Wastewater Speaks: Evaluating SARS-CoV-2 Surveillance, Sampling Methods, and Seasonal Infection Trends on a University Campus
Abstract
:1. Introduction
2. Materials and Methods
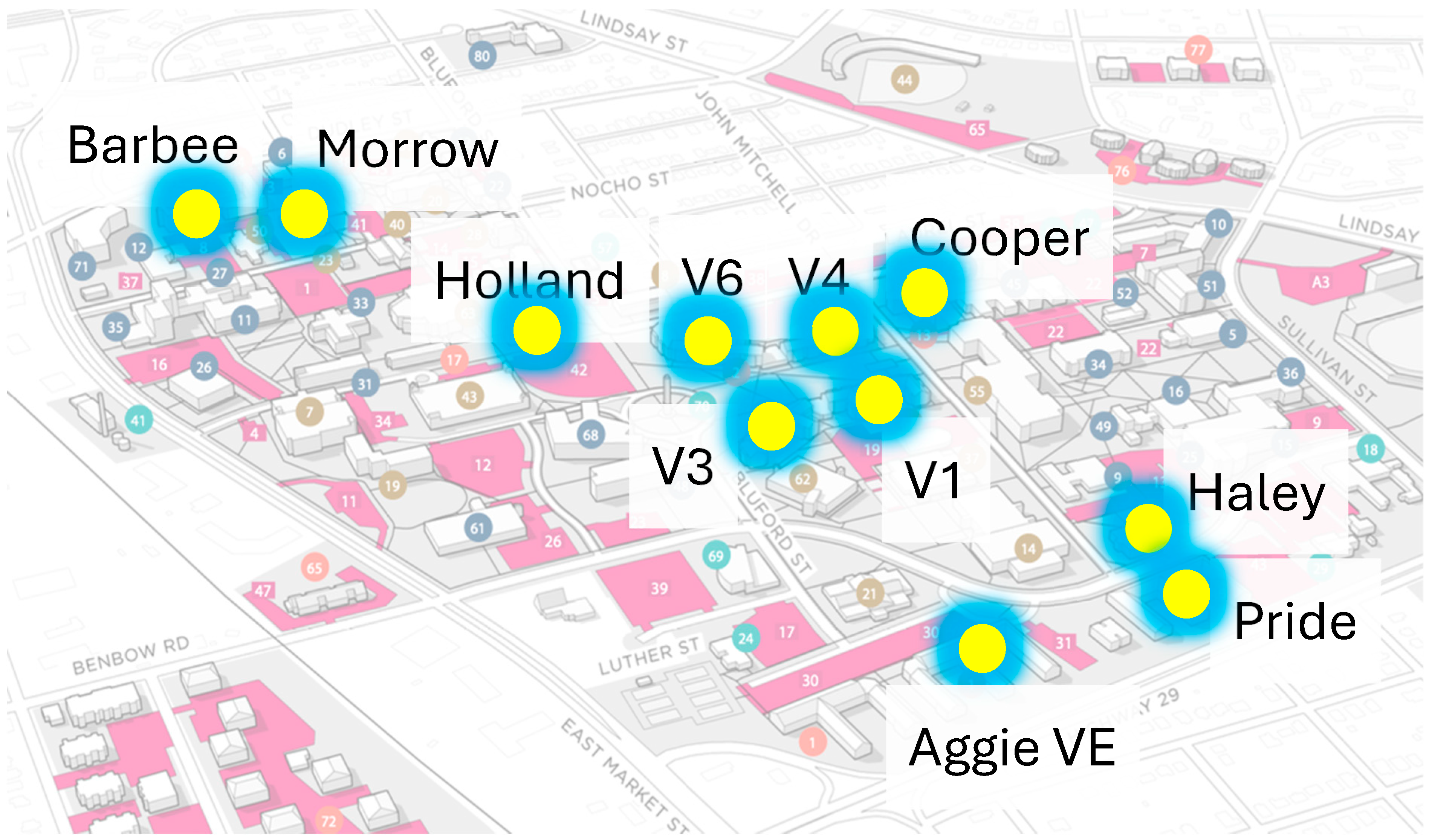
2.1. Sampling Sites
2.2. Clinical Testing
2.3. Wastewater Collection and Pre-Treatment
2.4. Viral RNA Extraction
2.5. Real Time-Quantitative Polymerase Chain Reaction
2.6. Quality Control
2.7. Statistical Analysis
3. Results and Discussion
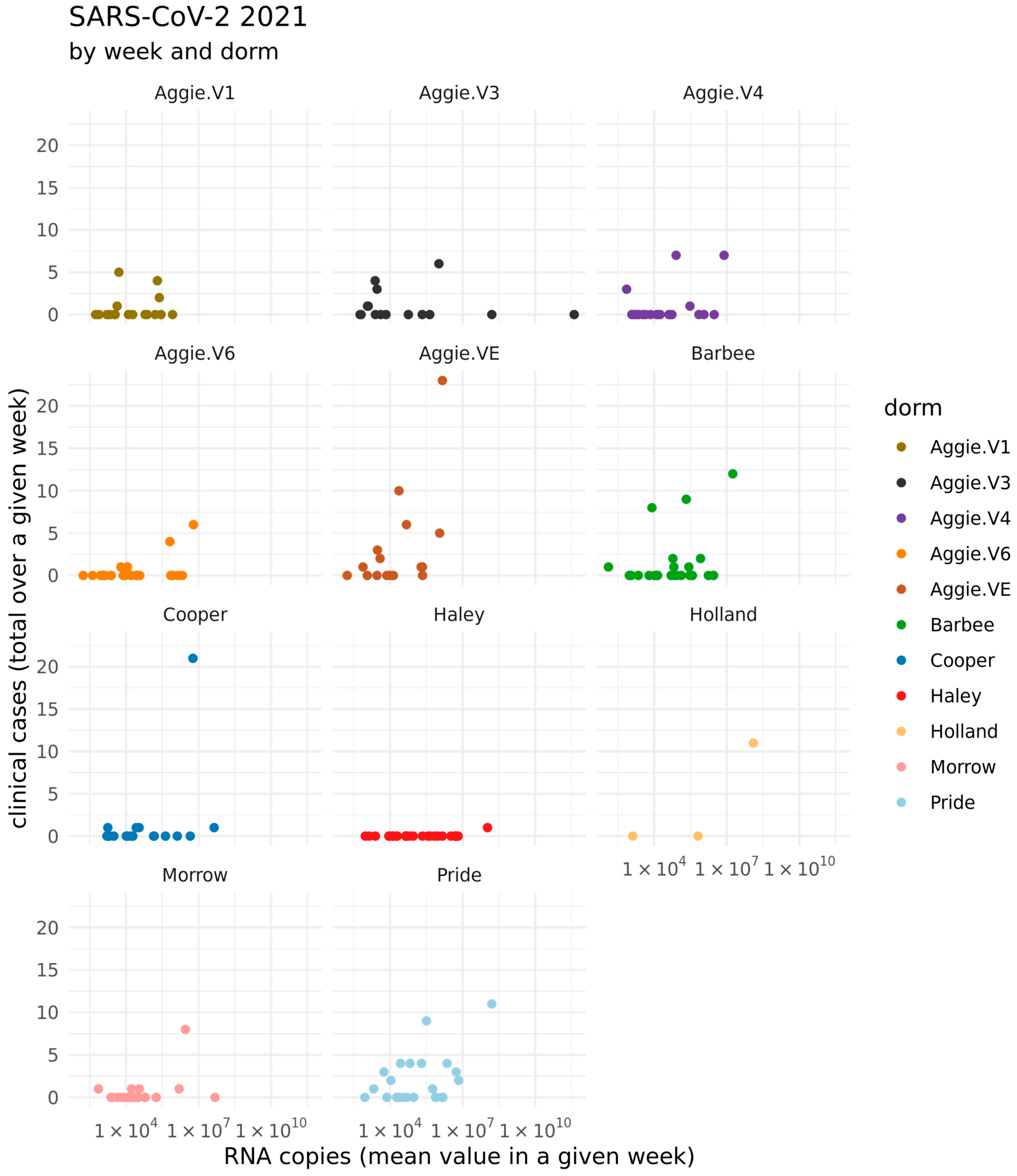
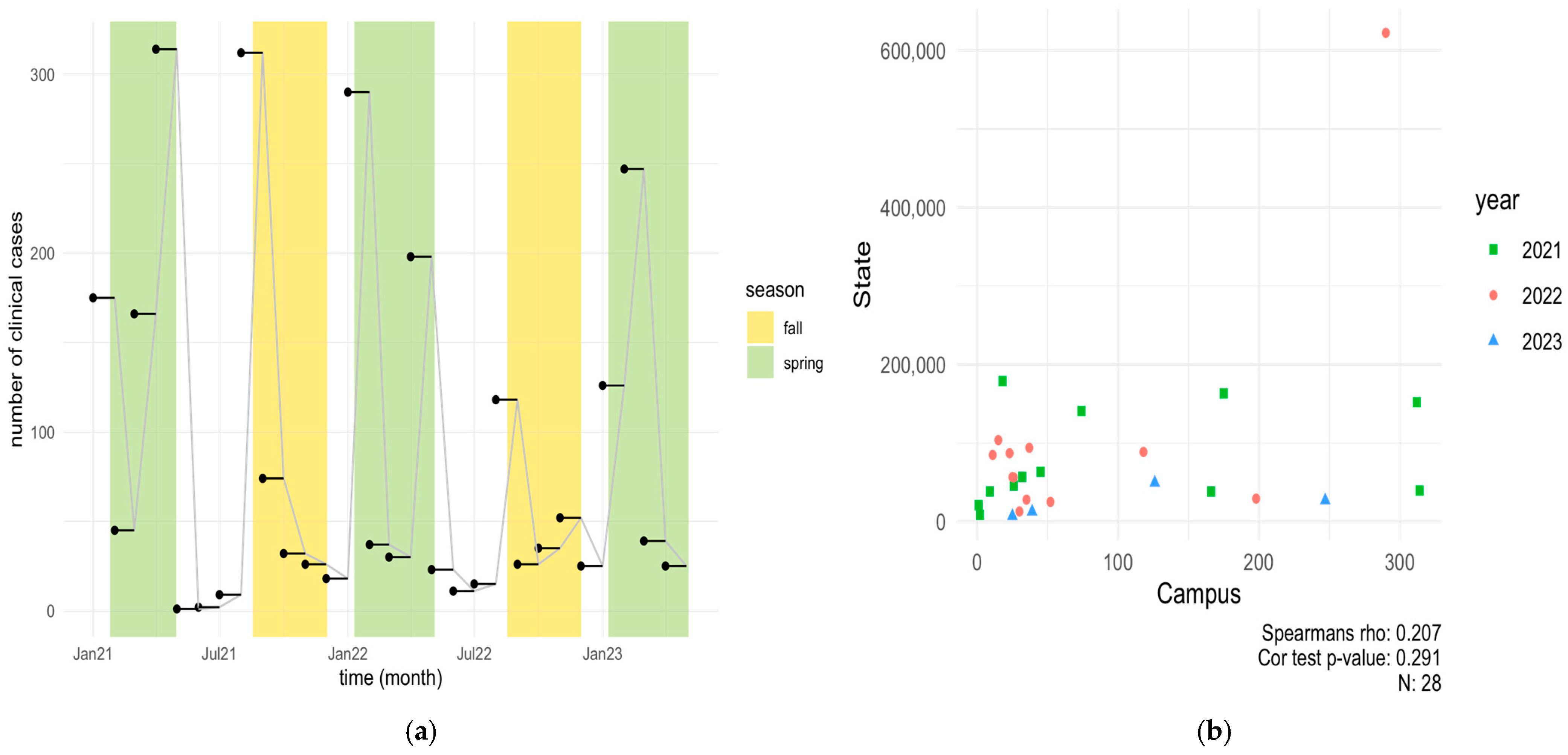
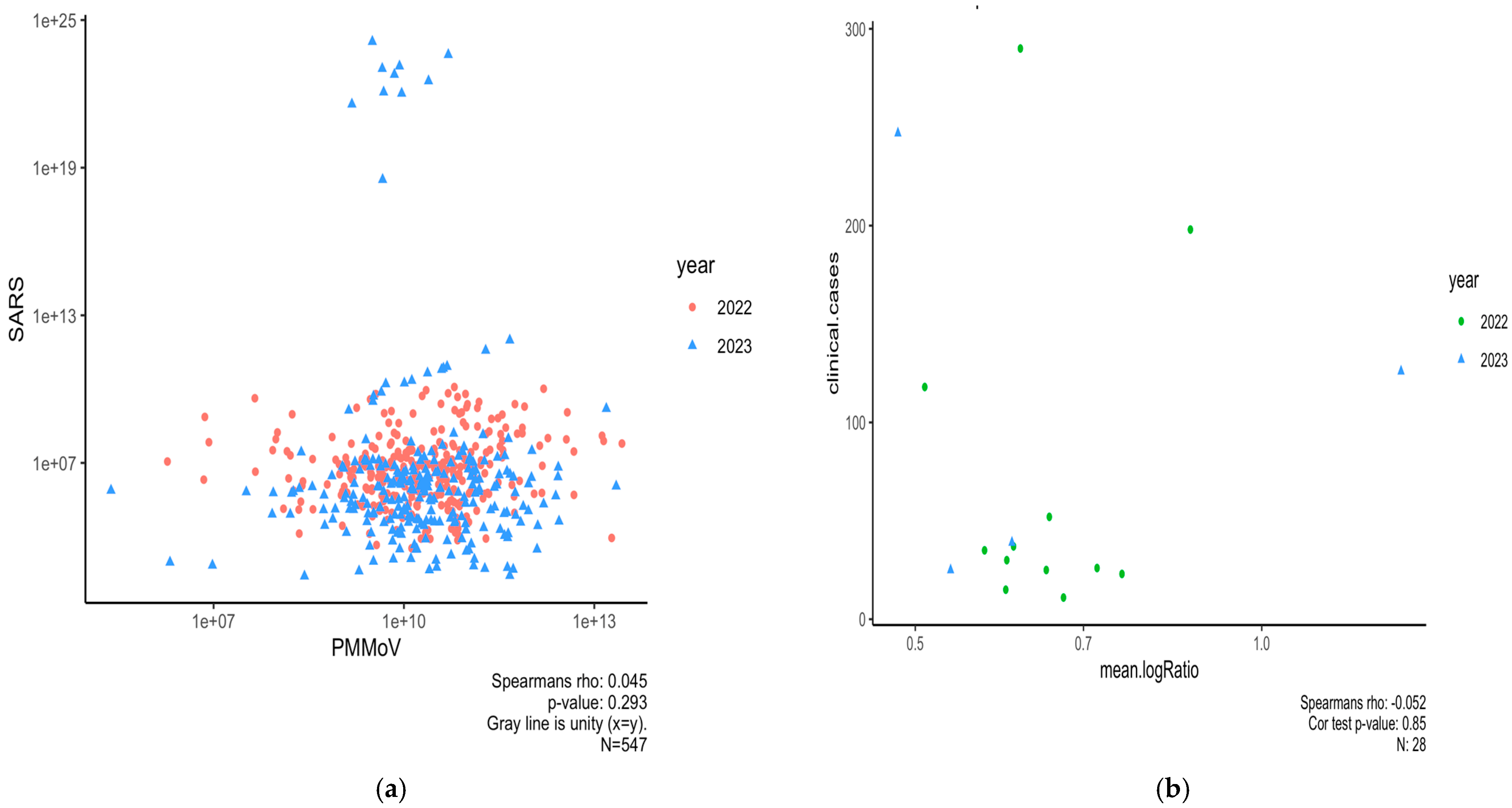
3.1. Clinical COVID-19 Positive Cases on NCAT Campus (2021–2023) and Correlation with State of NC
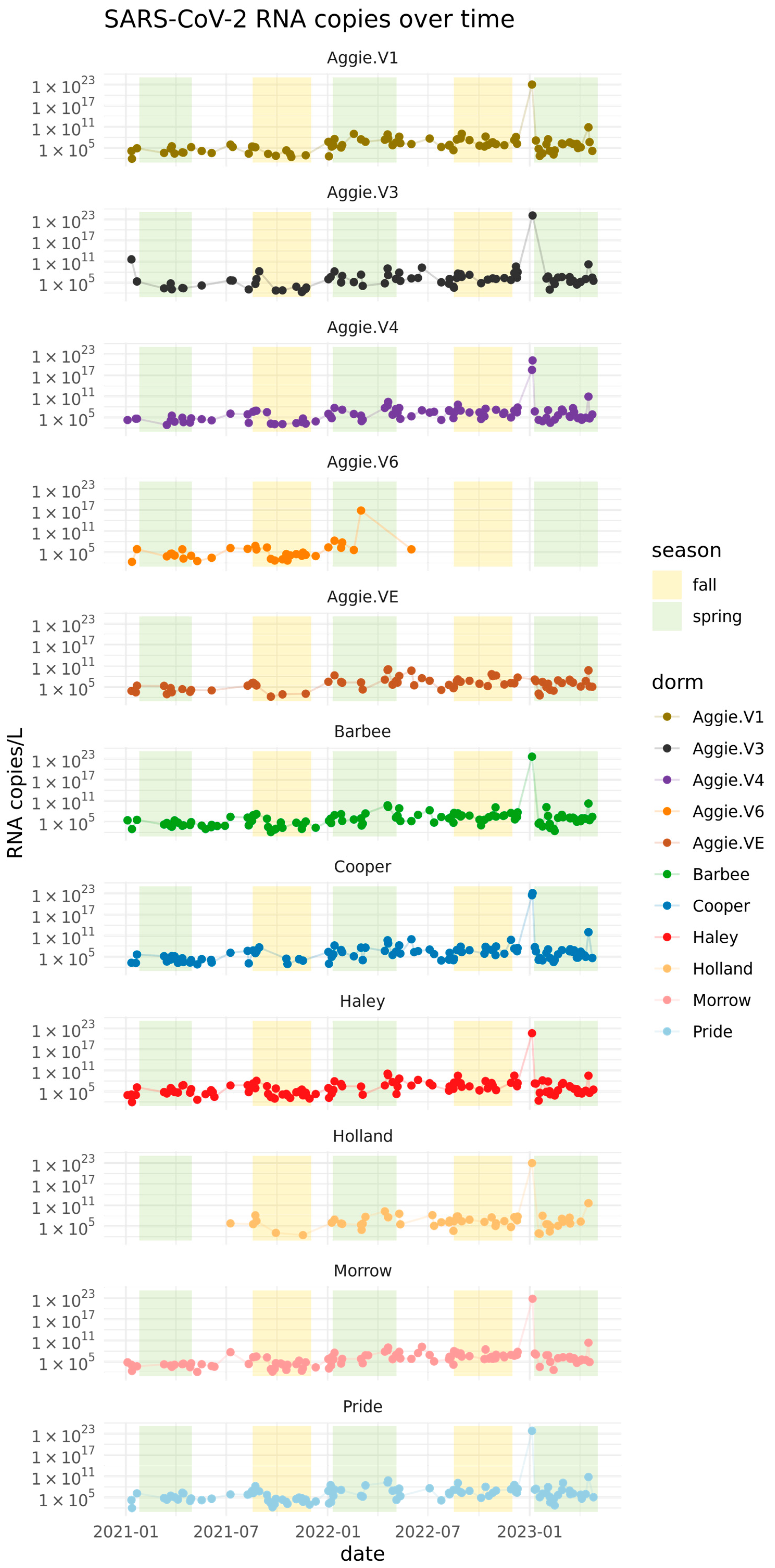
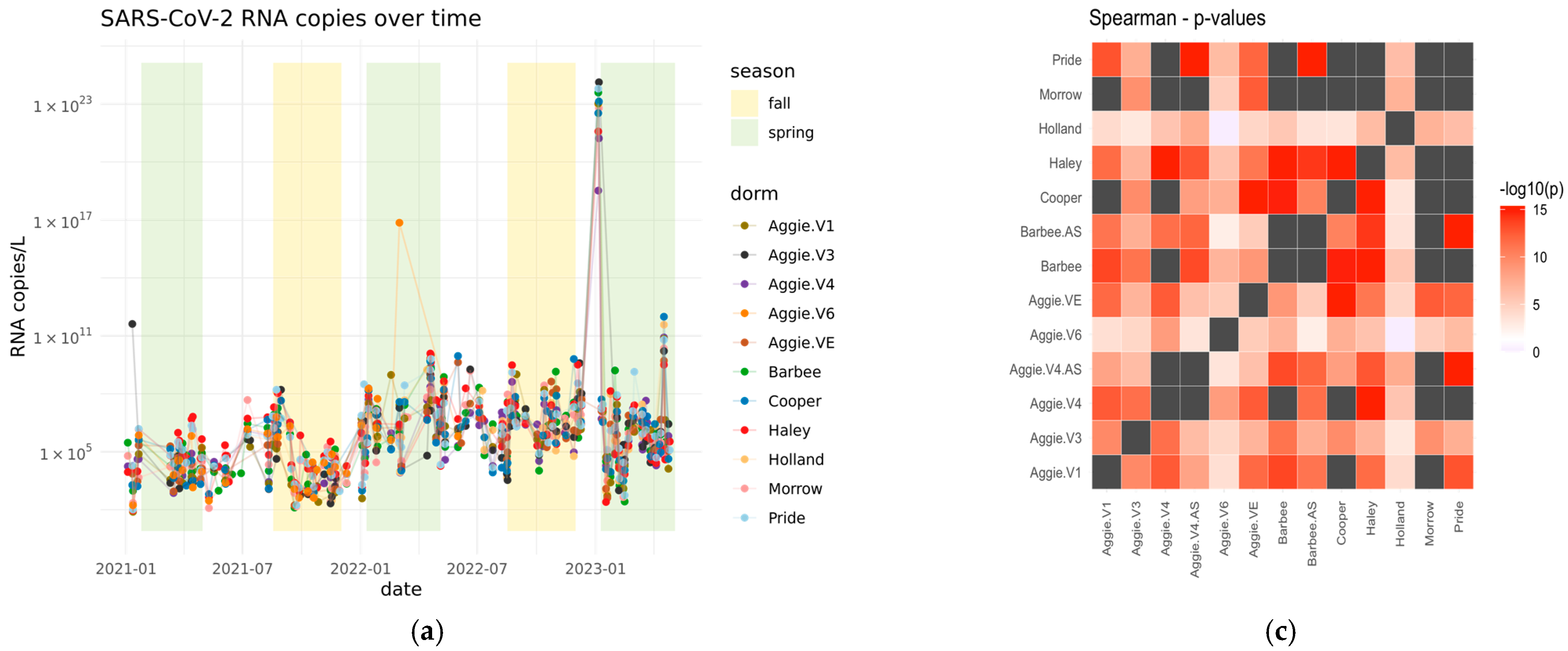
3.2. Quantification of SARS-CoV-2 Viral RNA in Wastewater in Spring 2021–2023
3.3. Normalizing SARS-CoV-2 RNA Concentrations by PMMoV Does Not Substantially Impact Results
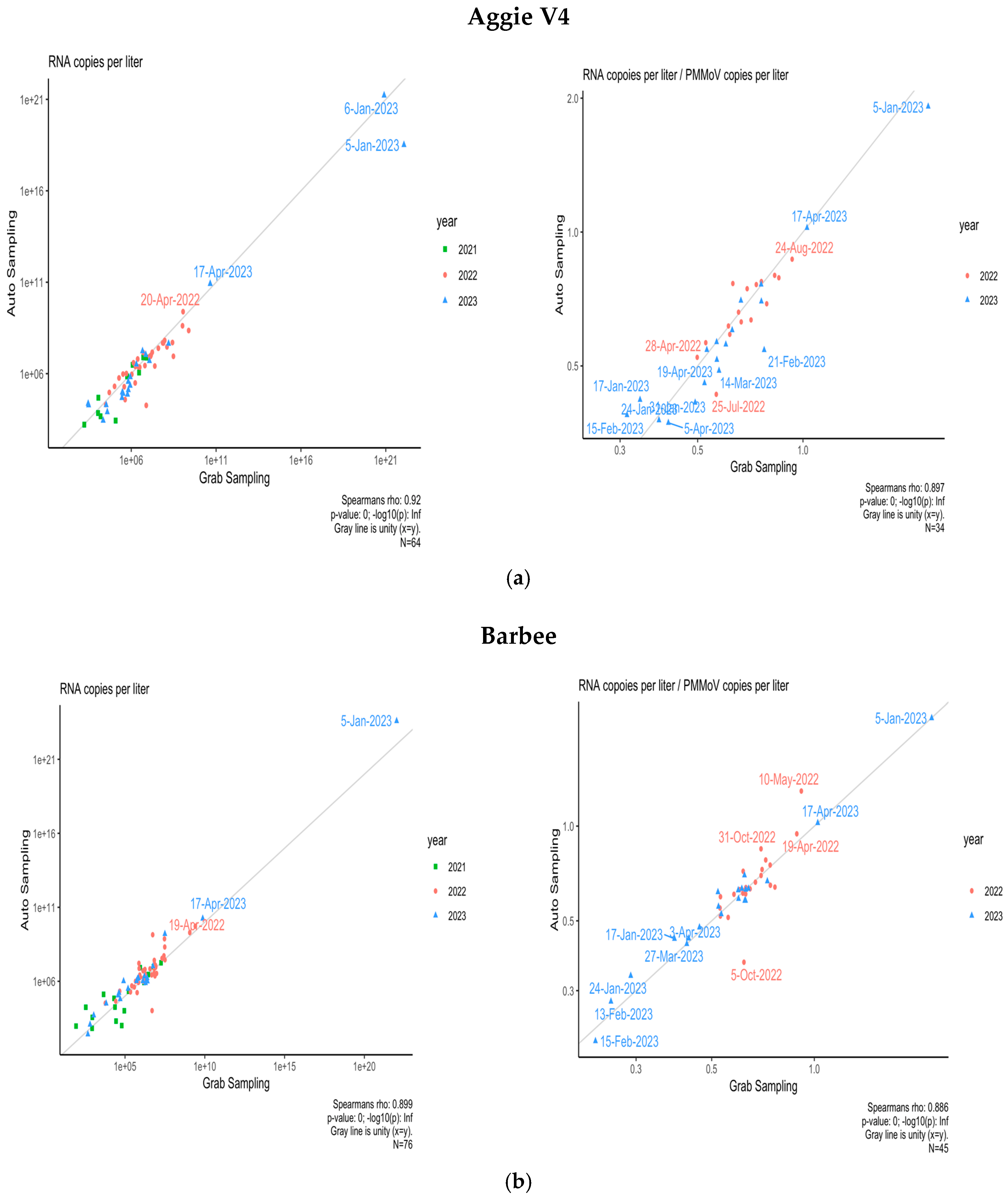
3.4. Comparison of Composite and Grab Sampling Methods on NCA&T Campus
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| WBE | Wastewater-Based Epidemiology |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| CDC | Center for disease control and prevention |
| ACE2 | Angiotensin-converting enzyme 2 |
| LoD | Limit of Detection |
| LoQ | Limit of Quantification |
| PMMoV | Pepper mild mottle virus |
| PCR | Polymerase Chain reaction |
Appendix A
| Sample Site | Type of Sample | Semester | Frequency of Sampling |
|---|---|---|---|
| Aggie Village 4 | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Aggie Village 3 | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Aggie Village 1 | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Aggie Village E | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Morrow | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Holland | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Pride | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Barbee | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Cooper | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Haley | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Aggie Village 6 | Grab | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Aggie Village 4-AS | Composite | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week | ||
| Barbee AS | Composite | Spring | twice a week |
| Summer | once a week | ||
| Fall | twice a week |
| Name. | Sequence | Probe/Quencher |
|---|---|---|
| 2019-nCoV_N1-F | 5′-GACCCCAAAATCAGCGAAAT-3′ | |
| 2019-nCoV_N1-R | 5′-TCTGGTTACTGCCAGTTGAATCTG-3′ | |
| 2019-nCoV_N1-P | 5′-ACCCCGCATTACGTTTGGTGGACC-3′ | FAM/ZEN/IBFQ |
| Level | Level | Score Mean Difference | Std Err Dif | Z | p-Value | Hodges–Lehmann | Lower CL | Upper CL |
|---|---|---|---|---|---|---|---|---|
| Haley | Aggie V6 | 22.9564 | 8.232900 | 2.78837 | 0.0053 * | 2.13656 | 0.67685 | 3.79031 |
| Cooper | Aggie V6 | 21.8878 | 7.613014 | 2.87504 | 0.0040 * | 2.26015 | 0.68478 | 4.03072 |
| Pride | Aggie V6 | 21.8085 | 7.613014 | 2.86464 | 0.0042 * | 2.39968 | 0.78832 | 3.98360 |
| Barbee | Aggie V6 | 19.5053 | 8.137181 | 2.39705 | 0.0165 * | 1.83228 | 0.34738 | 3.39687 |
| Morrow | Aggie V6 | 18.7431 | 7.993824 | 2.34469 | 0.0190 * | 1.84367 | 0.22610 | 3.50926 |
| Holland | Aggie V6 | 18.7084 | 5.549995 | 3.37088 | 0.0007 * | 3.55921 | 1.54755 | 5.09805 |
| Holland | Barbee | 17.0216 | 7.855377 | 2.16688 | 0.0302 * | 1.48582 | 0.13591 | 2.80863 |
| Aggie VE | Aggie V6 | 16.9639 | 6.443238 | 2.63283 | 0.0085 * | 2.19304 | 0.62432 | 4.07702 |
| Holland | Aggie V4 | 14.4970 | 7.243058 | 2.00151 | 0.0453 * | 1.48457 | 0.02790 | 2.78236 |
| Holland | Haley | 13.4924 | 7.928587 | 1.70175 | 0.0888 | 1.19719 | −0.21118 | 2.51796 |
| Holland | Aggie V1 | 13.0466 | 6.928378 | 1.88307 | 0.0597 | 1.24316 | −0.05696 | 2.80203 |
| Holland | Aggie V3 | 11.3852 | 6.689999 | 1.70182 | 0.0888 | 1.15060 | −0.18957 | 2.69423 |
| Holland | Cooper | 9.3176 | 7.456880 | 1.24953 | 0.2115 | 1.00426 | −0.48465 | 2.47990 |
| Holland | Aggie VE | 9.2815 | 6.589955 | 1.40843 | 0.1590 | 1.03178 | −0.40502 | 2.49874 |
| Cooper | Barbee | 7.4897 | 8.338446 | 0.89822 | 0.3691 | 0.46202 | −0.58276 | 1.52714 |
| Pride | Morrow | 7.4134 | 8.272762 | 0.89612 | 0.3702 | 0.48360 | −0.61921 | 1.56708 |
| Pride | Barbee | 7.3735 | 8.338446 | 0.88427 | 0.3765 | 0.51342 | −0.55939 | 1.58334 |
| Haley | Barbee | 5.8914 | 8.583283 | 0.68638 | 0.4925 | 0.38685 | −0.70313 | 1.42052 |
| Pride | Aggie V1 | 5.3294 | 7.815766 | 0.68188 | 0.4953 | 0.40307 | −0.84494 | 1.67871 |
| Cooper | Aggie V1 | 4.9956 | 7.815766 | 0.63917 | 0.5227 | 0.46081 | −0.86416 | 1.64649 |
| Pride | Aggie V4 | 4.9839 | 7.983121 | 0.62431 | 0.5324 | 0.38248 | −0.71471 | 1.54344 |
| Cooper | Aggie V4 | 4.0988 | 7.983121 | 0.51344 | 0.6076 | 0.29361 | −0.78093 | 1.46005 |
| Cooper | Aggie V3 | 3.8197 | 7.699417 | 0.49610 | 0.6198 | 0.32714 | −0.98463 | 1.53169 |
| Haley | Aggie V4 | 3.7471 | 8.282570 | 0.45241 | 0.6510 | 0.23286 | −0.81702 | 1.33025 |
| Aggie VE | Aggie V1 | 3.5788 | 7.249087 | 0.49369 | 0.6215 | 0.30862 | −1.02709 | 1.62380 |
| Haley | Aggie V1 | 3.2007 | 8.147566 | 0.39284 | 0.6944 | 0.25417 | −1.00324 | 1.50186 |
| Pride | Aggie V3 | 3.0254 | 7.699417 | 0.39294 | 0.6944 | 0.29335 | −0.98389 | 1.57729 |
| Aggie VE | Aggie V4 | 3.0096 | 7.488452 | 0.40190 | 0.6878 | 0.25393 | −0.95633 | 1.55410 |
| Aggie VE | Aggie V3 | 2.4574 | 7.072502 | 0.34745 | 0.7282 | 0.19152 | −1.13620 | 1.44823 |
| Pride | Haley | 1.9501 | 8.382832 | 0.23263 | 0.8160 | 0.13229 | −0.96052 | 1.25973 |
| Pride | Aggie VE | 1.9003 | 7.654120 | 0.24827 | 0.8039 | 0.14785 | −1.16108 | 1.32746 |
| Haley | Aggie V3 | 1.7844 | 8.058638 | 0.22143 | 0.8248 | 0.14705 | −1.15556 | 1.43785 |
| Cooper | Aggie VE | 1.7568 | 7.654120 | 0.22953 | 0.8185 | 0.12360 | −1.13386 | 1.32175 |
| Aggie V3 | Aggie V1 | 1.3233 | 7.310055 | 0.18103 | 0.8563 | 0.12271 | −1.20002 | 1.43147 |
| Pride | Cooper | 0.8265 | 8.103497 | 0.10200 | 0.9188 | 0.04909 | −1.14950 | 1.17855 |
| Morrow | Barbee | 0.3629 | 8.486108 | 0.04276 | 0.9659 | 0.02088 | −1.03813 | 1.06013 |
| Aggie V4 | Aggie V1 | 0.2865 | 7.669566 | 0.03735 | 0.9702 | 0.01429 | −1.27179 | 1.30055 |
| Haley | Aggie VE | −0.2952 | 8.025833 | −0.03678 | 0.9707 | −0.02714 | −1.30187 | 1.18195 |
| Morrow | Aggie V1 | −0.3545 | 8.017814 | −0.04421 | 0.9647 | −0.02727 | −1.31468 | 1.14226 |
| Aggie V4 | Aggie V3 | −0.8049 | 7.539826 | −0.10676 | 0.9150 | −0.07449 | −1.39833 | 1.18004 |
| Haley | Cooper | −1.1432 | 8.382832 | −0.13637 | 0.8915 | −0.09410 | −1.19203 | 1.02958 |
| Barbee | Aggie V1 | −1.3370 | 8.095375 | −0.16516 | 0.8688 | −0.09270 | −1.26328 | 1.10596 |
| Barbee | Aggie V4 | −2.2249 | 8.235193 | −0.27017 | 0.7870 | −0.13667 | −1.18708 | 0.97092 |
| Morrow | Aggie V3 | −2.2253 | 7.918567 | −0.28102 | 0.7787 | −0.19695 | −1.40998 | 1.03661 |
| Barbee | Aggie V3 | −2.4007 | 8.002350 | −0.30000 | 0.7642 | −0.14466 | −1.39156 | 0.98458 |
| Morrow | Aggie V4 | −2.4872 | 8.164964 | −0.30462 | 0.7607 | −0.14717 | −1.22786 | 0.99928 |
| Morrow | Aggie VE | −4.2911 | 7.881054 | −0.54448 | 0.5861 | −0.36435 | −1.58536 | 0.81397 |
| Barbee | Aggie VE | −5.0778 | 7.967675 | −0.63730 | 0.5239 | −0.37470 | −1.52178 | 0.77014 |
| Morrow | Haley | −5.4868 | 8.526738 | −0.64348 | 0.5199 | −0.36425 | −1.44335 | 0.70572 |
| Morrow | Cooper | −6.9224 | 8.272762 | −0.83677 | 0.4027 | −0.42659 | −1.54284 | 0.60190 |
| Pride | Holland | −10.8584 | 7.456880 | −1.45616 | 0.1453 | −1.02272 | −2.36280 | 0.42079 |
| Aggie V6 | Aggie V1 | −14.8219 | 6.906496 | −2.14609 | 0.0319 * | −2.03269 | −3.70189 | −0.24053 |
| Aggie V6 | Aggie V3 | −15.7721 | 6.581385 | −2.39647 | 0.0166 * | −1.98747 | −3.90295 | −0.40234 |
| Morrow | Holland | −15.8066 | 7.745967 | −2.04062 | 0.0413 * | −1.45810 | −2.84156 | −0.03704 |
| Aggie V6 | Aggie V4 | −19.3955 | 7.329062 | −2.64638 | 0.0081 * | −1.91501 | −3.50322 | −0.49975 |
References
- Fung, T.S.; Liu, D.X. Similarities and Dissimilarities of COVID-19 and Other Coronavirus Diseases. Annu. Rev. Microbiol. 2021, 75, 19–47. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef]
- Azkur, A.K.; Akdis, M.; Azkur, D.; Sokolowska, M.; Veen, W.; Brüggen, M.; O’Mahony, L.; Gao, Y.; Nadeau, K.; Akdis, C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy 2020, 75, 1564–1581. [Google Scholar] [CrossRef] [PubMed]
- Bar-Or, I.; Weil, M.; Indenbaum, V.; Bucris, E.; Bar-Ilan, D.; Elul, M.; Levi, N.; Aguvaev, I.; Cohen, Z.; Shirazi, R.; et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci. Total Environ. 2021, 789, 148002. [Google Scholar] [CrossRef]
- Bivins, A.; Greaves, J.; Fischer, R.; Yinda, K.C.; Ahmed, W.; Kitajima, M.; Munster, V.J.; Bibby, K. Persistence of SARS-CoV-2 in Water and Wastewater. Environ. Sci. Technol. Lett. 2020, 7, 937–942. [Google Scholar] [CrossRef]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef] [PubMed]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simón, P.; Allende, A.; Sánchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef]
- Sherchan, S.P.; Shahin, S.; Ward, L.M.; Tandukar, S.; Aw, T.G.; Schmitz, B.; Ahmed, W.; Kitajima, M. First detection of SARS-CoV-2 RNA in wastewater in North America: A study in Louisiana, USA. Sci. Total Environ. 2020, 743, 140621. [Google Scholar] [CrossRef]
- Han, P.; Ivanovski, S. Saliva—Friend and Foe in the COVID-19 Outbreak. Diagnostics 2020, 10, 290. [Google Scholar] [CrossRef]
- Graham, K.E.; Loeb, S.K.; Wolfe, M.K.; Catoe, D.; Sinnott-Armstrong, N.; Kim, S.; Yamahara, K.M.; Sassoubre, L.M.; Mendoza Grijalva, L.M.; Roldan-Hernandez, L.; et al. SARS-CoV-2 RNA in Wastewater Settled Solids Is Associated with COVID-19 Cases in a Large Urban Sewershed. Environ. Sci. Technol. 2021, 55, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Choi, P.M.; Tscharke, B.J.; Donner, E.; O’Brien, J.W.; Grant, S.C.; Kaserzon, S.L.; Mackie, R.; O’Malley, E.; Crosbie, N.D.; Thomas, K.V.; et al. Wastewater-based epidemiology biomarkers: Past, present and future. TrAC Trends Anal. Chem. 2018, 105, 453–469. [Google Scholar] [CrossRef]
- Gonzalez, R.; Curtis, K.; Bivins, A.; Bibby, K.; Weir, M.H.; Yetka, K.; Thompson, H.; Keeling, D.; Mitchell, J.; Gonzalez, D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020, 186, 116296. [Google Scholar] [CrossRef]
- Asghar, H.; Diop, O.M.; Weldegebriel, G.; Malik, F.; Shetty, S.; El Bassioni, L.; Akande, A.O.; Al Maamoun, E.; Zaidi, S.; Adeniji, A.J.; et al. Environmental Surveillance for Polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014, 210, S294–S303. [Google Scholar] [CrossRef]
- Hellmér, M.; Paxéus, N.; Magnius, L.; Enache, L.; Arnholm, B.; Johansson, A.; Bergström, T.; Norder, H. Detection of pathogenic viruses in sewage provided early warnings of hepatitis A virus and norovirus outbreaks. Appl. Environ. Microbiol. 2014, 80, 6771–6781. [Google Scholar] [CrossRef] [PubMed]
- Peccia, J.; Zulli, A.; Brackney, D.E.; Grubaugh, N.D.; Kaplan, E.H.; Casanovas-Massana, A.; Ko, A.I.; Malik, A.A.; Wang, D.; Wang, M.; et al. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. MedRxiv 2020. [Google Scholar] [CrossRef]
- Jones, D.L.; Baluja, M.Q.; Graham, D.W.; Corbishley, A.; McDonald, J.E.; Malham, S.K.; Hillary, L.S.; Connor, T.R.; Gaze, W.H.; Moura, I.B.; et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020, 749, 141364. [Google Scholar] [CrossRef]
- Sharkey, M.E.; Kumar, N.; Mantero, A.M.A.; Babler, K.M.; Boone, M.M.; Cardentey, Y.; Cortizas, E.M.; Grills, G.S.; Herrin, J.; Kemper, J.M.; et al. Lessons learned from SARS-CoV-2 measurements in wastewater. Sci. Total Environ. 2021, 798, 149177. [Google Scholar] [CrossRef]
- Cevik, M.; Tate, M.; Lloyd, O.; Maraolo, A.E.; Schafers, J.; Ho, A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe 2021, 2, e13–e22. [Google Scholar] [CrossRef]
- Fongaro, G.; Stoco, P.H.; Souza, D.S.M.; Grisard, E.C.; Magri, M.E.; Rogovski, P.; Schörner, M.A.; Barazzetti, F.H.; Christoff, A.P.; De Oliveira, L.F.V.; et al. The presence of SARS-CoV-2 RNA in human sewage in Santa Catarina, Brazil, November 2019. Sci. Total Environ. 2021, 778, 146198. [Google Scholar] [CrossRef]
- La Rosa, G.; Iaconelli, M.; Mancini, P.; Bonanno Ferraro, G.; Veneri, C.; Bonadonna, L.; Lucentini, L.; Suffredini, E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020, 736, 139652. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef]
- Wu, F.; Xiao, A.; Zhang, J.; Moniz, K.; Endo, N.; Armas, F.; Bonneau, R.; Brown, M.A.; Bushman, M.; Chai, P.R.; et al. SARS-CoV-2 titers in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wolfe, M.K.; Archana, A.; Catoe, D.; Coffman, M.M.; Dorevich, S.; Graham, K.E.; Kim, S.; Grijalva, L.M.; Roldan-Hernandez, L.; Silverman, A.I.; et al. Scaling of SARS-CoV-2 RNA in Settled Solids from Multiple Wastewater Treatment Plants to Compare Incidence Rates of Laboratory-Confirmed COVID-19 in Their Sewersheds. Environ. Sci. Technol. Lett. 2021, 8, 398–404. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, M.K.; Topol, A.; Knudson, A.; Simpson, A.; White, B.; Vugia, D.J.; Yu, A.T.; Li, L.; Balliet, M.; Stoddard, P.; et al. High-Frequency, High-Throughput Quantification of SARS-CoV-2 RNA in Wastewater Settled Solids at Eight Publicly Owned Treatment Works in Northern California Shows Strong Association with COVID-19 Incidence. mSystems 2021, 6, e0082921. [Google Scholar] [CrossRef]
- Adhikari, S.; Halden, R.U. Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ. Int. 2022, 163, 107217. [Google Scholar] [CrossRef]
- Ahmed, W.; Tscharke, B.; Bertsch, P.M.; Bibby, K.; Bivins, A.; Choi, P.; Clarke, L.; Dwyer, J.; Edson, J.; Nguyen, T.M.H.; et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: A temporal case study. Sci. Total Environ. 2021, 761, 144216. [Google Scholar] [CrossRef]
- Gushgari, A.J.; Venkatesan, A.K.; Chen, J.; Steele, J.C.; Halden, R.U. Long-term tracking of opioid consumption in two United States cities using wastewater-based epidemiology approach. Water Res. 2019, 161, 171–180. [Google Scholar] [CrossRef]
- Vo, V.; Tillett, R.L.; Chang, C.-L.; Gerrity, D.; Betancourt, W.Q.; Oh, E.C. SARS-CoV-2 variant detection at a university dormitory using wastewater genomic tools. Sci. Total Environ. 2022, 805, 149930. [Google Scholar] [CrossRef]
- Betancourt, W.Q.; Schmitz, B.W.; Innes, G.K.; Prasek, S.M.; Pogreba Brown, K.M.; Stark, E.R.; Foster, A.R.; Sprissler, R.S.; Harris, D.T.; Sherchan, S.P.; et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021, 779, 146408. [Google Scholar] [CrossRef]
- Gibas, C.; Lambirth, K.; Mittal, N.; Juel, M.A.I.; Barua, V.B.; Roppolo Brazell, L.; Hinton, K.; Lontai, J.; Stark, N.; Young, I.; et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021, 782, 146749. [Google Scholar] [CrossRef] [PubMed]
- Harris-Lovett, S.; Nelson, K.L.; Beamer, P.; Bischel, H.N.; Bivins, A.; Bruder, A.; Butler, C.; Camenisch, T.D.; De Long, S.K.; Karthikeyan, S.; et al. Wastewater Surveillance for SARS-CoV-2 on College Campuses: Initial Efforts, Lessons Learned, and Research Needs. Int. J. Environ. Res. Public Health 2021, 18, 4455. [Google Scholar] [CrossRef] [PubMed]
- Karthikeyan, S.; Nguyen, A.; McDonald, D.; Zong, Y.; Ronquillo, N.; Ren, J.; Zou, J.; Farmer, S.; Humphrey, G.; Henderson, D.; et al. Rapid, Large-Scale Wastewater Surveillance and Automated Reporting System Enable Early Detection of Nearly 85% of COVID-19 Cases on a University Campus. mSystems 2021, 6, e00793-21. [Google Scholar] [CrossRef]
- Scott, L.C.; Aubee, A.; Babahaji, L.; Vigil, K.; Tims, S.; Aw, T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021, 200, 111374. [Google Scholar] [CrossRef] [PubMed]
- North Carolina Respiratory Virus Summary Dashboard [Internet]. Available online: https://covid19.ncdhhs.gov/dashboard (accessed on 2 April 2025).
- Wizard® Enviro Total Nucleic Acid Kit, Promega 2020 [Internet]. 2020. Available online: https://www.promega.com/-/media/files/resources/protocols/technical-manuals/500/wizard-enviro-total-nucleic-acid-kit-protocol-tm662.pdf?rev=b99cc4a13f404cd99f39ec700efd6847&sc_lang=en (accessed on 2 April 2025).
- U.S. Centers for Disease Control and Prevention. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT–PCR Diagnostic Panel [Internet]. 2021. Available online: https://www.fda.gov/media/134922/download?fbclid=IwAR1DdEweazD3ixmrpZMc07VXM0_n1qx455rGV7E0fAEcA1QZf3Peh0Qxypo (accessed on 2 April 2025).
- Ai, Y.; Davis, A.; Jones, D.; Lemeshow, S.; Tu, H.; He, F.; Ru, P.; Pan, X.; Bohrerova, Z.; Lee, J. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021, 801, 149757. [Google Scholar] [CrossRef]
- Zhan, Q.; Babler, K.M.; Sharkey, M.E.; Amirali, A.; Beaver, C.C.; Boone, M.M.; Comerford, S.; Cooper, D.; Cortizas, E.M.; Currall, B.B.; et al. Relationships between SARS-CoV-2 in Wastewater and COVID-19 Clinical Cases and Hospitalizations, with and without Normalization against Indicators of Human Waste. ACS EST Water 2022, 2, 1992–2003. [Google Scholar] [CrossRef]
- Wastewater Monitoring Dashboard [Internet]. Available online: https://covid19.ncdhhs.gov/dashboard/wastewater-monitoring (accessed on 2 April 2025).
- Alvarez, E.; Bielska, I.A.; Hopkins, S.; Belal, A.A.; Goldstein, D.M.; Slick, J.; Pavalagantharajah, S.; Wynfield, A.; Dakey, S.; Gedeon, M.-C.; et al. Limitations of COVID-19 testing and case data for evidence-informed health policy and practice. Health Res. Policy Syst. 2023, 21, 11. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, P.; Zhang, H.; Ibaraki, M.; VanTassell, J.; Geith, K.; Cavallo, M.; Kann, R.; Saber, L.; Kraft, C.S.; et al. Early warning of a COVID-19 surge on a university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Sci. Total Environ. 2022, 821, 153291. [Google Scholar] [CrossRef]
- Williams, B.B.; Newborn, A.; Karamat, A.; Zamcho, F.; Salerno, J.L.; Gillevet, P.M.; Farris, D.; Wintermeyer, S.F.; Van Aken, B. Detection of SARS-CoV-2 RNA in wastewater from dormitory buildings in a university campus: Comparison with individual testing results. Water Sci. Technol. 2023, 88, 2364–2377. [Google Scholar] [CrossRef]
- Foladori, P.; Cutrupi, F.; Segata, N.; Manara, S.; Pinto, F.; Malpei, F.; Bruni, L.; La Rosa, G. SARS-CoV-2 from faeces to wastewater treatment: What do we know? A review. Sci. Total Environ. 2020, 743, 140444. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020, 172, 115473. [Google Scholar] [CrossRef] [PubMed]
- Swift, C.L.; Isanovic, M.; Correa Velez, K.E.; Norman, R.S. SARS-CoV-2 concentration in wastewater consistently predicts trends in COVID-19 case counts by at least two days across multiple WWTP scales. Environ. Adv. 2023, 11, 100347. [Google Scholar] [CrossRef] [PubMed]
- Kumblathan, T.; Liu, Y.; Uppal, G.K.; Hrudey, S.E.; Li, X.-F. Wastewater-Based Epidemiology for Community Monitoring of SARS-CoV-2: Progress and Challenges. ACS Environ. Au 2021, 1, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Anglemyer, A.; Moore, T.H.; Parker, L.; Chambers, T.; Grady, A.; Chiu, K.; Parry, M.; Wilczynska, M.; Flemyng, E.; Bero, L. Digital contact tracing technologies in epidemics: A rapid review. Cochrane Database Syst. Rev. 2020, 2020, 8. [Google Scholar] [CrossRef]
- Feng, S.; Roguet, A.; McClary-Gutierrez, J.S.; Newton, R.J.; Kloczko, N.; Meiman, J.G.; McLellan, S.L. Evaluation of Sampling, Analysis, and Normalization Methods for SARS-CoV-2 Concentrations in Wastewater to Assess COVID-19 Burdens in Wisconsin Communities. ACS EST Water 2021, 1, 1955–1965. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Towhid, S.T.; Mercier, É.; Hegazy, N.; Tian, X.; Bhatnagar, K.; Zhang, Z.; Naughton, C.C.; MacKenzie, A.E.; Graber, T.E.; et al. COVID-19 wastewater surveillance in rural communities: Comparison of lagoon and pumping station samples. Sci. Total Environ. 2021, 801, 149618. [Google Scholar] [CrossRef]
- Wu, F.; Xiao, A.; Zhang, J.; Moniz, K.; Endo, N.; Armas, F.; Bonneau, R.; Brown, M.A.; Bushman, M.; Chai, P.R.; et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022, 805, 150121. [Google Scholar] [CrossRef]
- Duvallet, C.; Wu, F.; McElroy, K.A.; Imakaev, M.; Endo, N.; Xiao, A.; Zhang, J.; Floyd-O’Sullivan, R.; Powell, M.M.; Mendola, S.; et al. Nationwide Trends in COVID-19 Cases and SARS-CoV-2 RNA Wastewater Concentrations in the United States. ACS EST Water 2022, 2, 1899–1909. [Google Scholar] [CrossRef]
- Greenwald, H.D.; Kennedy, L.C.; Hinkle, A.; Whitney, O.N.; Fan, V.B.; Crits-Christoph, A.; Harris-Lovett, S.; Flamholz, A.I.; Al-Shayeb, B.; Liao, L.D.; et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X 2021, 12, 100111. [Google Scholar] [CrossRef] [PubMed]
- Maal-Bared, R.; Qiu, Y.; Li, Q.; Gao, T.; Hrudey, S.E.; Bhavanam, S.; Ruecker, N.J.; Ellehoj, E.; Lee, B.E.; Pang, X. Does normalization of SARS-CoV-2 concentrations by Pepper Mild Mottle Virus improve correlations and lead time between wastewater surveillance and clinical data in Alberta (Canada): Comparing twelve SARS-CoV-2 normalization approaches. Sci. Total Environ. 2023, 856, 158964. [Google Scholar] [CrossRef] [PubMed]
- Sakarovitch, C.; Schlosser, O.; Courtois, S.; Proust-Lima, C.; Couallier, J.; Pétrau, A.; Litrico, X.; Loret, J.-F. Monitoring of SARS-CoV-2 in wastewater: What normalisation for improved understanding of epidemic trends? J. Water Health 2022, 20, 712–726. [Google Scholar] [CrossRef]
- Bethea, M.; Forman, D.T. Beta 2-microglobulin: Its significance and clinical usefulness. Ann. Clin. Lab. Sci. 1990, 20, 163–168. [Google Scholar] [PubMed]
- Palumbo, A.; Avet-Loiseau, H.; Oliva, S.; Lokhorst, H.M.; Goldschmidt, H.; Rosinol, L.; Richardson, P.; Caltagirone, S.; Lahuerta, J.J.; Facon, T.; et al. Revised International Staging System for Multiple Myeloma: A Report From International Myeloma Working Group. J. Clin. Oncol. 2015, 33, 2863–2869. [Google Scholar] [CrossRef]
- Ugan, Y.; Korkmaz, H.; Dogru, A.; Koca, Y.S.; Balkarlı, A.; Aylak, F.; Tunc, S.E. The significance of urinary beta-2 microglobulin level for differential diagnosis of familial Mediterranean fever and acute appendicitis. Clin. Rheumatol. 2016, 35, 1669–1672. [Google Scholar] [CrossRef]
- Hsu, S.-Y.; Bayati, M.; Li, C.; Hsieh, H.-Y.; Belenchia, A.; Klutts, J.; Zemmer, S.A.; Reynolds, M.; Semkiw, E.; Johnson, H.-Y.; et al. Biomarkers selection for population normalization in SARS-CoV-2 wastewater-based epidemiology. Water Res. 2022, 223, 118985. [Google Scholar] [CrossRef]
- Augusto, M.R.; Claro, I.C.M.; Siqueira, A.K.; Sousa, G.S.; Caldereiro, C.R.; Duran, A.F.A.; De Miranda, T.B.; Bomediano Camillo, L.D.M.; Cabral, A.D.; De Freitas Bueno, R. Sampling strategies for wastewater surveillance: Evaluating the variability of SARS-COV-2 RNA concentration in composite and grab samples. J. Environ. Chem. Eng. 2022, 10, 107478. [Google Scholar] [CrossRef]
- Kmush, B.L.; Monk, D.; Green, H.; Sachs†, D.A.; Zeng, T.; Larsen, D.A. Comparability of 24-hour composite and grab samples for detection of SARS-2-CoV RNA in wastewater. FEMS Microbes 2022, 3, xtac017. [Google Scholar] [CrossRef]
- Curtis, K.; Keeling, D.; Yetka, K.; Larson, A.; Gonzalez, R. Wastewater SARS-CoV-2 RNA Concentration and Loading Variability from Grab and 24-Hour Composite Samples. MedRxiv 2020. [Google Scholar] [CrossRef]
- Rafiee, M.; Isazadeh, S.; Mohseni-Bandpei, A.; Mohebbi, S.R.; Jahangiri-rad, M.; Eslami, A.; Dabiri, H.; Roostaei, K.; Tanhaei, M.; Amereh, F. Moore swab performs equal to composite and outperforms grab sampling for SARS-CoV-2 monitoring in wastewater. Sci. Total Environ. 2021, 790, 148205. [Google Scholar] [CrossRef] [PubMed]
- Dhiyebi, H.A.; Abu Farah, J.; Ikert, H.; Srikanthan, N.; Hayat, S.; Bragg, L.M.; Qasim, A.; Payne, M.; Kaleis, L.; Paget, C.; et al. Assessment of seasonality and normalization techniques for wastewater-based surveillance in Ontario, Canada. Front. Public Health 2023, 11, 1186525. [Google Scholar] [CrossRef] [PubMed]
- Breadner, P.R.; Dhiyebi, H.A.; Fattahi, A.; Srikanthan, N.; Hayat, S.; Aucoin, M.G.; Boegel, S.J.; Bragg, L.M.; Craig, P.M.; Xie, Y.; et al. A comparative analysis of the partitioning behaviour of SARS-CoV-2 RNA in liquid and solid fractions of wastewater. Sci. Total Environ. 2023, 895, 165095. [Google Scholar] [CrossRef]
- Joung, M.J.; Mangat, C.; Mejia, E.; Nagasawa, A.; Nichani, A.; Perez-Iratxeta, C.; Peterson, S.; Champredon, D. Coupling wastewater-based epidemiological surveillance and modelling of SARS-COV-2/COVID-19: Practical applications at the Public Health Agency of Canada. Can. Commun. Dis. Rep. 2023, 49, 166–174. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.-J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef] [PubMed]
- Girón-Guzmán, I.; Sánchez, G.; Pérez-Cataluña, A. Tracking epidemic viruses in wastewaters. Microb. Biotechnol. 2024, 17, e70020. [Google Scholar] [CrossRef]
- Carmo Dos Santos, M.; Cerqueira Silva, A.C.; Dos Reis Teixeira, C.; Pinheiro Macedo Prazeres, F.; Fernandes Dos Santos, R.; De Araújo Rolo, C.; De Souza Santos, E.; Santos Da Fonseca, M.; Oliveira Valente, C.; Saraiva Hodel, K.V.; et al. Wastewater surveillance for viral pathogens: A tool for public health. Heliyon 2024, 10, e33873. [Google Scholar] [CrossRef]
- Verani, M.; Pagani, A.; Federigi, I.; Lauretani, G.; Atomsa, N.T.; Rossi, V.; Viviani, L.; Carducci, A. Wastewater-Based Epidemiology for Viral Surveillance from an Endemic Perspective: Evidence and Challenges. Viruses 2024, 16, 482. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhatia, S.; Maswanganye, T.N.; Jeje, O.; Winston, D.; Lamssali, M.; Deng, D.; Blakley, I.; Fodor, A.A.; Jeffers-Francis, L. Wastewater Speaks: Evaluating SARS-CoV-2 Surveillance, Sampling Methods, and Seasonal Infection Trends on a University Campus. Microorganisms 2025, 13, 924. https://doi.org/10.3390/microorganisms13040924
Bhatia S, Maswanganye TN, Jeje O, Winston D, Lamssali M, Deng D, Blakley I, Fodor AA, Jeffers-Francis L. Wastewater Speaks: Evaluating SARS-CoV-2 Surveillance, Sampling Methods, and Seasonal Infection Trends on a University Campus. Microorganisms. 2025; 13(4):924. https://doi.org/10.3390/microorganisms13040924
Chicago/Turabian StyleBhatia, Shilpi, Tinyiko Nicole Maswanganye, Olusola Jeje, Danielle Winston, Mehdi Lamssali, Dongyang Deng, Ivory Blakley, Anthony A. Fodor, and Liesl Jeffers-Francis. 2025. "Wastewater Speaks: Evaluating SARS-CoV-2 Surveillance, Sampling Methods, and Seasonal Infection Trends on a University Campus" Microorganisms 13, no. 4: 924. https://doi.org/10.3390/microorganisms13040924
APA StyleBhatia, S., Maswanganye, T. N., Jeje, O., Winston, D., Lamssali, M., Deng, D., Blakley, I., Fodor, A. A., & Jeffers-Francis, L. (2025). Wastewater Speaks: Evaluating SARS-CoV-2 Surveillance, Sampling Methods, and Seasonal Infection Trends on a University Campus. Microorganisms, 13(4), 924. https://doi.org/10.3390/microorganisms13040924








