Simple Summary
In this study, we demonstrated the serotypes of three Pasteurellaeae species including Pasteurella multocida (PM), Glaesserella parasuis (GPS), and Actinobacillus pleuropneumoniae (APP), which are prevalent on Chinese pig farms. We also investigated the resistance profiles of these three bacterial species against different antimicrobial agents that are used for treating infections caused by them in pigs. We identified several virulence factor genes that were highly frequently presented and were conserved among these three Pasteurellaeae species. This study provides valuable insights into the epidemiology of these bacterial pathogens, which have significant implications for swine health and antibiotic stewardship.
Abstract
Pasteurella multocida (PM), Glaesserella parasuis (GPS), and Actinobacillus pleuropneumoniae (APP) are among the species with the top five isolation rates on Chinese pig farms annually. To understand the antimicrobial susceptibility and genotypes of these three pathogens that are currently prevalent on pig farms, we investigated 151 bacterial strains (64 PM, 48 GPS, and 39 APP) isolated from 4190 samples from farms in 12 Chinese provinces between 2021 and 2023. The prevalent serotypes were PM type D (50.0%), GPS type 5/12 (47.92%), and APP type 7 (35.90%). A relatively high proportion of PM and APP were resistant to ampicillin (PM, 93.75%; APP, 71.79%), tilmicosin (PM, 64.06%; APP, 58.97%), tetracycline (PM, 43.75%; APP, 61.54%), and enrofloxacin (PM, 34.38%; APP, 10.26%). Ampicillin, tetracycline, and enrofloxacin exhibited low MIC90 values against GPS (8 µg/mL), while sulfamethoxazole-trimethoprim had a high MIC90 value (512 µg/mL). A total of 18 genes conferring resistance to various antimicrobial classes were identified, and tet(L), tet(M), tet(A), blaTEM, sul2, aph(3′)-Ia, dfrA12, qnrS1, strA, sul3, and mef(B) exhibited a high frequency of identification (≥70%). The analysis of regular virulence factor genes showed that several genes, including fimB, fimA, fimD, fimF, and fepG, were found in all PM, GPS, and APP strains. However, certain genes exhibited species-specific preferences, even if they belonged to the same category.
1. Introduction
The family Pasteurellaceae comprises various pathogens that present significant threats to animal and/or public health and safety. Among them, Pasteurella multocida, Glaesserella parasuis (formerly Haemophilus parasuis), and Actinobacillus pleuropneumoniae are common pathogens in the global pig farming industry, causing respiratory diseases in pigs of different ages and leading to high morbidity, mortality, and economic losses [1]. Of these three Pasteurellaceae species, P. multocida is a Gram-negative pathogen responsible for swine atrophic rhinitis and pneumonic pasteurellosis [2,3]. P. multocida strains can be classified into five capsular serogroups (A, B, D, E, and F) [4]. Current reports indicate that types A and D are the predominant P. multocida serogroups among pigs, with sporadic cases of serogroup F strains reported [5,6].
G. parasuis is also a Gram-negative bacillus that depends on nicotinamide adenine dinucleotide (NAD) and causes Glässer’s disease in young pigs, presenting a total of fifteen serotypes (serotypes 1–15) [7]. Among them, serotypes 1, 5, 10, and 12 are highly pathogenic, serotypes 2, 4, 8, and 15 are moderately pathogenic, and serotypes 3, 6, 7, 9, and 11 are non-pathogenic, commonly found in healthy swine upper respiratory tracts [8]. Currently, serotypes 1, 5, 7, and 10 are the major infectious serotypes worldwide [9]. A. pleuropneumoniae can be divided into fifteen serotypes [10]. Among them, serotypes 1, 7, and 9 are the dominant serotypes in China [1]. The virulence of different serotypes primarily depends on the expression levels of RTX cytotoxins ApxI, ApxII, ApxIII, and ApxIV [11]. In addition to the toxins, all three of these Pasteurellaceae pathogens also encode multiple virulence factors, contributing to their pathogenesis and fitness. These include adhesion factors such as the type 1 fimbriae (FimB, FimE, FimA, FimI, FimC, FimD, FimF, FimG, and FimH), flagella (FlgJ, FlgH, FlgG, FlgE, FlgD, and FlgM), pili (YkgK/ecpR, YagZ/EcpA, YagY/EcpB, YagX/EcpC, YagW/EcpD, YagV/ecpE, EbpA, EbpB, and EbpC), and non-fimbrial adhesin (EfaA), proteins involved in iron uptake, transportation, and metabolism (FepA, FepB, FepC, FepD, FepG, IucA, IucB, IucC, IucD, IroB, IroC, IroD, IroE, and IroN), proteins contributing to serratiochelin and analog production (EntA, EntB, EntC, EntD, EntE, and EntF), the type II secretion system (T2SS; GspB, GspC, GspD, GspE, GspF, GspG, GspH, GspI, GspJ, GspK, GspL, and GspM), and outer membrane protein (OmpA) [12,13,14,15].
Currently, antimicrobial therapy plays a crucial role in the prevention and control of the aforementioned three swine Pasteurellaceae pathogens. Sulfonamides, lincomycin, and penicillins are commonly used for preventive treatments against swine pasteurellosis, while tetracyclines are recommended for treating infections caused by A. pleuropneumoniae [3,10]. For swine G. parasuis infections, florfenicols and rifampicin are recommended options [9]. However, recent studies have indicated that strains isolated from pig farms in recent years exhibit reduced sensitivity to the aforementioned drugs [1,16]. For instance, it was found that clinically isolated strains in Denmark displayed a 7.6% resistance to tetracyclines [17], while studies in Hungary revealed that clinically isolated strains exhibited a 55.6% resistance to tetracyclines and a resistance ranging from 5% to 52.5% to enrofloxacin [18]. Reports have indicated that between 2008 and 2010, G. parasuis in southern China showed resistance rates of 58%, 45.5%, 41.1%, and 29.4% to trimethoprim/sulfamethoxazole, enrofloxacin, tetracyclines, and penicillins, respectively [19]. A number of antimicrobial resistance genes (ARGs) have been reported to be responsible for the acquisition of these resistant phenotypes, including tet(A), tet(B), tet(C), tet(G), tet(H), tet(L), tet(M), and tet(O) for tetracycline resistance, blaCMY-2, blaOXA-2, blaPSE-2, blaROB-1, and blaTEM-1 for penicillin resistance, strA, strB, aadA1, aadA14, and aadA25 for streptomycin resistance, erm(A), erm(C), erm(T), and erm(42) for macrolide resistance, and qnrA1, qnrB6, and aac(6′)-Ib-cr for quinolone resistance [20].
China is the largest country in swine breeding and pork consumption globally. P. multocida, G. parasuis, and A. pleuropneumoniae stand as prevalent pathogens on Chinese pig farms, showing top five isolation rates in China annually [1]. Consequently, it is crucial that we comprehend the epidemic distribution characteristics of these three pathogenic bacteria within Chinese pig farming. However, further investigation is needed into the antimicrobial resistance profiles, as well as the serotypes and genotypes, of these three important swine Pasteurellaceae pathogens prevalent in China, as the epidemiological and resistant data for them are still limited. Therefore, this study conducted isolation identification, antimicrobial susceptibility testing, and whole-genome sequencing on the aforementioned three pathogens from pig farms in twelve provinces in China over the past three years. We aimed to gain insights into the current prevalence, dominant serotypes/genotypes, and antimicrobial resistance profiles of these three swine Pasteurellaceae pathogens.
2. Materials and Methods
2.1. Sample Collection and Bacterial Isolation
Between January 2021 and December 2023, a total of 4190 lung tissues from pigs that had succumbed to respiratory symptoms were submitted by pig farms from 12 provinces in China to the Veterinary Diagnostic Laboratory of Huazhong Agricultural University (HZAUVDL) in Wuhan, China, for examination for P. multocida, G. parasuis, and A. pleuropneumoniae. Bacterial strains were isolated from the samples under sterile conditions using an inoculation loop and cultured on tryptic soy agar (TSA; BD, Franklin Lakes, NJ, USA) supplemented with 10 µg/mL of Nicotinamide adenine dinucleotide (NAD; Sigma, St. Louis, MO, USA) and 5% newborn bovine serum (NBS; Boster Bio., Pleasanton, CA, USA) at 37 °C for 24 h. A single bacterial colony was selected from each sample for purification, followed by identification and capsular serotyping using specific gene PCR amplification methods [1,6]. The isolated and identified bacterial strains were preserved in 600 µL of tryptic soy broth (TSB; BD, Franklin Lakes, NJ, USA) containing 10 µg/mL of NAD (Sigma, St. Louis, MO, USA) and 5% NBS (Boster Bio., Pleasanton, CA, USA), mixed with 400 µL of sterile glycerol, and stored at −20 °C for future use.
2.2. Antimicrobial Susceptibility Testing
The protocol for the broth microdilution method, as recommended by the United States Clinical and Laboratory Standards Institute (CLSI document VET01S), was utilized to determine the minimal inhibitory concentrations (MICs) of the tested antimicrobial agents against different P. multocida and/or A. pleuropneumoniae strains [21]. As there is no CLSI method available for broth microdilution susceptibility testing in G. parasuis, previously recommended methods were followed [22,23]. The antimicrobial agents examined in this study included sulfamethoxazole-trimethoprim (sulfonamides), ceftiofur (cephalosporins), ampicillin (penicillin), tetracycline (tetracyclines), tilmicosin (macrolides), enrofloxacin (fluoroquinolones), and gentamicin (aminoglycosides). All agents were procured commercially from MedChemExpress (Monmouth Junction, NJ, USA), and their solutions for antimicrobial susceptibility testing (AST) were prepared following CLSI document VET01S guidelines (for P. multocida and A. pleuropneumoniae) or the referenced articles (for G. parasuis) [22]. The AST results for P. multocida and/or A. pleuropneumoniae were interpreted using the CLSI breakpoints [21]. In cases where CLSI breakpoints were unavailable, the MIC data were not categorized as susceptible, intermediate, or resistant. Each antimicrobial agent underwent testing in triplicate, and the entire experiment was conducted independently three times. Staphylococcus aureus ATCC® 29213 and/or Escherichia coli ATCC 25922 were utilized as quality control strains. Multidrug-resistant (MDR) strains are defined as bacteria resistant to more than three classes of antibiotics [24].
2.3. Illumina Sequencing and Bioinformatical Analysis
Bacterial genomic DNA was extracted using the Cetyltrimethyl Ammonium Bromide (CTAB) method and assessed for DNA concentrations, quality, and integrity using a Qubit Fluorometer (Invitrogen, Waltham, MA, USA) and a NanoDrop Spectrophotometer (Thermo Scientific, Waltham, MA, USA). Subsequently, DNA libraries were prepared with the NEBNext®Ultra™ II DNA Library Prep Kit (New England Biolabs, Ipswich, MA, USA) and sequenced on an Illumina Miseq™ platform (Illumina, San Diego, CA, USA). Raw reads underwent processing with Adapter Removal [25] to eliminate adapter contaminations and the SOAPec program (version 2.03, https://anaconda.org/bioconda/soapec, accessed on 12 November 2024) to filter out low-quality data. Clean reads were then de novo assembled using SOAPdenovo2 [26], and the assembled contigs were used for further bioinformatic analysis.
Antimicrobial resistance genes (ARGs) were identified using the Comprehensive Antibiotic Resistance Database (CARD) [27], while virulence factor genes were identified with the Virulence Factor Database (VFDB) [28]. Whole-genome single-nucleotide polymorphisms (WG-SNPs) were determined through genome-wide pairwise comparison using MAFFT (version 7.222) [29]. A phylogenetic tree was constructed using Parsnp software (version 1.2) [30]. Sequence data were deposited in NCBI under BioProjects PRJNA1082799 (P. multocida), PRJNA1080278 (G. parasuis), and PRJNA1079884 (A. pleuropneumoniae).
2.4. Statistical Analyses
Statistical analyses were performed to compare the resistant rates of a specific antimicrobial agent between two bacterial species using a Chi-squared test. p < 0.05 was considered to be significant.
3. Results
3.1. Bacterial Isolation and Distribution of Serotypes/Genotypes
A total of 151 strains from the three Pasteurellaceae species were isolated from the 4190 pig samples delivered. The overall isolation rate was 3.60% (151/4190). Among these strains, 64 were identified as P. multocida (1.53%, 64/4190), 48 were G. parasuis (1.15%, 48/4190), and 39 were A. pleuropneumoniae (0.93%, 39/4190). Detailed information regarding sample collection and bacterial isolation is provided in Table 1.

Table 1.
Isolation of the three Pasteurellaeae species from pigs in China between 2021 and 2023.
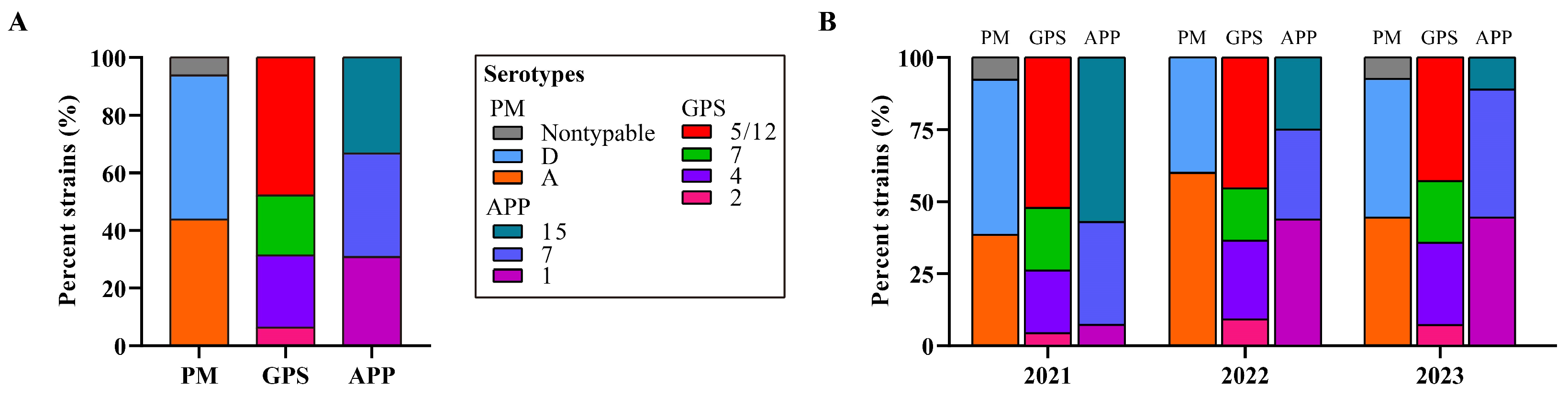
Bacterial typing revealed two known capsular types for the 64 P. multocida strains, with type D being predominant (50.0%, 32/64), followed by type A (43.75%, 28/64). Notably, four P. multocida strains (6.25%, 4/64) were untypable (Figure 1). Among the 48 G. parasuis strains, serotype 5/12 was most common (47.92%, 23/48), followed by serotype 4 (25.0%, 12/28), capsular serotype 7 (20.83%, 10/48), and capsular serotype 2 (6.25%, 3/48) (see Figure 1). Three capsular serotypes were identified for the 39 A. pleuropneumoniae strains, including serotype 7 (35.90%, 14/39), serotype 1 (30.77%, 12/39), and serotype 15 (30.33%, 13/39) (see Figure 1).
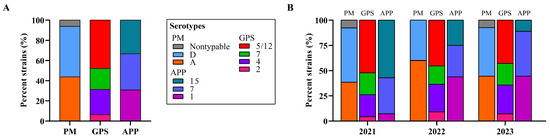
Figure 1.
Distribution of bacterial serotypes of the three Pasteurellaeae species. (A) A column chart showing the distribution of serotypes of the three Pasteurellaeae species determined in this study. (B) A column chart showing the distribution of serotypes of the three Pasteurellaeae species in different years. PM, Pasteurella multocida; GPS, Glaesserella parasuis; APP, Actinobacillus pleuropneumoniae.
3.2. Antimicrobial-Resistant Phenotypes
The distribution of MIC values obtained for the 151 strains of the three Pasteurellaceae species is detailed in Table 2. The results of the antimicrobial susceptibility testing indicated that among P. multocida strains, the proportions resistant to ampicillin, tilmicosin, tetracycline, and enrofloxacin were 93.75% (60/64), 64.06% (41/64), 43.75% (28/64), and 34.38% (22/64), respectively (Table 2). The MIC90 values for sulfamethoxazole-trimethoprim, ceftriaxone, and gentamicin were 512 µg/mL, 32 µg/mL, and 32 µg/mL, respectively (Table 2). Regarding A. pleuropneumoniae strains, the rates of resistance against ampicillin, tilmicosin, tetracycline, and enrofloxacin were 71.79% (28/39), 58.97% (23/39), 61.54% (24/39), and 10.26% (4/39), respectively (Table 2). The MIC90 values for sulfamethoxazole-trimethoprim, ceftriaxone, and gentamicin were 512 μg/mL, 32 μg/mL, and 32 μg/mL, respectively (Table 2). However, there was no significant difference in the resistant rates of the above-mentioned agents between P. multocida and A. pleuropneumoniae (p > 0.05).

Table 2.
Minimal inhibitory concentration (MIC) values of the tested antibiotics against the three Pasteurellaeae species from pigs in China between 2021 and 2023.
Due to the absence of CLSI breakpoints for G. parasuis, the isolates could not be categorized as susceptible, intermediate, or resistant, and, consequently, the proportions of resistant isolates could not be determined. Among the antimicrobial agents tested, ampicillin, tetracycline, and enrofloxacin exhibited the lowest MIC90 values (8 µg/mL), while sulfamethoxazole-trimethoprim had the highest MIC90 value (µg/mL) (Table 2). Notably, the MIC90 values for the various tested antimicrobial agents against P. multocida, G. parasuis, and A. pleuropneumoniae strains isolated in this study were uniform (Table 2). However, ceftriaxone displayed a higher MIC50 value for P. multocida (32 µg/mL) compared to G. parasuis (32 µg/mL) and A. pleuropneumoniae (2 µg/mL) (Table 2). Moreover, tetracycline demonstrated a lower MIC50 value for P. multocida (1 µg/mL) than for G. parasuis (8 µg/mL) and A. pleuropneumoniae (8 µg/mL). Gentamicin exhibited a higher MIC50 value for G. parasuis (32 µg/mL) compared to P. multocida (4 µg/mL) and A. pleuropneumoniae (4 µg/mL), whereas tilmicosin displayed a lower MIC50 value for G. parasuis (16 µg/mL) than for P. multocida (128 µg/mL) and A. pleuropneumoniae (128 µg/mL). Enrofloxacin showed a lower MIC50 value for A. pleuropneumoniae (0.25 µg/mL) than for P. multocida (0.5 µg/mL) and G. parasuis (0.5 µg/mL) (Table 2).
3.3. Distribution of Antimicrobial Resistance Genes and Their Associations with the Resistant Phenotypes
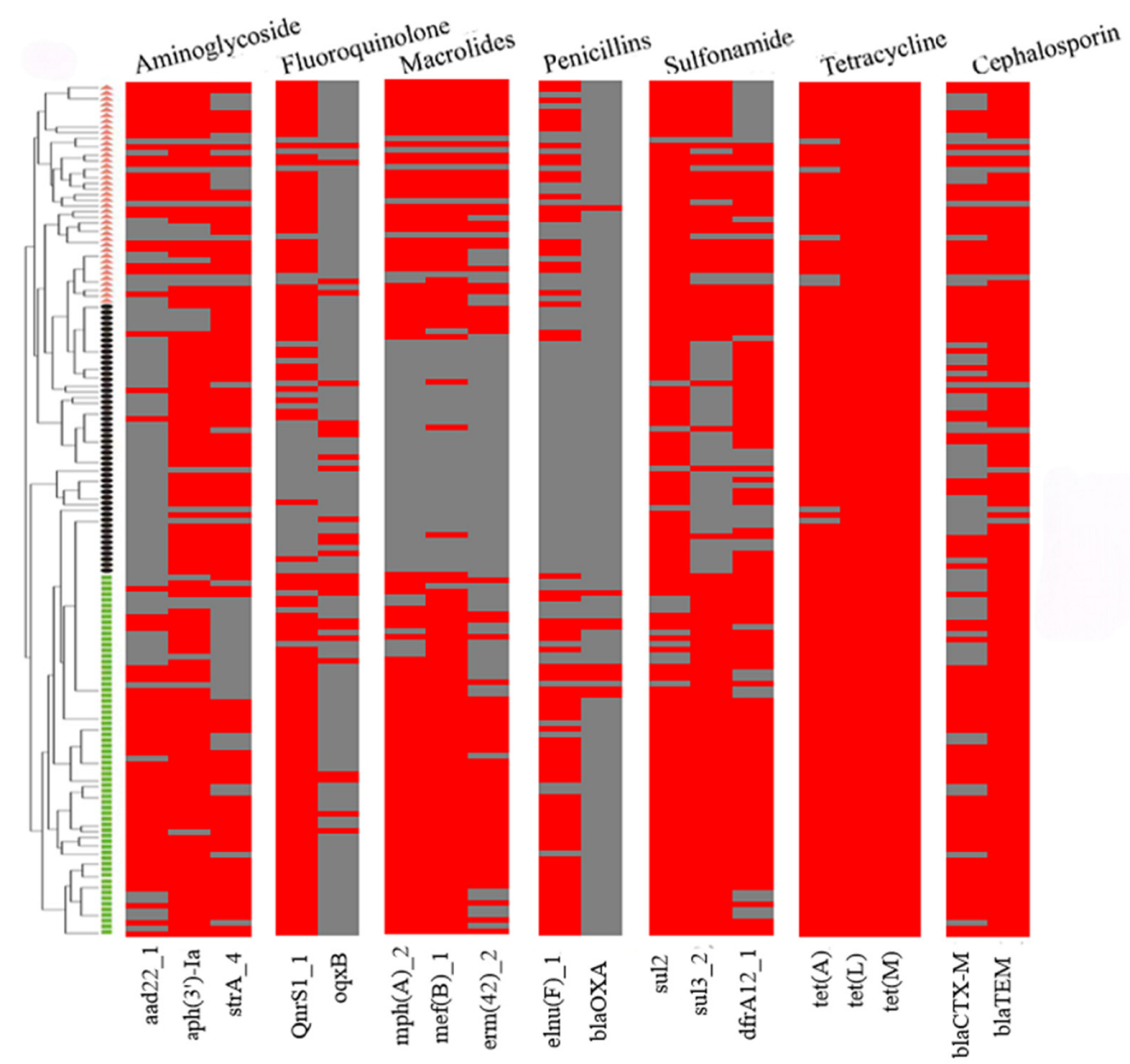
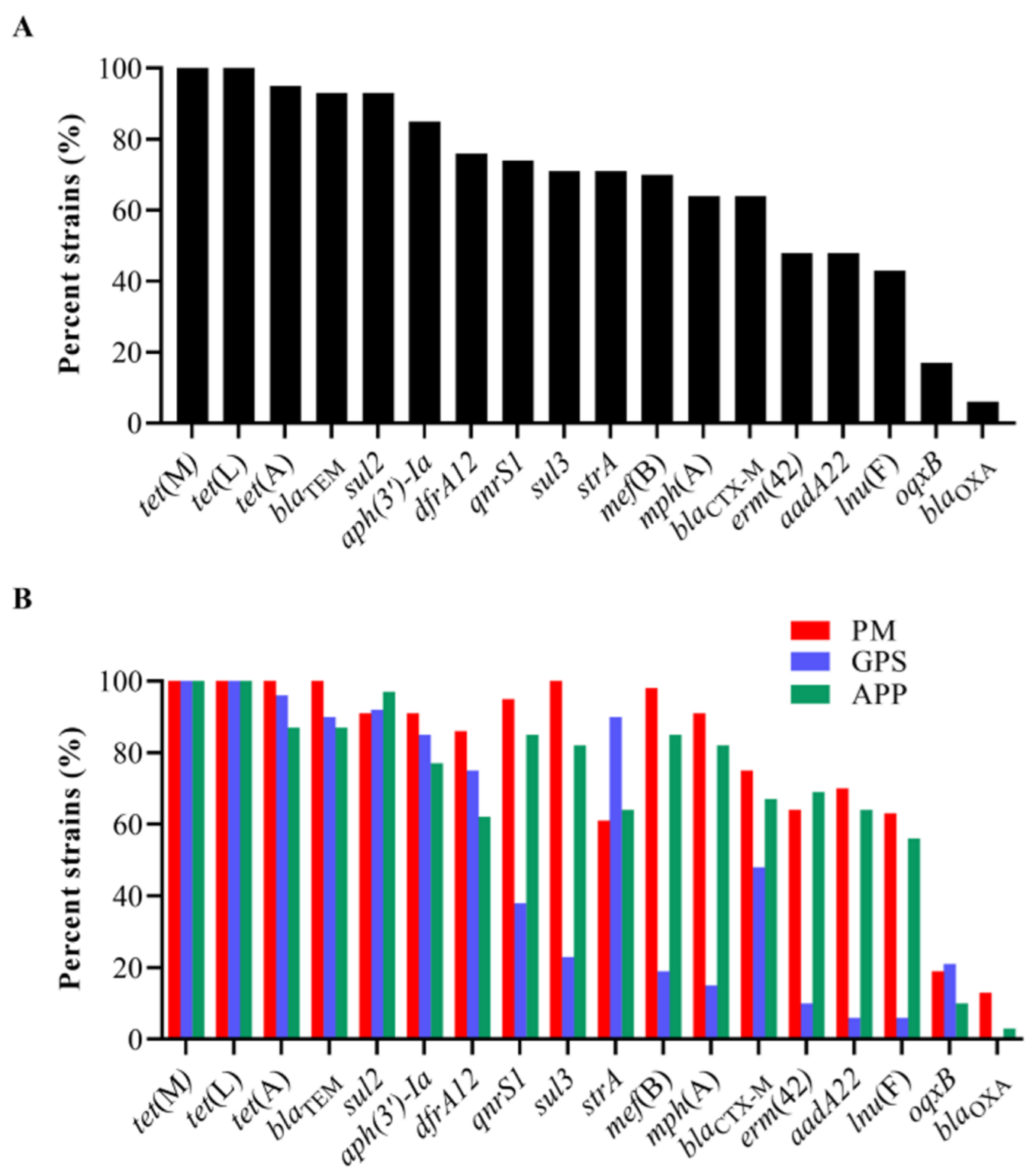
The examination of ARGs identified 18 genes that confer resistance to various classes of antibiotics, including sulfonamides (sul2, sul3, and dfrA12), cephalosporins (blaCTX-M and blaTEM), penicillin (lnu(F) and blaOXA), tetracycline (tet(A), tet(L), and tet(M)), macrolides (mph(A), mef(B), and erm(42)), quinolones (qnrS1 and oqxB), and aminoglycosides (aadA22, aph(3′)-Ia, and strA) in the 151 strains of the three Pasteurellaceae species (Figure 2). Among them, tet(L) (100%, 151/151), tet(M) (100%, 151/151), tet(A) (95%, 144/151), blaTEM (93%, 141/151), sul2 (93%, 140/151), aph(3′)-Ia (85%, 129/151), dfrA12 (76%, 115/151), qnrS1 (74%, 112/151), strA (71%, 107/151), sul3 (71%, 107/151), and mef(B) (70%, 105/151) exhibited a high frequency of identification, while oqxB (17%, 26/151) and blaOXA (6%, 9/151) showed a low frequency of identification (Figure 3A).
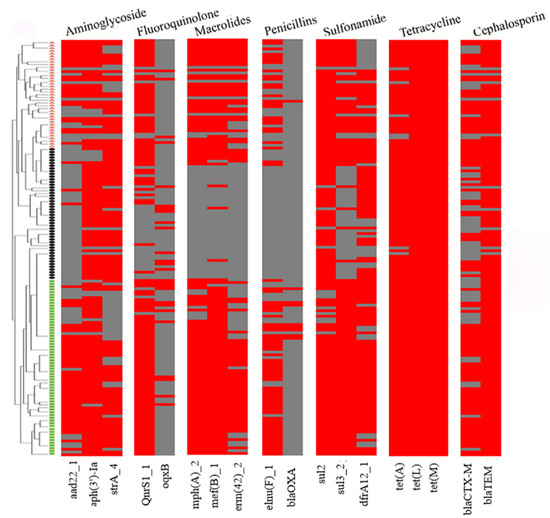
Figure 2.
A heatmap showing the distribution of antimicrobial resistance genes among the three Pasteurellaeae species. Pasteurella multocida strains are indicated using small green boxes; Glaesserella parasuis strains are indicated using small black pies; Actinobacillus pleuropneumoniae strains are indicated using orange small triangles. Strips in red refer to the presence of an antimicrobial resistance gene while those in gray refer to the absence of an antimicrobial resistance gene.
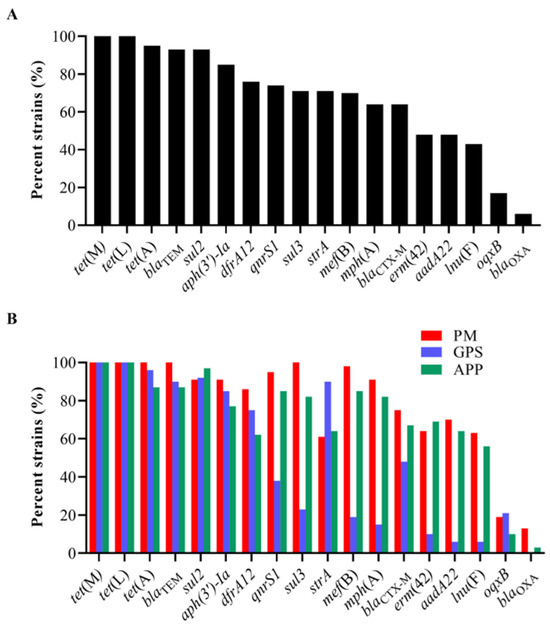
Figure 3.
Distribution of antimicrobial resistance genes (ARGs) among the three Pasteurellaeae species. (A) A column chart showing the detection rates of different ARGs among the 151 Pasteurellaeae strains. (B) A column chart showing the detection rates of different ARGs among Pasteurella multocida, Glaesserella parasuis, and Actinobacillus pleuropneumoniae.
All P. multocida, G. parasuis, and A. pleuropneumoniae strains showed high proportions (≥60%) of carrying several ARGs, including tet(L), tet(M), tet(A), blaTEM, dfrA12, sul2, strA, and aph(3′)-Ia (Figure 3B). However, the examination of several ARGs in G. parasuis was remarkably lower than those in P. multocida and/or A. pleuropneumoniae (Figure 3B). These included aadA22 (GPS vs. PM/APP: 6% vs. 70%/64%), qnrS1 (GPS vs. PM/APP: 38% vs. 95%/85%), mph(A) (GPS vs. PM/APP: 15% vs. 91%/82%), mef(B) (GPS vs. PM/APP: 19% vs. 98%/85%), erm(42) (GPS vs. PM/APP: 10% vs. 64%/69%), lnu(F) (GPS vs. PM/APP: 6% vs. 63%/56%), sul3 (GPS vs. PM/APP: 23% vs. 100%/82%), and blaCTX-M (GPS vs. PM/APP: 48% vs. 75%/67%). The presence of oqxB and blaOXA in these three Pasteurellaceae species was low. Notably, a weak association between the presence of ARGs and the resistant phenotypes was observed in this study. For example, although 100% of the P. multocida (PM) and A. pleuropneumoniae (APP) carried tetracycline-resistant genes tet(L) and tet(M), less than 65% of them demonstrated phenotypes of tetracycline resistance (PM: 43.75%, APP: 61.54%) (Figure 3B, Table 2). Similarly, over 80% of the P. multocida and A. pleuropneumoniae carried macrolide-resistant genes mph(A) and mef(B); only approximately 60% of the P. multocida and A. pleuropneumoniae were resistant to tilmicosin (PM: 64.06%, APP: 58.97%) (Figure 3B, Table 2).
3.4. Distribution of Genes Associated with Bacterial Fitness and Virulence
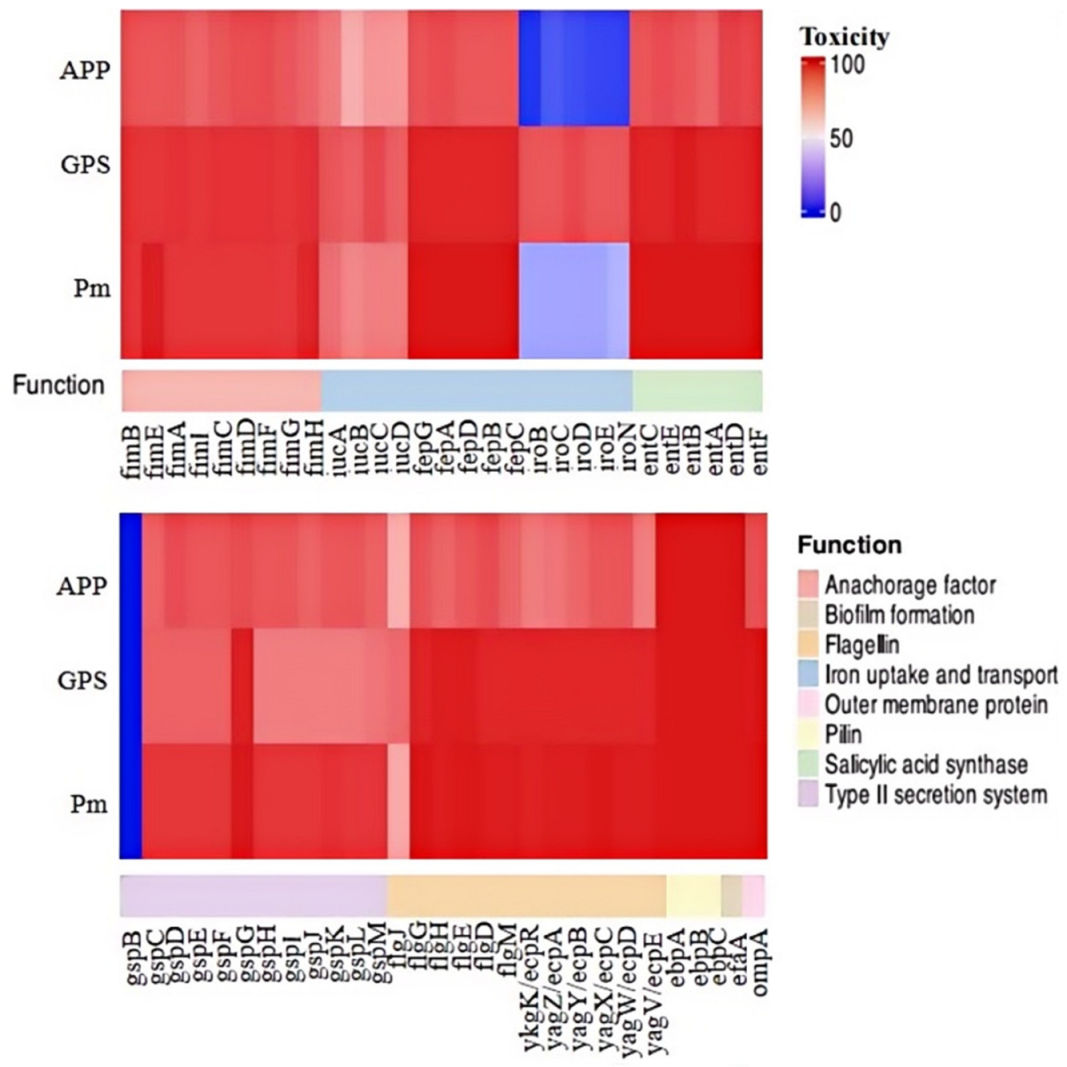
We proceeded to analyze the distribution of 58 genes associated with bacterial fitness and virulence across the 151 strains of the three Pasteurellaceae species (Figure 4). These 58 genes encode bacterial type 1 fimbriae (fimB, fimE, fimA, fimI, fimC, fimD, fimF, fimG, and fimH), proteins involved in iron uptake, transportation, and metabolism (fepA, fepB, fepC, fepD, fepG, iucA, iucB, iucC, iucD, iroB, iroC, iroD, iroE, and iroN), proteins contributing to serratiochelin and analog production (entA, entB, entC, entD, entE, and entF), the type II secretion system (T2SS; gspB, gspC, gspD, gspE, gspF, gspG, gspH, gspI, gspJ, gspK, gspL, and gspM), flagella (flgJ, flgH, flgG, flgE, flgD, and flgM), pili (ykgK/ecpR, yagZ/ecpA, yagY/ecpB, yagX/ecpC, yagW/ecpD, yagV/ecpE, ebpA, ebpB, and ebpC), non-fimbrial adhesin (efaA), and outer membrane protein (ompA). The results indicate that several of these genes, including fimB, fimA, fimD, fimF, and fepG, were found in all P. multocida, G. parasuis, and A. pleuropneumoniae strains examined in this study (Figure 4). Overall, genes related to type 1 fimbriae biosynthesis were highly prevalent among the three Pasteurellaceae species investigated. However, certain genes exhibited species-specific preferences, even if they belonged to the same category. For instance, iroBCDEN genes were identified in over 85% of the G. parasui strains but were present in less than 35% of the P. multocida strains and fewer than 20% of the A. pleuropneumoniae strains (Figure 4).
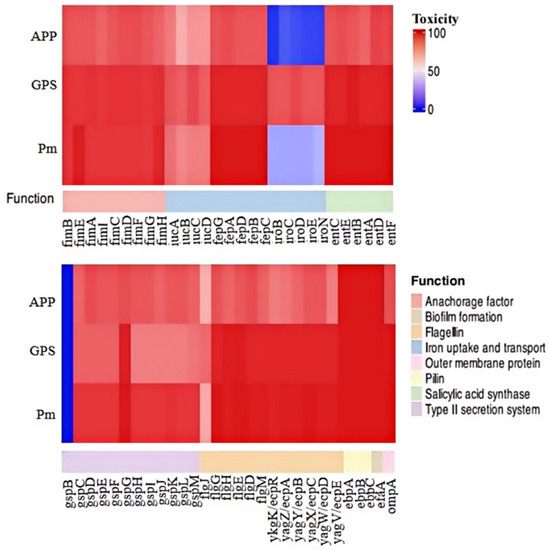
Figure 4.
A heatmap showing the distribution of virulence factor genes among the three Pasteurellaeae species. PM, Pasteurella multocida; GPS, Glaesserella parasuis; APP, Actinobacillus pleuropneumoniae.
4. Discussion
Respiratory infections pose significant challenges in the Chinese pig industry, with P. multocida, G. parasuis, and A. pleuropneumoniae identified as the primary causative bacteria behind swine respiratory illnesses [31,32]. In this study, we conducted bacterial isolation of three Pasteurellaceae species—P. multocida, G. parasuis, and A. pleuropneumoniae—from the lungs of pigs afflicted with respiratory disorders across various Chinese provinces. These three Pasteurellaceae species, recognized as the primary agents behind swine respiratory ailments, rank among the top five frequently isolated bacteria on pig farms in China [1,31]. Notably, the overall isolation rates of these Pasteurellaceae species in our study are comparatively lower than those reported in other research conducted in China [1,32]. Several factors may contribute to these lower isolation rates, with sample freshness emerging as a significant consideration. Although all samples were promptly processed upon arrival, prolonged transportation distances could impact sample freshness and the successful isolation of the three bacterial species. Moreover, it is plausible that P. multocida, G. parasuis, and/or A. pleuropneumoniae might not be solely responsible for the symptoms observed in the diseased pigs whose lung samples were analyzed. This is particularly pertinent given that numerous other pathogens such as Streptococcus suis, Mycoplasma pneumoniae, porcine reproductive and respiratory syndrome virus (PRRSV), and porcine circovirus (PCV) are also capable of causing swine respiratory infections and are prevalent on Chinese pig farms [32].
Our investigation revealed that P. multocida serogroups A and D, G. parasuis serotypes 5/12 and 4, and A. pleuropneumoniae serotypes 7 and 1 were predominantly prevalent on pig farms in China, findings that align closely with those of other studies conducted in China [6,32]. Notably, these serotypes are also documented as commonly occurring types on pig farms in various other Asian countries [33,34,35]. It is important to also acknowledge the identification of several other serotypes. For instance, while G. parasuis serotype 7 may not be as frequently identified as serotypes 5/12 and 4 on farms, multiple studies have emphasized serovar 7 as a significant disease-associated serotype of G. parasuis [35,36,37]. Moreover, a small proportion of P. multocida strains were untypable, indicating capsular serotypes that do not fall within categories A, B, D, E, or F [38]. The presence of untypable capsular serotypes in swine P. multocida has been reported in studies conducted in China and other countries [31,38,39]. The existence of untypable capsules in P. multocida could be attributed to either a lack of available tests [31] or could potentially signify the emergence of novel capsular types.
Our antimicrobial susceptibility testing results revealed concerning findings regarding the resistance profiles of P. multocida and A. pleuropneumoniae strains. For instance, ampicillin and enrofloxacin are commonly employed in treating pasteurellosis in pigs [3]. However, a significant proportion (93.75%) of the P. multocida strains tested in this study exhibited resistance to ampicillin, while 34.38% showed resistance to enrofloxacin. Notably, the resistance rates of P. multocida strains against these two antimicrobial agents, as well as tilmicosin, were notably higher compared to those in a recent study [16], suggesting that P. multocida strains prevalent on different farms may display varying resistance patterns. While several studies have indicated that a substantial number of A. pleuropneumoniae clinical strains from Europe and North America are susceptible to ampicillin and enrofloxacin [40,41], our data in this study, along with findings from other studies, highlighted significant proportions of A. pleuropneumoniae clinical strains from pigs in Taiwan and Australia that were resistant to ampicillin and other β-lactams [20,42]. These results hint at the existence of complex resistance profiles among A. pleuropneumoniae strains. Furthermore, the resistance rates of A. pleuropneumoniae to other agents such as tilmicosin and tetracycline were comparable to those reported in other studies [41]. Although the resistant rates of G. parasuis against these tested agents could not be determined due to the lack of available breakpoints, our broth microdilution assays revealed that many agents demonstrated high MIC90 values against a high proportion of the G. parasuis isolates, which is consistent with previous studies from different countries [20,23], and these results indicate worrisome resistance conditions. As for the clinical management of infections caused by bacteria including the three Pasteurellaceae species investigated in this study, antimicrobial stewardship, bio-security improvement, and vaccination represent practical strategies to combat the increasing threat of bacterial antimicrobial resistance [43]. In addition, continuously monitoring the resistant phenotypes of clinical isolates is also important.
Our analysis did not reveal a significant correlation between the presence of ARGs and resistant phenotypes in the three Pasteurellaceae species. A previously published article also noted that ARGs in porcine P. multocida were not linked to its antimicrobial susceptibility pattern [44]. Similarly, in another study, the susceptibility to antimicrobials in A. pleuropneumoniae did not strongly align with genotypic patterns [42]. This suggests that the development of antimicrobial resistance in Pasteurellaceae species may involve mechanisms beyond ARGs [45]. In addition to ARGs, we also analyzed the distribution of VFGs among the three Pasteurellaceae species. Many genes encoding proteins involved in bacterial adherence (such as type 1 fimbriae, flagella, pili, non-fimbrial adhesin, or OmpA) were found to be widely carried by these species. Previous genome-based studies have demonstrated similarities in these proteins across the three Pasteurellaceae species [12,13,14,15]. Consequently, some of these proteins could serve as potential immunogenic candidates to combat infections caused by P. multocida, G. parasuis, and/or A. pleuropneumoniae, as bacterial adherence-associated proteins are considered promising targets for intervention strategies [46,47].
We should acknowledge that our study has several limitations. The samples used for bacterial isolation were delivered by farm owners, which may limit our collection of bacterial strains from more regions. In addition, the period is also relatively short, which may restrict our further understanding of the comprehensive change dynamics behind the antimicrobial resistance trends in these three Pasteurellaceae species. We intend to continuously monitor the antimicrobial resistance profiles of different bacterial species prevalent on Chinese pig farms, and we hope to provide a comprehensive picture to reflect the bacterial antimicrobial resistance in swine in future.
5. Conclusions
In conclusion, our study reported on the isolation, prevalent serotypes, antimicrobial susceptibility, and presence of ARGs and VFGs in three Pasteurellaceae species—P. multocida, G. parasuis, and A. pleuropneumoniae—from pigs in China. While these species exhibited diverse serotypes, certain types were detected at high frequencies. Our antimicrobial susceptibility testing highlighted significant levels of antimicrobial resistance in these Pasteurellaceae species, with a notable proportion of strains showing resistance to therapeutic agents. Although our whole-genome sequencing analysis identified various ARGs, a strong correlation between phenotypic and genotypic susceptibility to antimicrobials was not observed. The distribution of VFGs, particularly those involved in bacterial adherence, was widespread among the three species, suggesting potential targets for intervention strategies.
Author Contributions
F.L.: Methodology, Investigation, Writing—original draft. X.Z., G.C., Y.Z., Q.C. and L.L.: Methodology, Investigation. H.C.: Writing—review and editing, Supervision. Z.P.: Writing—review and editing. C.T.: Writing—review and editing, Supervision, Project administration, Funding acquisition, Conceptualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key R&D Program of China (2022YFD1800900), and the China Agriculture Research System of MOF and MARA (CARS-35). The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Institutional Review Board Statement
This study has been approved by the Ethic Committee of Huazhong Agricultural University. The approval number is HZAUSW-2024-0043, approval date is 9 May 2024.
Informed Consent Statement
Not applicable.
Data Availability Statement
Sequence data were deposited in NCBI under BioProjects PRJNA1082799 (P. multocida), PRJNA1080278 (G. parasuis), and PRJNA1079884 (A. pleuropneumoniae).
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| APP | Actinobacillus pleuropneumoniae |
| ARG | Antimicrobial resistance genes |
| AST | Antimicrobial susceptibility testing |
| GPS | Glaesserella parasuis |
| MIC | Minimum inhibitory concentration |
| PM | Pasteurella multocida |
| VFG | Virulence factor gene |
References
- Zhang, B.; Ku, X.; Yu, X.; Sun, Q.; Wu, H.; Chen, F.; Zhang, X.; Guo, L.; Tang, X.; He, Q. Prevalence and antimicrobial susceptibilities of bacterial pathogens in Chinese pig farms from 2013 to 2017. Sci. Rep. 2019, 9, 9908. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.A.; Ho, M. Pasteurella multocida: From zoonosis to cellular microbiology. Clin. Microbiol. Rev. 2013, 26, 631–655. [Google Scholar] [CrossRef]
- Register, K.B.; Brockmeier, S.L. Pasteurellosis. In Diseases of Swine, 11th ed.; Wiley: Hoboken, NJ, USA, 2019; pp. 884–897. [Google Scholar]
- Townsend, K.M.; Boyce, J.D.; Chung, J.Y.; Frost, A.J.; Adler, B. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 2001, 39, 924–929. [Google Scholar] [CrossRef]
- Peng, Z.; Wang, X.; Zhou, R.; Chen, H.; Wilson, B.A.; Wu, B. Pasteurella multocida: Genotypes and Genomics. Microbiol. Mol. Biol. Rev. 2019, 83, 10–1108. [Google Scholar] [CrossRef]
- Liu, S.; Lin, L.; Yang, H.; Wu, W.; Guo, L.; Zhang, Y.; Wang, F.; Wang, X.; Song, W.; Hua, L.; et al. Pasteurella multocida capsular: Lipopolysaccharide types D:L6 and A:L3 remain to be the main epidemic genotypes of pigs in China. Anim. Dis. 2021, 1, 26. [Google Scholar] [CrossRef]
- Kielstein, P.; Rapp-Gabrielson, V.J. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J. Clin. Microbiol. 1992, 30, 862–865. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, H.; Blackall, P.J.; Yin, Z.; Wang, L.; Liu, Z.; Jin, M. Serological characterization of Haemophilus parasuis isolates from China. Vet. Microbiol. 2005, 111, 231–236. [Google Scholar] [CrossRef]
- Aragon, V.; Segalés, J.; (Dan) Tucker, A.W. Glässer’s Disease. In Diseases of Swine, 11th ed.; Wiley: Hoboken, NJ, USA, 2019; pp. 844–853. [Google Scholar]
- Gottschalk, M.; Broes, A. Actinobacillosis. In Diseases of Swine, 11th ed.; Wiley: Hoboken, NJ, USA, 2019; pp. 749–766. [Google Scholar]
- Soto Perezchica, M.M.; Guerrero Barrera, A.L.; Avelar Gonzalez, F.J.; Quezada Tristan, T.; Macias Marin, O. Actinobacillus pleuropneumoniae, surface proteins and virulence: A review. Front. Vet. Sci. 2023, 10, 1276712. [Google Scholar] [CrossRef] [PubMed]
- May, B.J.; Zhang, Q.; Li, L.L.; Paustian, M.L.; Whittam, T.S.; Kapur, V. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 2001, 98, 3460–3465. [Google Scholar] [CrossRef]
- Xu, Z.; Yue, M.; Zhou, R.; Jin, Q.; Fan, Y.; Bei, W.; Chen, H. Genomic characterization of Haemophilus parasuis SH0165, a highly virulent strain of serovar 5 prevalent in China. PLoS ONE 2011, 6, e19631. [Google Scholar] [CrossRef]
- Xu, Z.; Zhou, Y.; Li, L.; Zhou, R.; Xiao, S.; Wan, Y.; Zhang, S.; Wang, K.; Li, W.; Li, L.; et al. Genome biology of Actinobacillus pleuropneumoniae JL03, an isolate of serotype 3 prevalent in China. PLoS ONE 2008, 3, e1450. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liang, W.; Liu, W.; Wu, B.; Tang, B.; Tan, C.; Zhou, R.; Chen, H. Genomic characterization of Pasteurella multocida HB01, a serotype A bovine isolate from China. Gene 2016, 581, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhu, Z.; Shang, Y.; Song, W.; Yang, J.; Bi, H.; Wang, Z.; Xie, R.; Zhao, M.; Hua, L.; et al. Discovery of the tigecycline resistance gene cluster tmexCD3-toprJ1 in Pasteurella multocida strains isolated from pigs in China. Vet. Microbiol. 2024, 292, 110046. [Google Scholar] [CrossRef] [PubMed]
- Holmer, I.; Salomonsen, C.M.; Jorsal, S.E.; Astrup, L.B.; Jensen, V.F.; Høg, B.B.; Pedersen, K. Antibiotic resistance in porcine pathogenic bacteria and relation to antibiotic usage. BMC Vet. Res. 2019, 15, 449. [Google Scholar] [CrossRef]
- Somogyi, Z.; Mag, P.; Simon, R.; Kerek, Á.; Makrai, L.; Biksi, I.; Jerzsele, Á. Susceptibility of Actinobacillus pleuropneumoniae, Pasteurella multocida and Streptococcus suis Isolated from Pigs in Hungary between 2018 and 2021. Antibiotics 2023, 12, 1298. [Google Scholar] [CrossRef]
- Xu, C.; Zhang, J.; Zhao, Z.; Guo, L.; Zhang, B.; Feng, S.; Zhang, L.; Liao, M. Antimicrobial susceptibility and PFGE genotyping of Haemophilus parasuis isolates from pigs in South China (2008–2010). J. Vet. Med. Sci. 2011, 73, 1061–1065. [Google Scholar] [CrossRef]
- Dayao, D.; Gibson, J.S.; Blackall, P.J.; Turni, C. Antimicrobial resistance genes in Actinobacillus pleuropneumoniae, Haemophilus parasuis and Pasteurella multocida isolated from Australian pigs. Aust. Vet. J. 2016, 94, 227–231. [Google Scholar] [CrossRef]
- CLSI VET01SEd7E; CLSI: Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 7th ed. CLSI: Malvern, PA, USA, 2024.
- Prüller, S.; Turni, C.; Blackall, P.J.; Beyerbach, M.; Klein, G.; Kreienbrock, L.; Strutzberg-Minder, K.; Kaspar, H.; Meemken, D.; Kehrenberg, C. Towards a Standardized Method for Broth Microdilution Susceptibility Testing of Haemophilus parasuis. J. Clin. Microbiol. 2017, 55, 264–273. [Google Scholar] [CrossRef]
- Brogden, S.; Pavlović, A.; Tegeler, R.; Kaspar, H.; De Vaan, N.; Kehrenberg, C. Antimicrobial susceptibility of Haemophilus parasuis isolates from Germany by use of a proposed standard method for harmonized testing. Vet. Microbiol. 2018, 217, 32–35. [Google Scholar] [CrossRef]
- Feßler, A.T.; Wang, Y.; Burbick, C.R.; Diaz-Campos, D.; Fajt, V.R.; Lawhon, S.D.; Li, X.Z.; Lubbers, B.V.; Maddock, K.; Miller, R.A.; et al. Antimicrobial susceptibility testing in veterinary medicine: Performance, interpretation of results, best practices and pitfalls. One Health Adv. 2023, 1, 26. [Google Scholar] [CrossRef]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Alcock, B.P.; Huynh, W.; Chalil, R.; Smith, K.W.; Raphenya, A.R.; Wlodarski, M.A.; Edalatmand, A.; Petkau, A.; Syed, S.A.; Tsang, K.K.; et al. CARD 2023: Expanded curation, support for machine learning, and resistome prediction at the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2023, 51, D690–D699. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Treangen, T.J.; Ondov, B.D.; Koren, S.; Phillippy, A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Zhao, Z.; Hu, J.; Wu, B.; Cai, X.; He, Q.; Chen, H. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J. Clin. Microbiol. 2009, 47, 951–958. [Google Scholar] [CrossRef]
- Sun, Q.; Yu, X.; He, D.; Ku, X.; Hong, B.; Zeng, W.; Zhang, H.; He, Q. Investigation and analysis of etiology associated with porcine respiratory disease complex in China from 2017 to 2021. Front. Vet. Sci. 2022, 9, 960033. [Google Scholar] [CrossRef]
- Kim, B.; Hur, J.; Lee, J.Y.; Choi, Y.; Lee, J.H. Molecular serotyping and antimicrobial resistance profiles of Actinobacillus pleuropneumoniae isolated from pigs in South Korea. Vet. Q. 2016, 36, 137–144. [Google Scholar] [CrossRef]
- Kim, J.; Kim, J.W.; Oh, S.I.; So, B.; Kim, W.I.; Kim, H.Y. Characterisation of Pasteurella multocida isolates from pigs with pneumonia in Korea. BMC Vet. Res. 2019, 15, 119. [Google Scholar] [CrossRef]
- Macedo, N.; Gottschalk, M.; Strutzberg-Minder, K.; Van, C.N.; Zhang, L.; Zou, G.; Zhou, R.; Marostica, T.; Clavijo, M.J.; Tucker, A.; et al. Molecular characterization of Glaesserella parasuis strains isolated from North America, Europe and Asia by serotyping PCR and LS-PCR. Vet. Res. 2021, 52, 68. [Google Scholar] [CrossRef] [PubMed]
- Schuwerk, L.; Hoeltig, D.; Waldmann, K.H.; Strutzberg-Minder, K.; Valentin-Weigand, P.; Rohde, J. Serotyping and pathotyping of Glaesserella parasuis isolated 2012–2019 in Germany comparing different PCR-based methods. Vet. Res. 2020, 51, 137. [Google Scholar] [CrossRef]
- Dazzi, C.C.; Guizzo, J.A.; Prigol, S.R.; Kreutz, L.C.; Driemeier, D.; Chaudhuri, S.; Schryvers, A.B.; Frandoloso, R. New Pathological Lesions Developed in Pigs by a “Non-virulent” Strain of Glaesserella parasuis. Front. Vet. Sci. 2020, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, H.; Liang, W.; Chen, Y.; Tang, X.; Chen, H.; Wu, B. A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China. Arch. Microbiol. 2018, 200, 107–118. [Google Scholar] [CrossRef]
- Truswell, A.; Laird, T.J.; Jones, S.; O’Dea, M.; Blinco, J.; Abraham, R.; Morison, D.; Jordan, D.; Hampson, D.J.; Pang, S.; et al. Antimicrobial Resistance of and Genomic Insights into Pasteurella multocida Strains Isolated from Australian Pigs. Microbiol. Spectr. 2023, 11, e0378422. [Google Scholar] [CrossRef]
- El Garch, F.; de Jong, A.; Simjee, S.; Moyaert, H.; Klein, U.; Ludwig, C.; Marion, H.; Haag-Diergarten, S.; Richard-Mazet, A.; Thomas, V.; et al. Monitoring of antimicrobial susceptibility of respiratory tract pathogens isolated from diseased cattle and pigs across Europe, 2009–2012: VetPath results. Vet. Microbiol. 2016, 194, 11–22. [Google Scholar] [CrossRef]
- Siteavu, M.I.; Drugea, R.I.; Pitoiu, E.; Ciobotaru-Pirvu, E. Antimicrobial Resistance of Actinobacillus pleuropneumoniae, Streptococcus suis, and Pasteurella multocida Isolated from Romanian Swine Farms. Microorganisms 2023, 11, 2410. [Google Scholar] [CrossRef] [PubMed]
- Ke, C.H.; Lai, P.Y.; Hsu, F.Y.; Hsueh, P.R.; Chiou, M.T.; Lin, C.N. Antimicrobial susceptibility and resistome of Actinobacillus pleuropneumoniae in Taiwan: A next-generation sequencing analysis. Vet. Q. 2024, 44, 1–13. [Google Scholar] [CrossRef]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef]
- Petrocchi-Rilo, M.; Gutiérrez-Martín, C.B.; Pérez-Fernández, E.; Vilaró, A.; Fraile, L.; Martínez-Martínez, S. Antimicrobial Resistance Genes in Porcine Pasteurella multocida Are Not Associated with Its Antimicrobial Susceptibility Pattern. Antibiotics 2020, 9, 614. [Google Scholar] [CrossRef]
- Michael, G.B.; Bossé, J.T.; Schwarz, S. Antimicrobial Resistance in Pasteurellaceae of Veterinary Origin. Microbiol. Spectr. 2018, 6, 10. [Google Scholar] [CrossRef] [PubMed]
- Krachler, A.M.; Orth, K. Targeting the bacteria-host interface: Strategies in anti-adhesion therapy. Virulence 2013, 4, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).