Comparative Genomic Profiles of Salmonella Typhimurium and Salmonella Dublin Bovine Isolates from the U.S. Indicate Possible Factors Associated with the Host Adaptation of Salmonella Dublin in the Region
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Hierarchical Clustering of Core Genome Multilocus Sequence (HierCC-cgMLST) Analysis
2.3. Antimicrobial, Virulence, Stress Genes, and Plasmid Identification Analysis
2.4. Statistical Analysis
3. Results
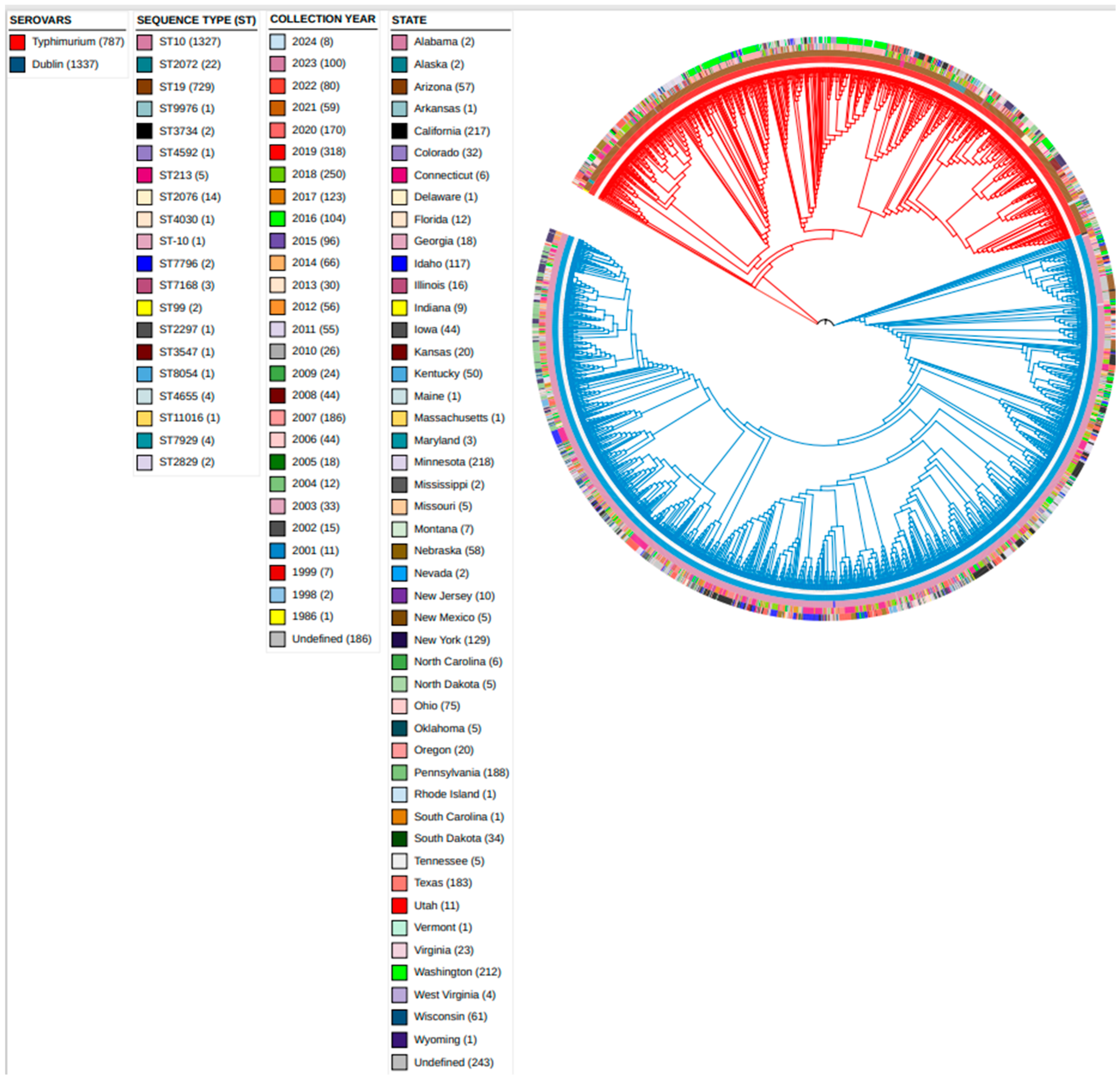
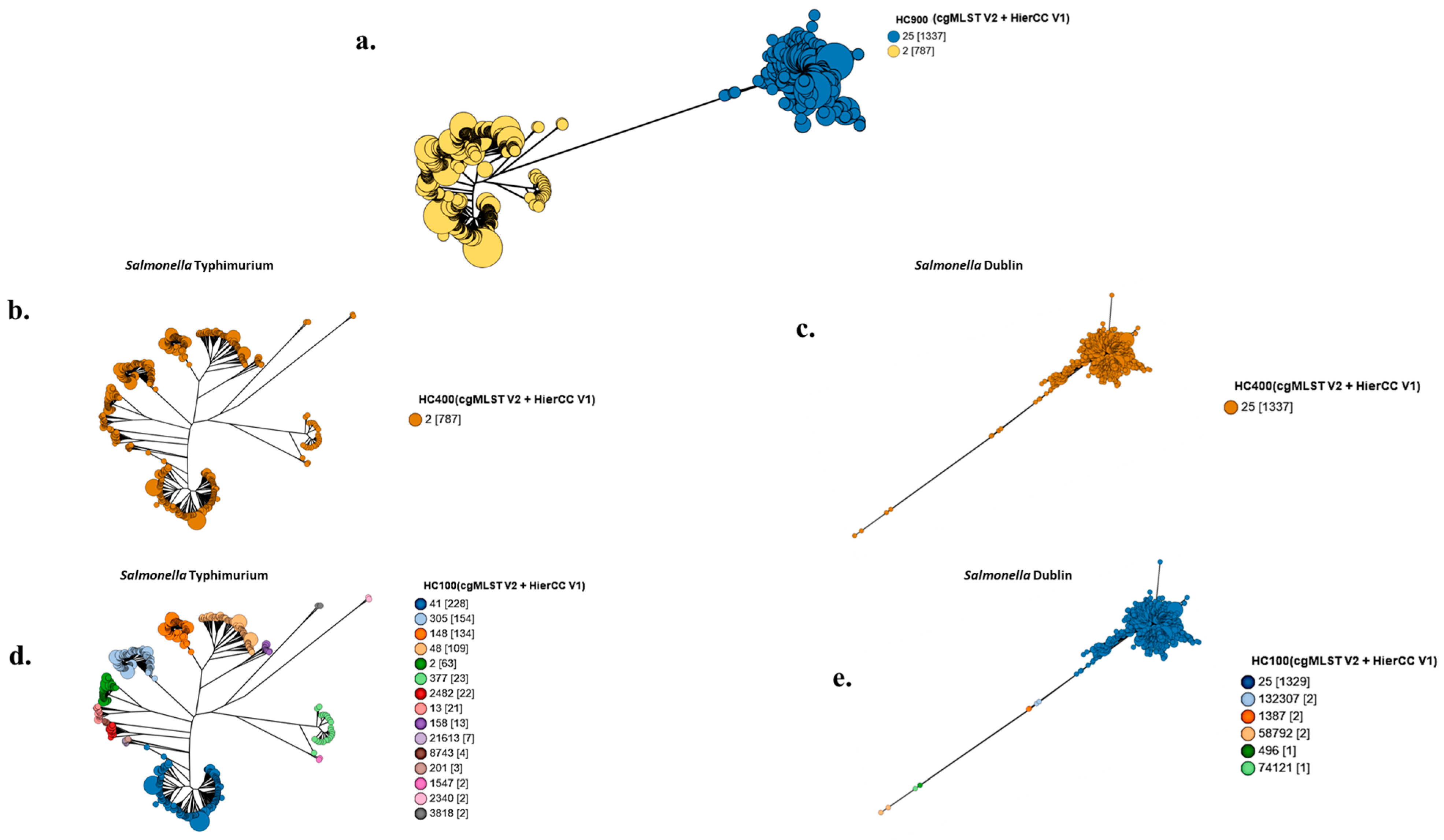
3.1. Grape Tree Phylogeny Based on HierCC-cgMLST
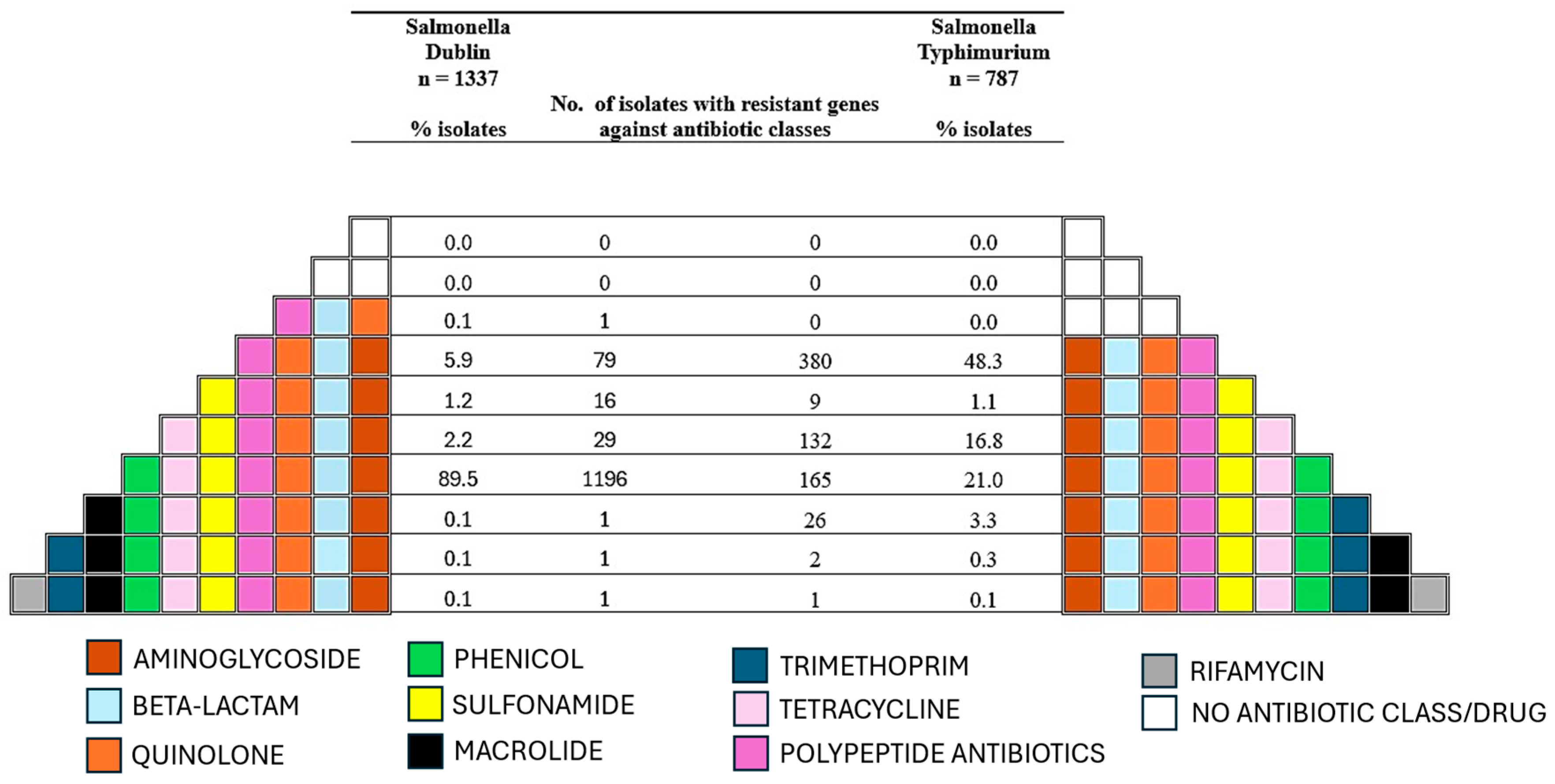
3.2. Antimicrobial Resistance Genes
3.3. Virulence Genes
3.4. Stress Genes
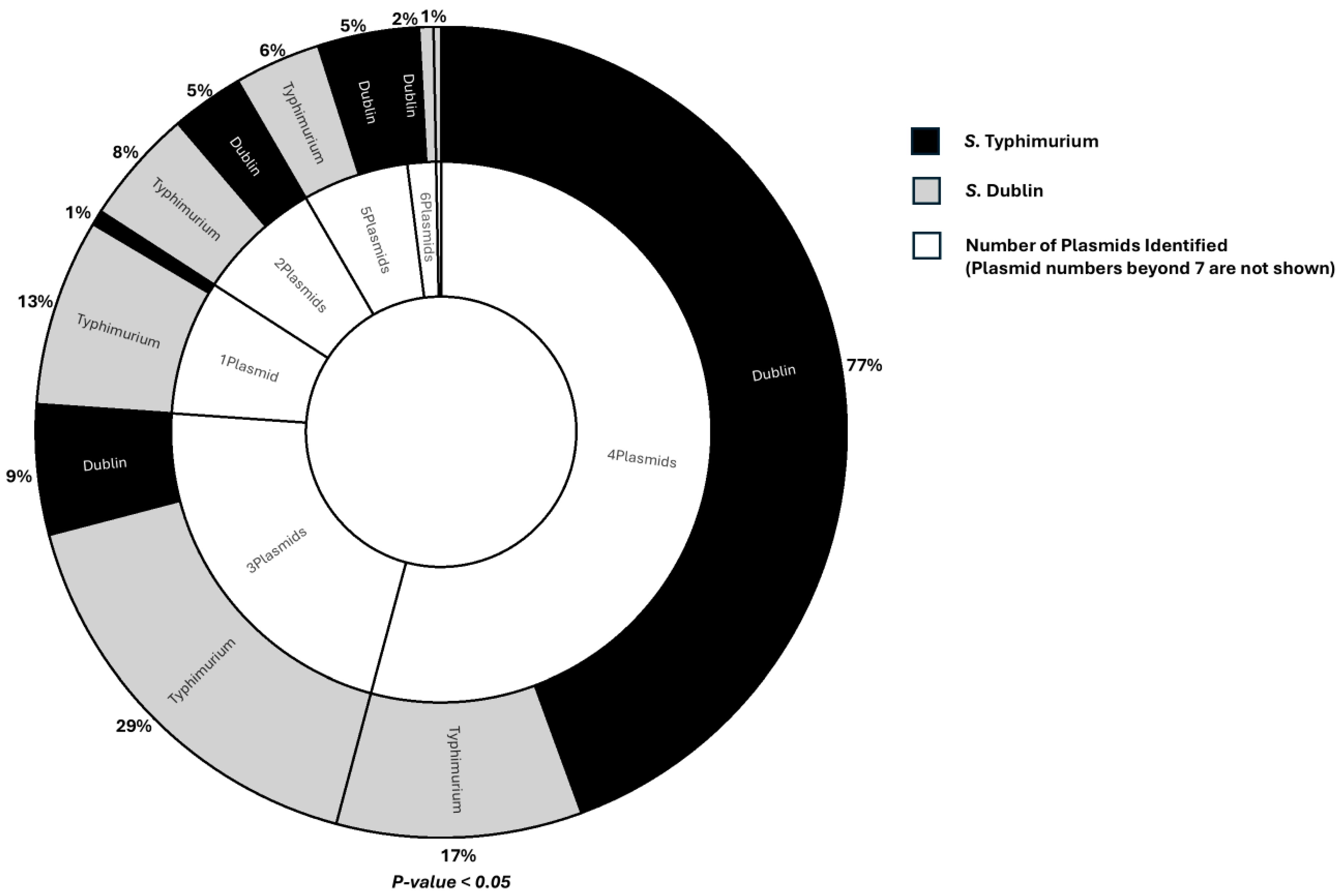
3.5. Plasmids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salaheen, S.; Kim, S.W.; Haley, B.J.; Van Kessel, J.A.S. Differences between the Global Transcriptomes of Salmonella enterica Serovars Dublin and Cerro Infecting Bovine Epithelial Cells. BMC Genom. 2022, 23, 498. [Google Scholar] [CrossRef] [PubMed]
- Velasquez-Munoz, A.; Castro-Vargas, R.; Cullens-Nobis, F.M.; Mani, R.; Abuelo, A. Review: Salmonella Dublin in Dairy Cattle. Front. Vet. Sci. 2024, 10, 1331767. [Google Scholar] [CrossRef] [PubMed]
- Costa, L.F.; Paixão, T.A.; Tsolis, R.M.; Bäumler, A.J.; Santos, R.L. Salmonellosis in Cattle: Advantages of Being an Experimental Model. Res. Vet. Sci. 2012, 93, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Fenske, G.J.; Thachil, A.; McDonough, P.L.; Glaser, A.; Scaria, J.; Delaye, L. Geography Shapes the Population Genomics of Salmonella enterica Dublin. Genome Biol. Evol. 2019, 11, 2220–2231. [Google Scholar] [CrossRef]
- Kingsley, R.A.; Kay, S.; Connor, T.; Barquist, L.; Sait, L.; Holt, K.E.; Sivaraman, K.; Wileman, T.; Goulding, D.; Clare, S.; et al. Genome and Transcriptome Adaptation Accompanying Emergence of the Definitive Type 2 Host-Restricted Salmonella enterica Serovar Typhimurium Pathovar. MBio 2013, 5, 10–1128. [Google Scholar] [CrossRef]
- Tanner, J.R.; Kingsley, R.A. Evolution of Salmonella within Hosts. Trends Microbiol. 2018, 26, 986–998. [Google Scholar] [CrossRef]
- Tasmin, R.; Hasan, N.A.; Grim, C.J.; Grant, A.; Choi, S.Y.; Alam, M.S.; Bell, R.; Cavanaugh, C.; Balan, K.V.; Babu, U.S.; et al. Genotypic and Phenotypic Characterization of Multidrug Resistant Salmonella Typhimurium and Salmonella Kentucky Strains Recovered from Chicken Carcasses. PLoS ONE 2017, 12, e0176938. [Google Scholar] [CrossRef]
- Hu, L.; Brown, E.W.; Zhang, G. Diversity of Antimicrobial Resistance, Stress Resistance, and Virulence Factors of Salmonella, Shiga Toxin-Producing Escherichia coli, and Listeria monocytogenes from Produce, Spices, and Tree Nuts by Whole Genome Sequencing. Front. Sustain. Food Syst. 2023, 7, 1281005. [Google Scholar] [CrossRef]
- Fang, Y.; Tran, F.; Stanford, K.; Yang, X. Stress Resistance and Virulence Gene Profiles Associated with Phylogeny and Phenotypes of Escherichia coli from Cattle. J. Food Prot. 2023, 86, 100122. [Google Scholar] [CrossRef]
- Beceiro, A.; Tomás, M.; Bou, G. Antimicrobial Resistance and Virulence: A Successful or Deleterious Association in the Bacterial World? Clin. Microbiol. Rev. 2013, 26, 185–230. [Google Scholar] [CrossRef]
- Klose, C.; Scuda, N.; Ziegler, T.; Eisenberger, D.; Hanczaruk, M.; Riehm, J.M. Whole-Genome Investigation of Salmonella Dublin Considering Mountain Pastures as Reservoirs in Southern Bavaria, Germany. Microorganisms 2022, 10, 885. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Lyu, N.; Li, Z.; Ma, S.; Cao, D.; Pan, Y.; Hu, Y.; Huang, H.; Gao, G.F.; et al. The Temporal Dynamics of Antimicrobial-Resistant Salmonella enterica and Predominant Serovars in China. Natl. Sci. Rev. 2023, 10, nwac269. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, G.L.; MacCannell, D.R.; Taylor, J.; Carleton, H.A.; Neuhaus, E.B.; Bradbury, R.S.; Posey, J.E.; Gwinn, M. Pathogen Genomics in Public Health. N. Engl. J. Med. 2019, 381, 2569–2580. [Google Scholar] [CrossRef]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Canese, K.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef]
- Sayers, E.W.; Cavanaugh, M.; Clark, K.; Pruitt, K.D.; Sherry, S.T.; Yankie, L.; Karsch-Mizrachi, I. GenBank 2024 Update. Nucleic Acids Res. 2024, 52, D134–D137. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X.; Zhu, B.; Lyu, N.; Liu, Y.; Ma, S.; Jia, S.; Wan, B.; Du, Y.; Zhang, G.; et al. Genomic Analysis of Almost 8,000 Salmonella Genomes Reveals Drivers and Landscape of Antimicrobial Resistance in China. Microbiol. Spectr. 2023, 11, e0208023. [Google Scholar] [CrossRef]
- Frentrup, M.; Zhou, Z.; Steglich, M.; Meier-Kolthoff, J.P.; Göker, M.; Riedel, T.; Bunk, B.; Spröer, C.; Overmann, J.; Blaschitz, M.; et al. A Publicly Accessible Database for Clostridioides Difficile Genome Sequences Supports Tracing of Transmission Chains and Epidemics. Microb. Genom. 2020, 6, mgen000410. [Google Scholar] [CrossRef] [PubMed]
- Bonifait, L.; Thépault, A.; Baugé, L.; Rouxel, S.; Le Gall, F.; Chemaly, M. Occurrence of Salmonella in the Cattle Production in France. Microorganisms 2021, 9, 872. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. Grapetree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- Achtman, M.; Zhou, Z.; Alikhan, N.F.; Tyne, W.; Parkhill, J.; Cormican, M.; Chiou, C.S.; Torpdahl, M.; Litrup, E.; Prendergast, D.M.; et al. Genomic Diversity of Salmonella enterica—The UoWUCC 10K Genomes Project. Wellcome Open Res. 2021, 5, 223. [Google Scholar] [CrossRef]
- Wheeler, T.J. Large-Scale Neighbor-Joining with NINJA. In Algorithms in Bioinformatics: 9th International Workshop, WABI 2009, Philadelphia, PA, USA, September 12–13, 2009. Proceedings 9; Springer: Berlin/Heidelberg, Germany, 2009; Volume 5724, pp. 375–389. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded Curation, Support for Machine Learning, and Resistome Prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog Facilitate Examination of the Genomic Links among Antimicrobial Resistance, Stress Response, and Virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-Annot, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef]
- Maguire, M.; DeLappe, N.; Clarke, C.; Touhy, A.; Carlino-MacDonald, U.; Hutson, A.; Cormican, M.; Brennan, W.; Devane, G.; Morris, D.; et al. Genomic and Phylogenetic Analysis of Hypervirulent Klebsiella pneumoniae ST23 in Ireland. Microb. Genom. 2025, 11, 001373. [Google Scholar] [CrossRef]
- Fu, Y.; Smith, J.C.; Shariat, N.W.; M’ikanatha, N.M.; Dudley, E.G. Evidence for Common Ancestry and Microevolution of Passerine-Adapted Salmonella enterica Serovar Typhimurium in the UK and USA. Microb. Genom. 2022, 8, 000775. [Google Scholar] [CrossRef]
- Srednik, M.E.; Lantz, K.; Hicks, J.A.; Morningstar-Shaw, B.R.; Mackie, T.A.; Schlater, L.K. Antimicrobial Resistance and Genomic Characterization of Salmonella Dublin Isolates in Cattle from the United States. PLoS ONE 2021, 16, e0249617. [Google Scholar] [CrossRef]
- Carhuaricra Huaman, D.E.; Luna Espinoza, L.R.; Rodríguez Cueva, C.L.; Duran Gonzales, C.G.; Rosadio Alcántara, R.H.; Setubal, J.C.; Maturrano Hernández, L. Genomic Characterization of Salmonella Typhimurium Isolated from Guinea Pigs with Salmonellosis in Lima, Peru. Microorganisms 2022, 10, 1726. [Google Scholar] [CrossRef]
- Ali, M.S.; Na, S.-H.; Moon, B.-Y.; Kang, H.Y.; Kang, H.-S.; Kim, S.-J.; Kim, T.-S.; Heo, Y.-E.; Hwang, Y.-J.; Yoon, S.S.; et al. Antimicrobial Resistance Profiles and Molecular Characteristics of Extended-Spectrum β -Lactamase-Producing Salmonella enterica Serovar Typhimurium Isolates from Food Animals During 2010–2021 in South Korea. Foodborne Pathog. Dis. 2024, 21, 634–642. [Google Scholar] [CrossRef]
- United States Food and Drug Administration. Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals. 2021. Available online: https://www.fda.gov/media/154820/download (accessed on 14 February 2025).
- Bren, L. Animal Health and Consumer Protection. FDA Consum. Mag. 2006, 42. Available online: https://www.fda.gov/files/about%20fda/published/Animal-Health-and-Consumer-Protection.pdf (accessed on 2 December 2024).
- Soares, V.M.; Pereira, J.G.; Barreto, F.; Jank, L.; Rau, R.B.; Ribeiro, C.B.D.; Dos Santos Castilhos, T.; Tomaszewski, C.A.; Hillesheim, D.R.; Mondadori, R.G.; et al. Residues of Veterinary Drugs in Animal Products Commercialized in the Border Region of Brazil, Argentina, and Uruguay. J. Food Prot. 2022, 85, 980–986. [Google Scholar] [CrossRef] [PubMed]
- United States Department of Agriculture. Antimicrobial Use and Stewardship on U.S. Feedlots. 2017. Available online: https://www.aphis.usda.gov/sites/default/files/amu-feedlots.pdf (accessed on 12 December 2024).
- Zhang, F.; Liu, F.; Sheng, X.; Liu, Q.; Cui, L.; Cao, Z.; Hu, T.; Li, D.; Dai, M. Bacitracin-Resistant Staphylococcus aureus Induced in Chicken Gut and in Vitro under Bacitracin Exposure. Microb. Pathog. 2024, 191, 106666. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, M.S.; Tolmasky, M.E. Aminoglycoside Modifying Enzymes. Drug Resist. Updat. 2010, 13, 151–171. [Google Scholar] [CrossRef]
- Kagambèga, A.; McMillan, E.A.; Bouda, S.C.; Hiott, L.M.; Ramadan, H.; Soro, D.K.; Sharma, P.; Gupta, S.K.; Barro, N.; Jackson, C.R.; et al. Resistance Genes, Plasmids, Multilocus Sequence Typing (MLST), and Phenotypic Resistance of Non-Typhoidal Salmonella (NTS) Isolated from Slaughtered Chickens in Burkina Faso. Antibiotics 2022, 11, 782. [Google Scholar] [CrossRef]
- Lin, X.; Kück, U. Cephalosporins as Key Lead Generation Beta-Lactam Antibiotics. Appl. Microbiol. Biotechnol. 2022, 106, 8007–8020. [Google Scholar] [CrossRef]
- Wang, S.; You, C.; Memon, F.Q.; Zhang, G.; Sun, Y.; Si, H. BaeR Participates in Cephalosporins Susceptibility by Regulating the Expression Level of Outer Membrane Proteins in Escherichia coli. J. Biochem. 2021, 169, 101–108. [Google Scholar] [CrossRef]
- Pavelquesi, S.L.S.; de Oliveira Ferreira, A.C.A.; Rodrigues, A.R.M.; de Souza Silva, C.M.; Orsi, D.C.; da Silva, I.C.R. Presence of Tetracycline and Sulfonamide Resistance Genes in Salmonella spp.: Literature Review. Antibiotics 2021, 10, 1314. [Google Scholar] [CrossRef]
- Lunara Santos Pavelquesi, S.; Carolina Almeida de Oliveira Ferreira, A.; Fernandes Silva Rodrigues, L.; Maria de Souza Silva, C.; Cristina Rodrigues da Silva, I.; Castilho Orsi, D. Prevalence and Antimicrobial Resistance of Salmonella spp. Isolated From Chilled Chicken Meat Commercialized at Retail in Federal District, Brazil. J. Food Prot. 2023, 86, 100130. [Google Scholar] [CrossRef]
- Schwan, C.L.; Lomonaco, S.; Bastos, L.M.; Cook, P.W.; Maher, J.; Trinetta, V.; Bhullar, M.; Phebus, R.K.; Gragg, S.; Kastner, J.; et al. Genotypic and Phenotypic Characterization of Antimicrobial Resistance Profiles in Non-Typhoidal Salmonella enterica Strains Isolated From Cambodian Informal Markets. Front. Microbiol. 2021, 12, 711472. [Google Scholar] [CrossRef]
- Vrancianu, C.O.; Popa, L.I.; Bleotu, C.; Chifiriuc, M.C. Targeting Plasmids to Limit Acquisition and Transmission of Antimicrobial Resistance. Front. Microbiol. 2020, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Zongo, P.D.; Cabanel, N.; Royer, G.; Depardieu, F.; Hartmann, A.; Naas, T.; Glaser, P.; Rosinski-Chupin, I. An Antiplasmid System Drives Antibiotic Resistance Gene Integration in Carbapenemase-Producing Escherichia coli Lineages. Nat. Commun. 2024, 15, 4093. [Google Scholar] [CrossRef] [PubMed]
- Allain, M.; Morel-Journel, T.; Condamine, B.; Gibeaux, B.; Gachet, B.; Gschwind, R.; Denamur, E.; Landraud, L. IncC Plasmid Genome Rearrangements Influence the Vertical and Horizontal Transmission Tradeoff in Escherichia coli. Antimicrob. Agents Chemother. 2024, 68, e00554-24. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yin, Z.; Zhao, Q.; Zhao, Y.; Zhang, D.; Jiang, X.; Wu, W.; Chen, W.; Wang, H.; Song, Y.; et al. Genomic Characterization of Novel IncFII-Type Multidrug Resistant Plasmids P0716-KPC and P12181-KPC from Klebsiella pneumonia. Sci. Rep. 2017, 7, 5830. [Google Scholar] [CrossRef]
- Furlan, J.P.R.; Lopes, R.; Stehling, E.G. Multidrug Resistance IncC Plasmid Carrying BlaCMY-97 in Shiga Toxin-Producing Escherichia coli ST215-H54 of Ovine Origin. Infect. Genet. Evol. 2021, 93, 104989. [Google Scholar] [CrossRef]
- Grevskott, D.H.; Salvà-Serra, F.; Moore, E.R.B.; Marathe, N.P. Complete Sequence of a New Conjugative Multidrug-Resistance Encoding IncFII/IncFIA/IncFIB Plasmid Carrying NDM-6 Metallo-β-Lactamase from Pathogenic Escherichia coli Sequence Type 167 Isolated from Sewage in Norway. J. Glob. Antimicrob. Resist. 2023, 33, 291–293. [Google Scholar] [CrossRef]
- Muthuirulandi Sethuvel, D.P.; Anandan, S.; Devanga Ragupathi, N.K.; Gajendiran, R.; Kuroda, M.; Shibayama, K.; Veeraraghavan, B. IncFII Plasmid Carrying Antimicrobial Resistance Genes in Shigella flexneri: Vehicle for Dissemination. J. Glob. Antimicrob. Resist. 2019, 16, 215–219. [Google Scholar] [CrossRef]
- Zalewska, M.; Błażejewska, A.; Gawor, J.; Adamska, D.; Goryca, K.; Szeląg, M.; Kalinowski, P.; Popowska, M. The IncC and IncX1 Resistance Plasmids Present in Multi-Drug Resistant Escherichia coli Strains Isolated from Poultry Manure in Poland. Environ. Sci. Pollut. Res. 2024, 31, 47727–47741. [Google Scholar] [CrossRef]
- Zhang, X.Z.; Lei, C.W.; Zeng, J.X.; Chen, Y.P.; Kang, Z.Z.; Wang, Y.L.; Ye, X.L.; Zhai, X.W.; Wang, H.N. An IncX1 Plasmid Isolated from Salmonella enterica subsp. enterica Serovar Pullorum Carrying BlaTEM-1B, Sul2, Arsenic Resistant Operons. Plasmid 2018, 100, 14–21. [Google Scholar] [CrossRef]
- Bhat, A.; Sharma, R.; Desigan, K.; Lucas, M.M.; Mishra, A.; Bowers, R.M.; Woyke, T.; Epstein, B.; Tiffin, P.; Pueyo, J.J.; et al. Horizontal Gene Transfer of the Mer Operon Is Associated with Large Effects on the Transcriptome and Increased Tolerance to Mercury in Nitrogen-Fixing Bacteria. BMC Microbiol. 2024, 24, 247. [Google Scholar] [CrossRef]
- Priya, A.K.; Muruganandam, M.; Ali, S.S.; Kornaros, M. Clean-Up of Heavy Metals from Contaminated Soil by Phytoremediation: A Multidisciplinary and Eco-Friendly Approach. Toxics 2023, 11, 422. [Google Scholar] [CrossRef] [PubMed]
- Bao, H.; Wang, S.; Zhao, J.H.; Liu, S.L. Salmonella Secretion Systems: Differential Roles in Pathogen-Host Interactions. Microbiol. Res. 2020, 241, 126591. [Google Scholar] [CrossRef]
- Ferrer-Bustins, N.; Yvon, C.; Martín, B.; Leclerc, V.; Leblanc, J.C.; Corominas, L.; Sabaté, S.; Tolosa-Muñoz, E.; Chacón-Villanueva, C.; Bover-Cid, S.; et al. Genomic Insights of Salmonella Isolated from Dry Fermented Sausage Production Chains in Spain and France. Sci. Rep. 2024, 14, 11660. [Google Scholar] [CrossRef] [PubMed]
- Karash, S.; Jiang, T.; Kwon, Y.M. Genome-Wide Characterization of Salmonella Typhimurium Genes Required for the Fitness under Iron Restriction. BMC Genom. Data 2022, 23, 55. [Google Scholar] [CrossRef]
- Lou, L.; Zhang, P.; Piao, R.; Wang, Y. Salmonella Pathogenicity Island 1 (SPI-1) and Its Complex Regulatory Network. Front. Cell. Infect. Microbiol. 2019, 9, 270. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Kang, X.; Chen, J.; Song, Y.; Jia, C.; Teng, L.; Tang, Y.; Jiang, Z.; Peng, X.; Tao, X.; et al. Genome Degradation Promotes Salmonella Pathoadaptation by Remodeling Fimbriae-Mediated Proinflammatory Response. Natl. Sci. Rev. 2023, 10, 228. [Google Scholar] [CrossRef]
- Cherrak, Y.; Flaugnatti, N.; Durand, E.; Journet, L.; Cascales, E. Structure and Activity of the Type VI Secretion System. Microbiol. Spectr. 2019, 7, 10.1128. [Google Scholar] [CrossRef]
- Motta, E.V.S.; Lariviere, P.J.; Jones, K.R.; Song, Y.; Moran, N.A. Type VI Secretion Systems Promote Intraspecific Competition and Host Interactions in a Bee Gut Symbiont. Proc. Natl. Acad. Sci. USA 2024, 121, e2414882121. [Google Scholar] [CrossRef]
- Merciecca, T.; Bornes, S.; Nakusi, L.; Theil, S.; Rendueles, O.; Forestier, C.; Miquel, S. Role of Klebsiella pneumoniae Type VI Secretion System (T6SS) in Long-Term Gastrointestinal Colonization. Sci. Rep. 2022, 12, 16968. [Google Scholar] [CrossRef]
- Pillay, T.D.; Hettiarachchi, S.U.; Gan, J.; Diaz-Del-olmo, I.; Yu, X.J.; Muench, J.H.; Thurston, T.L.M.; Pearson, J.S. Speaking the Host Language: How Salmonella Effector Proteins Manipulate the Host. Microbiology 2023, 169, 001342. [Google Scholar] [CrossRef]
- Svahn, A.J.; Suster, C.J.E.; Chang, S.L.; Rockett, R.J.; Sim, E.M.; Cliff, O.M.; Wang, Q.; Arnott, A.; Ramsperger, M.; Sorrell, T.C.; et al. Pangenome Analysis of a Salmonella enteritidis Population Links a Major Outbreak to a Gifsy-1-Like Prophage Containing Anti-Inflammatory Gene GogB. Microbiol. Spectr. 2023, 11, e0279122. [Google Scholar] [CrossRef] [PubMed]
- Hodges, L.M.; Cooper, A.; Koziol, A.; Carrillo, C.D. Characterization of MLST-99 Salmonella Typhimurium and the Monophasic Variant I:4,[5],12:I:-Isolated from Canadian Atlantic Coast Shellfish. Microbiology 2024, 170, 001456. [Google Scholar] [CrossRef] [PubMed]
- García-Soto, S.; Linde, J.; Methner, U. Epidemiological Analysis on the Occurrence of Salmonella enterica subspecies enterica Serovar Dublin in the German Federal State Schleswig-Holstein Using Whole-Genome Sequencing. Microorganisms 2023, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Campioni, F.; Vilela, F.P.; Cao, G.; Kastanis, G.; dos Prazeres Rodrigues, D.; Costa, R.G.; Tiba-Casas, M.R.; Yin, L.; Allard, M.; Falcão, J.P. Whole Genome Sequencing Analyses Revealed That Salmonella enterica Serovar Dublin Strains from Brazil Belonged to Two Predominant Clades. Sci. Rep. 2022, 12, 10555. [Google Scholar] [CrossRef]
- Li, M.; Wang, K.; Tang, A.; Tang, A.; Chen, A.; Huang, Z. Investigation of the Genes Involved in the Outbreaks of Escherichia coli and Salmonella spp. in the United States. Antibiotics 2021, 10, 1274. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bentum, K.E.; Kuufire, E.; Nyarku, R.; Osei, V.; Adu-Addai, B.; Frye, J.G.; Jackson, C.R.; Samuel, T.; Abebe, W. Comparative Genomic Profiles of Salmonella Typhimurium and Salmonella Dublin Bovine Isolates from the U.S. Indicate Possible Factors Associated with the Host Adaptation of Salmonella Dublin in the Region. Microorganisms 2025, 13, 886. https://doi.org/10.3390/microorganisms13040886
Bentum KE, Kuufire E, Nyarku R, Osei V, Adu-Addai B, Frye JG, Jackson CR, Samuel T, Abebe W. Comparative Genomic Profiles of Salmonella Typhimurium and Salmonella Dublin Bovine Isolates from the U.S. Indicate Possible Factors Associated with the Host Adaptation of Salmonella Dublin in the Region. Microorganisms. 2025; 13(4):886. https://doi.org/10.3390/microorganisms13040886
Chicago/Turabian StyleBentum, Kingsley E., Emmanuel Kuufire, Rejoice Nyarku, Viona Osei, Benjamin Adu-Addai, Jonathan G. Frye, Charlene R. Jackson, Temesgen Samuel, and Woubit Abebe. 2025. "Comparative Genomic Profiles of Salmonella Typhimurium and Salmonella Dublin Bovine Isolates from the U.S. Indicate Possible Factors Associated with the Host Adaptation of Salmonella Dublin in the Region" Microorganisms 13, no. 4: 886. https://doi.org/10.3390/microorganisms13040886
APA StyleBentum, K. E., Kuufire, E., Nyarku, R., Osei, V., Adu-Addai, B., Frye, J. G., Jackson, C. R., Samuel, T., & Abebe, W. (2025). Comparative Genomic Profiles of Salmonella Typhimurium and Salmonella Dublin Bovine Isolates from the U.S. Indicate Possible Factors Associated with the Host Adaptation of Salmonella Dublin in the Region. Microorganisms, 13(4), 886. https://doi.org/10.3390/microorganisms13040886







