water whenever possible. | |
| Continue regular use of gloves by clinic staff with frequent changing of gloves. | |
| Encourage patients to wash hands or use hand sanitizer frequently. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Russell, G.; Alegoz, R.; Hester, K.; Sierzega, K.L.; Szul, M.J.; Hubert, N.; Rylander, T.; Jensen, S.; Ciancio, M.J.; Martinez-Guryn, K.; et al. The Microbiome of an Outpatient Sports Medicine Clinic During a Global Pandemic: Effects of Implementation of a Microbiome-Specific Cleaning Program. Microorganisms 2025, 13, 737. https://doi.org/10.3390/microorganisms13040737
Russell G, Alegoz R, Hester K, Sierzega KL, Szul MJ, Hubert N, Rylander T, Jensen S, Ciancio MJ, Martinez-Guryn K, et al. The Microbiome of an Outpatient Sports Medicine Clinic During a Global Pandemic: Effects of Implementation of a Microbiome-Specific Cleaning Program. Microorganisms. 2025; 13(4):737. https://doi.org/10.3390/microorganisms13040737
Chicago/Turabian StyleRussell, Greer, Rabia Alegoz, Kelley Hester, Kayla L. Sierzega, Martin J. Szul, Nathaniel Hubert, Timothy Rylander, Sarah Jensen, Mae J. Ciancio, Kristina Martinez-Guryn, and et al. 2025. "The Microbiome of an Outpatient Sports Medicine Clinic During a Global Pandemic: Effects of Implementation of a Microbiome-Specific Cleaning Program" Microorganisms 13, no. 4: 737. https://doi.org/10.3390/microorganisms13040737
APA StyleRussell, G., Alegoz, R., Hester, K., Sierzega, K. L., Szul, M. J., Hubert, N., Rylander, T., Jensen, S., Ciancio, M. J., Martinez-Guryn, K., & Evans, C. C. (2025). The Microbiome of an Outpatient Sports Medicine Clinic During a Global Pandemic: Effects of Implementation of a Microbiome-Specific Cleaning Program. Microorganisms, 13(4), 737. https://doi.org/10.3390/microorganisms13040737





