Integrative Genomics and Metabolomics Analyses Provide New Insights into the Molecular Basis of Plant Growth Promotion by Pantoea agglomerans
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequences, Bacterial Strains, and Culture Media
2.2. Comparative and Pan-Genome Analysis
2.3. In Silico Identification of Genetic Factors Involved in Plant-Bacteria Interactions
2.4. Identification of Regulatory Proteins
2.5. Indole Auxin Production in Shake Flasks
2.6. Measurement of Indole-3-Acetic Acid (IAA) and Related Indolic Metabolites Production
2.7. Untargeted Metabolomics and Statistical Analysis
3. Results
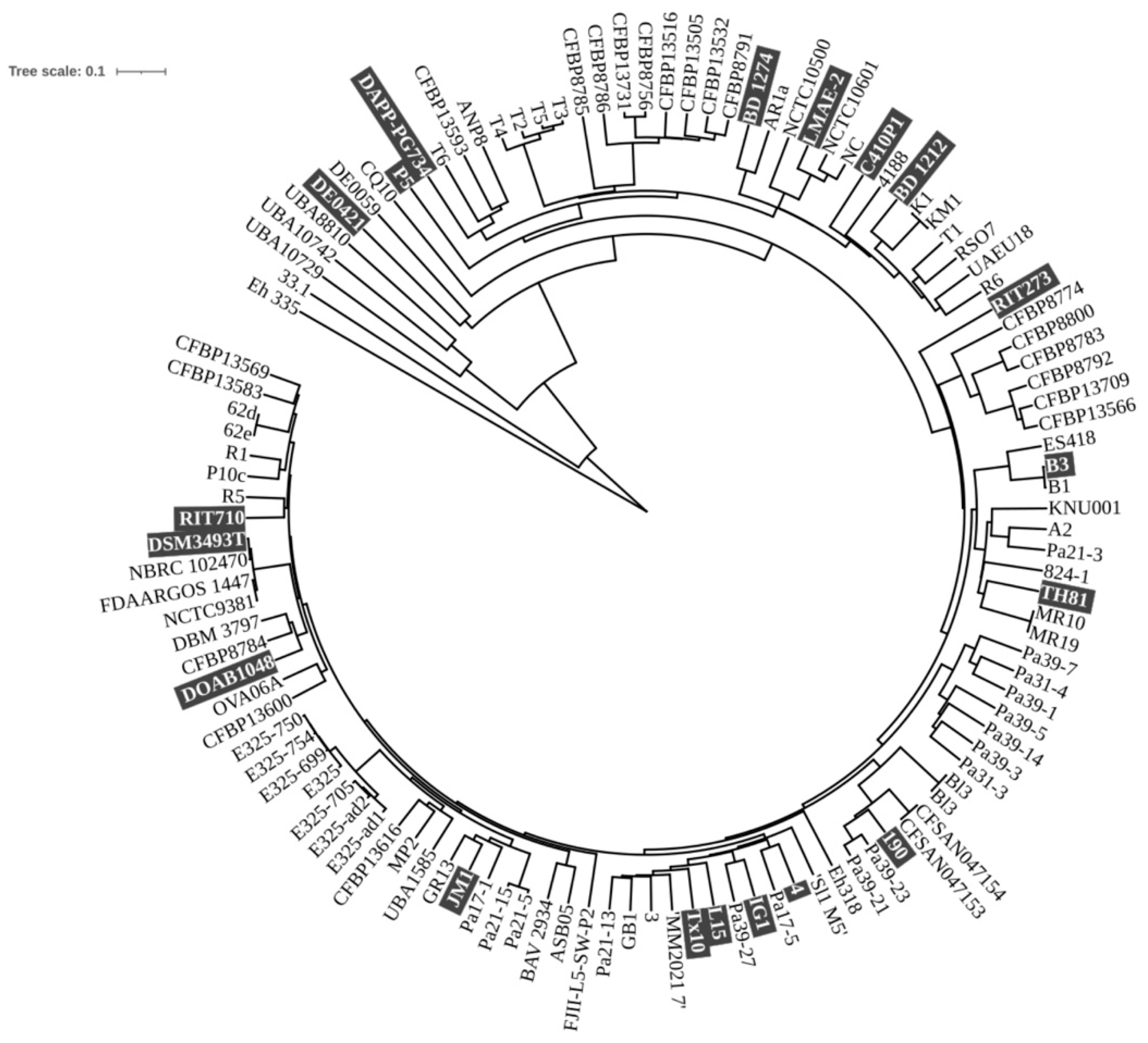
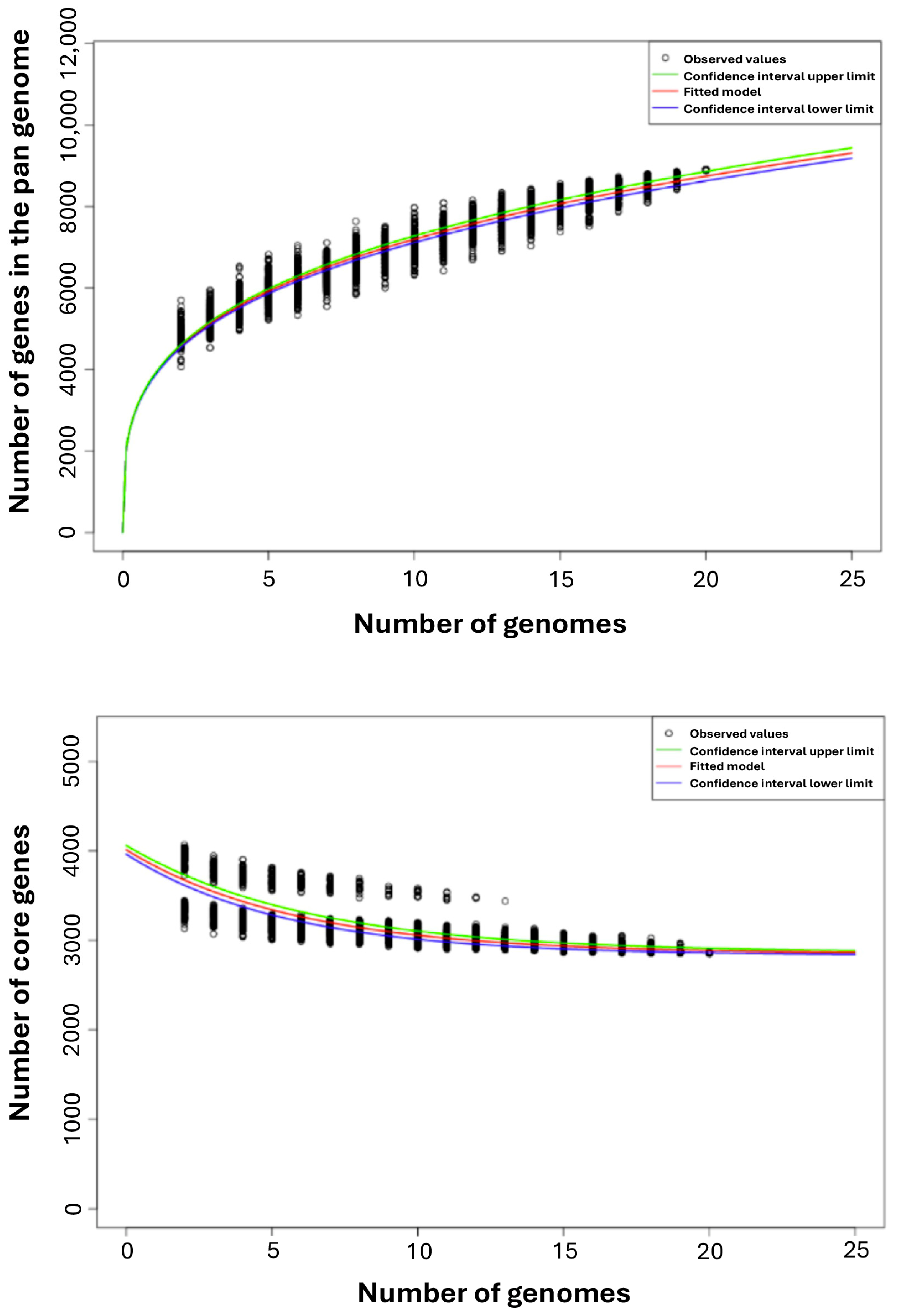
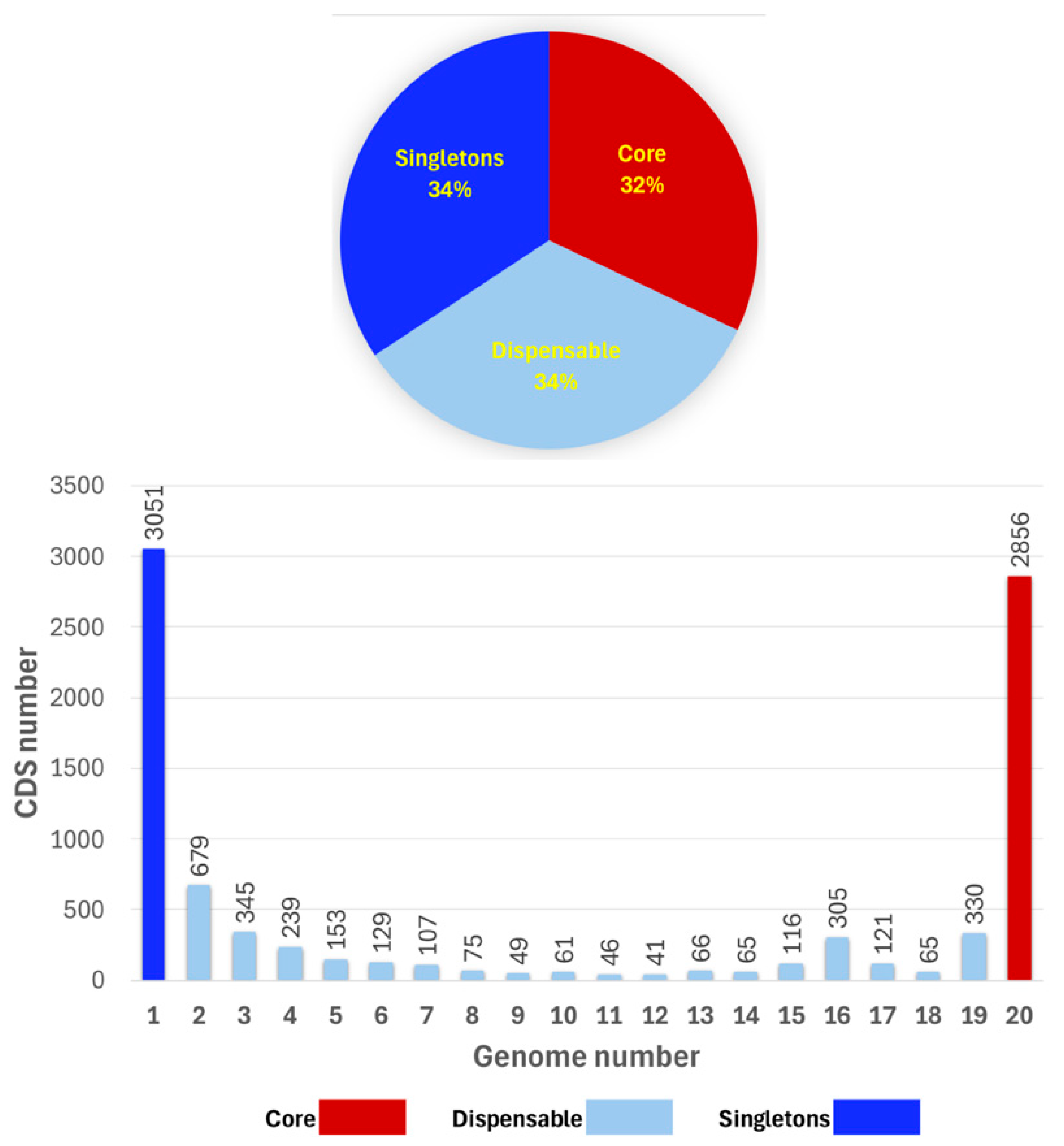
3.1. Pan- and Core Genome
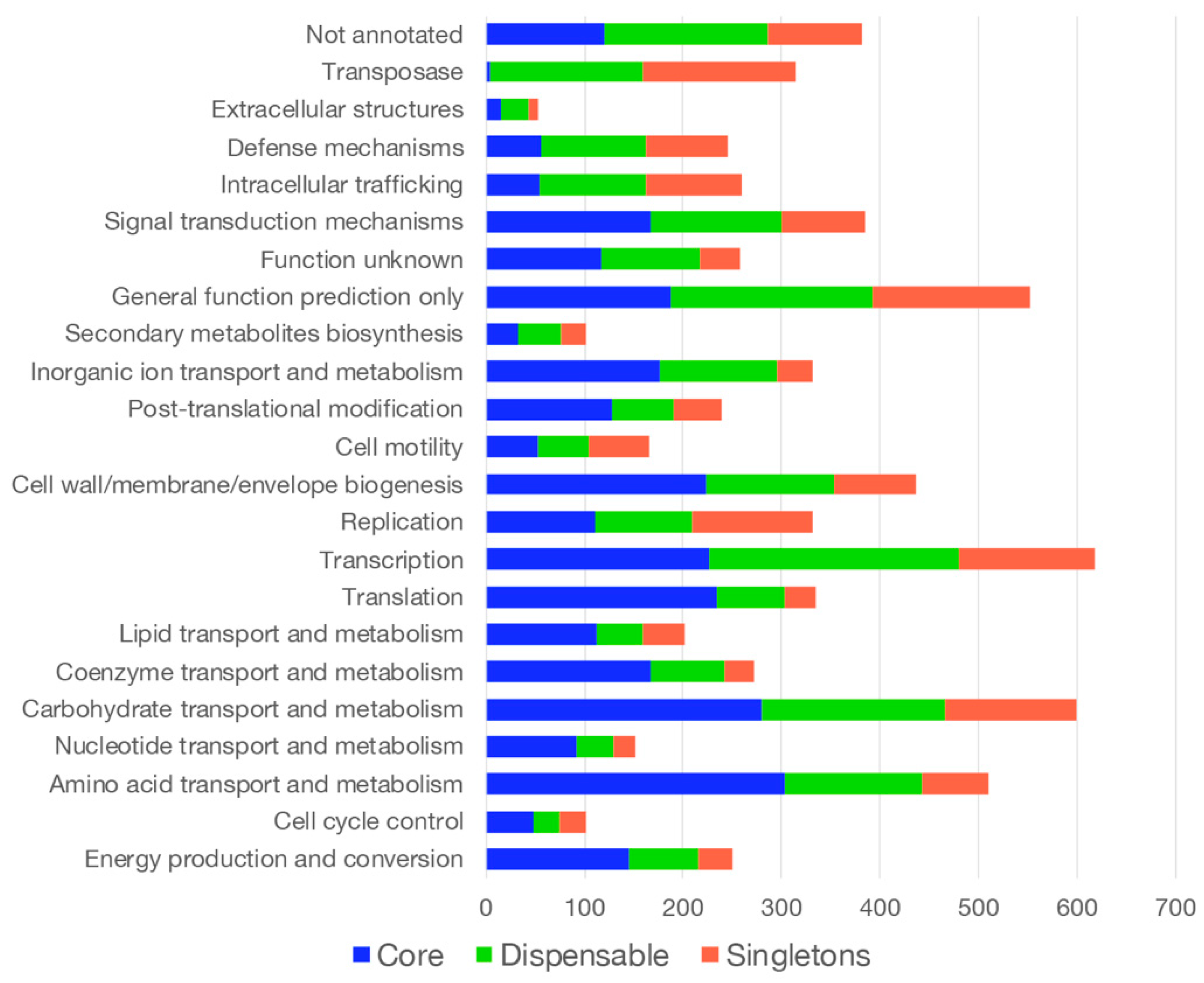
3.2. Functional Annotation of the Pan- and Core Genome Sequences
3.3. Genes Related to Plant Growth-Promotion
3.4. Nitrogen Metabolism
3.5. Sulfur Metabolism
3.6. Tolerance Against Heavy Metals
3.7. DNA Phosphorothioate (PT) Modification
3.8. The Oligopeptide (opp) Gene Cluster
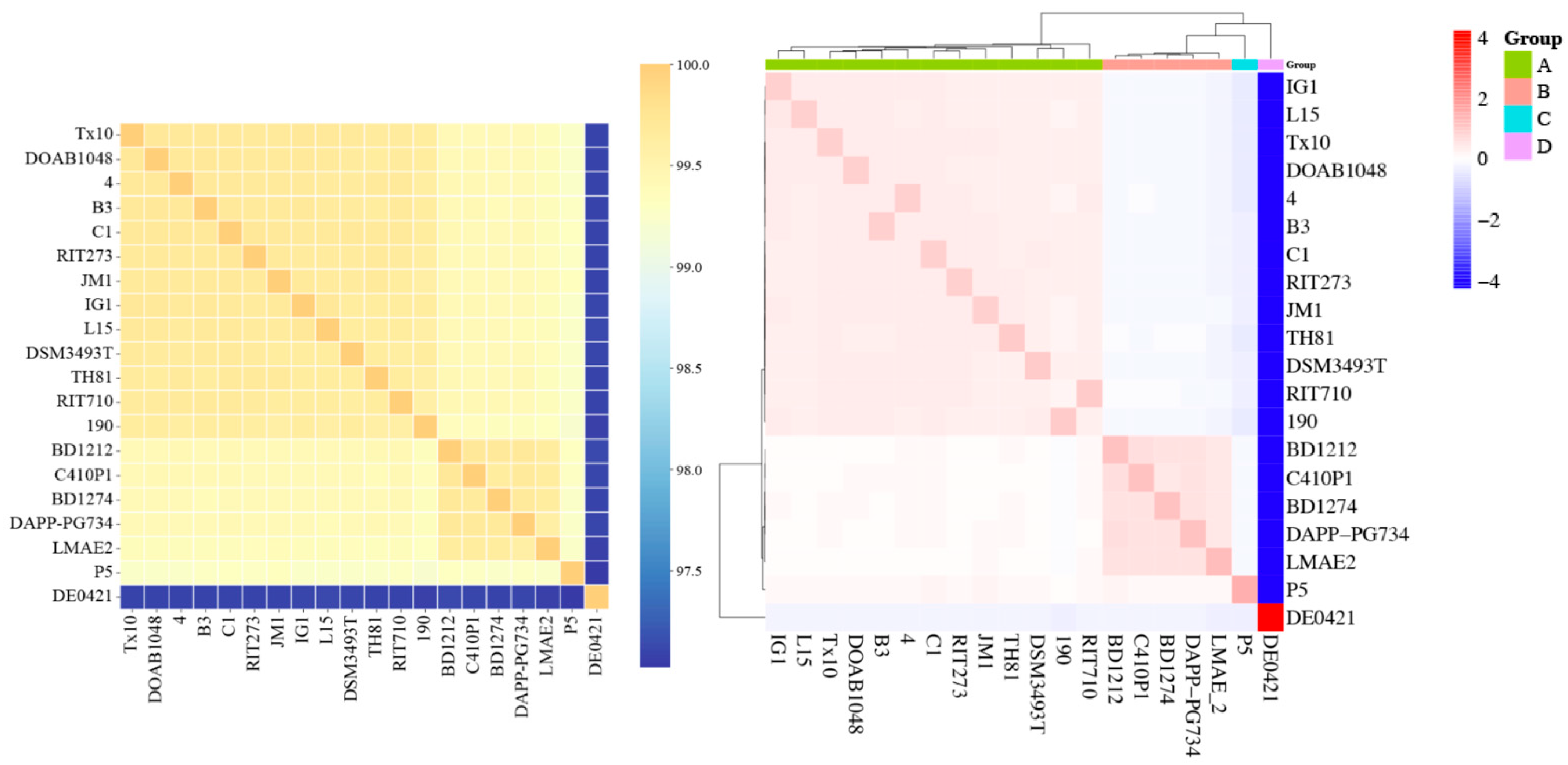
3.9. Whole-Genome Comparison Between P. agglomerans Strains C1 and DSM3493T
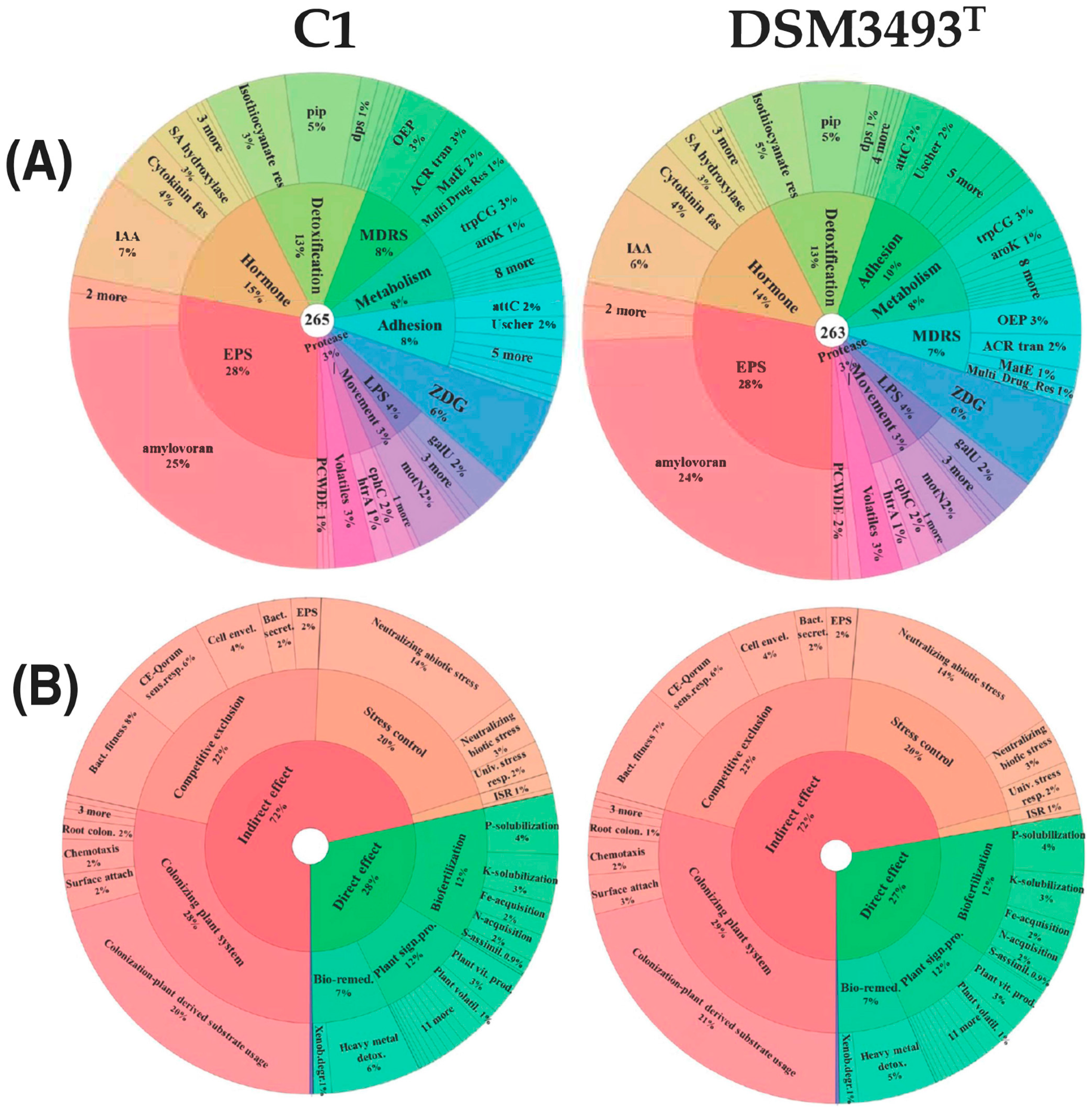
3.10. Genetic Traits Associated with Bacteria-Plant Interaction and Plant Growth Promotion
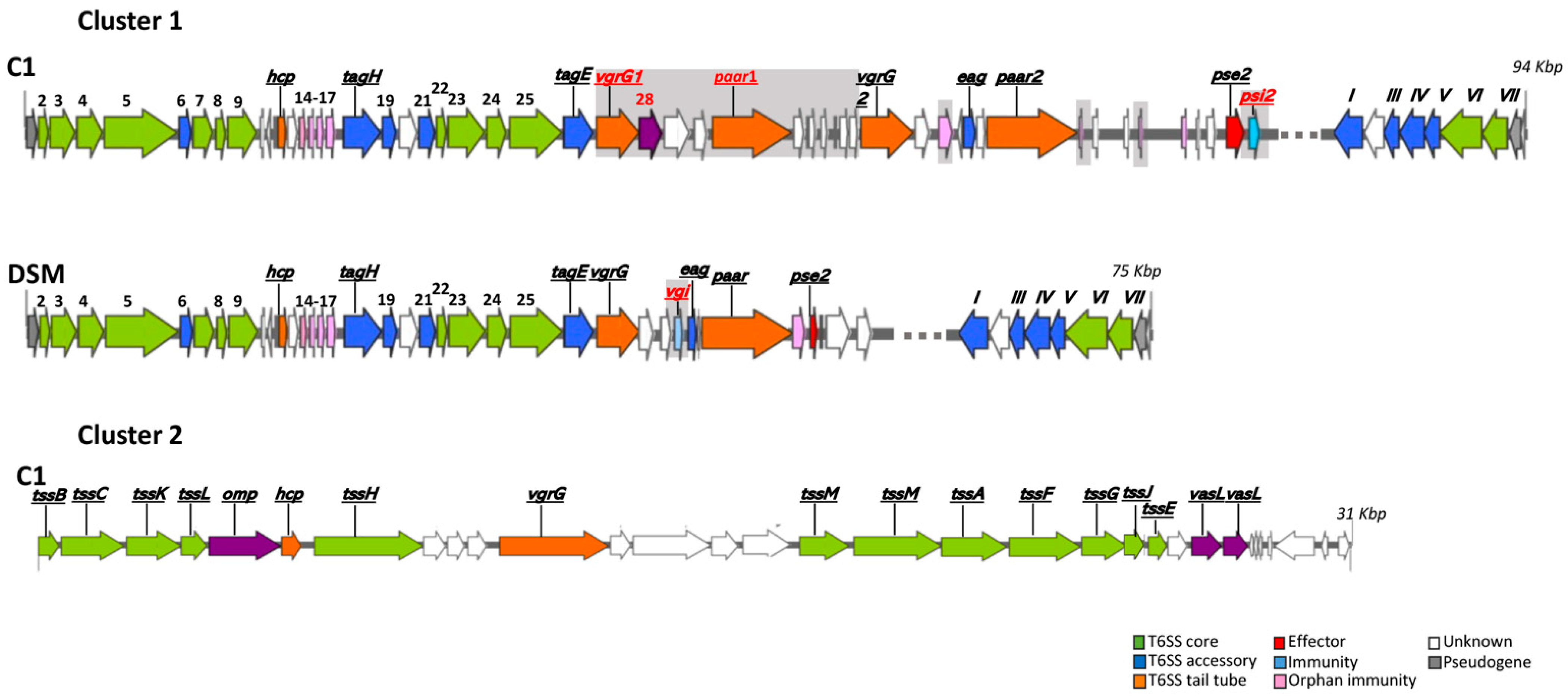
3.11. Type VI Secretion System
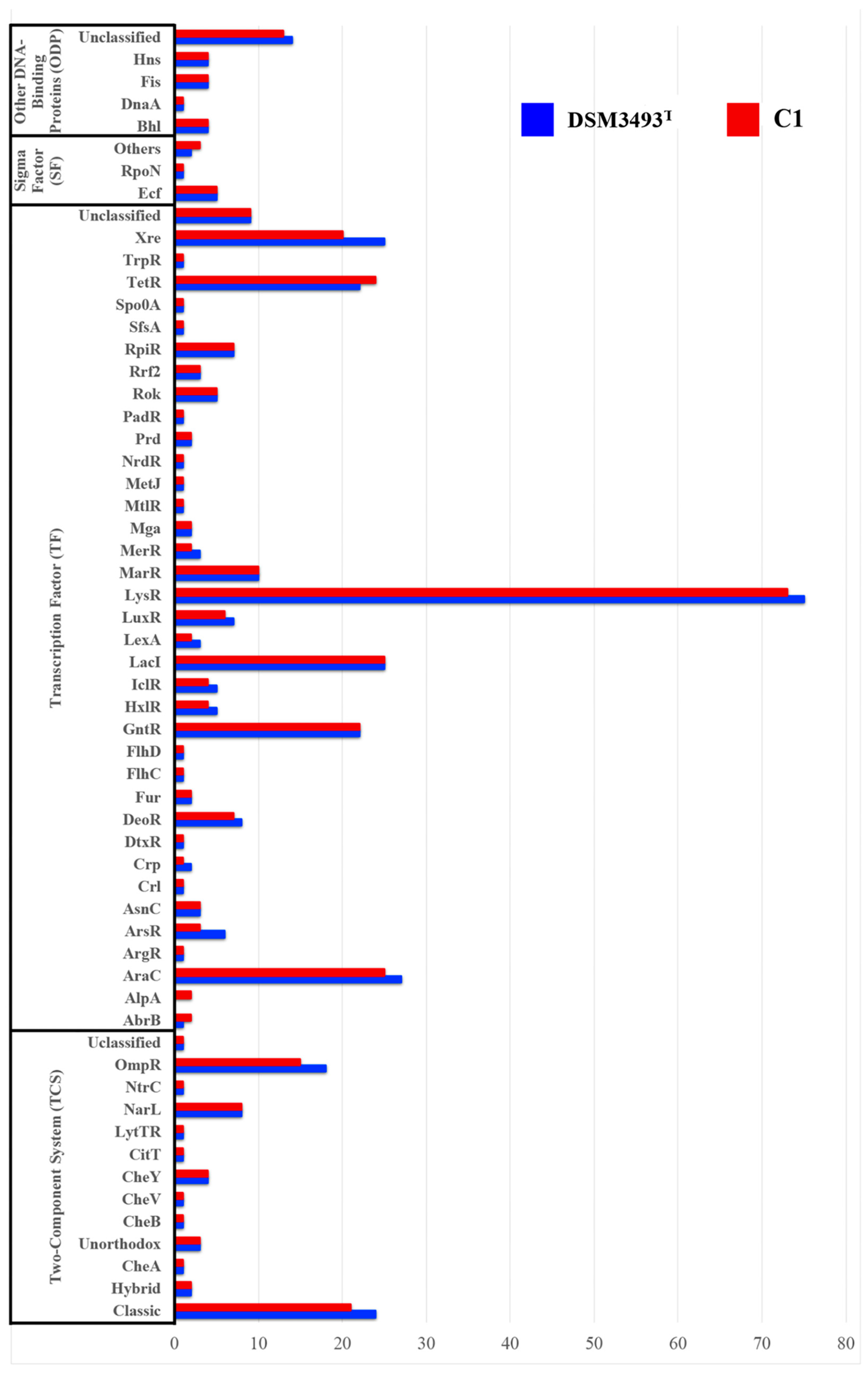
3.12. Regulatory Proteins (RP)
3.13. Auxin Production Under Different Culture Conditions
3.14. Comparative Untargeted Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PGPR | Plant Growth-Promoting Rhizobacteria |
| IAA | indole-3-acetic acid |
| IPDC | indole pyruvate decarboxylase |
| VY | vegetal peptone–yeast extract |
| EDGAR | Efficient database framework for comparative genome analyses using BLAST score ratios |
| AAI | Average Amino Acid Identity |
| SRVs | Score Ratio Values |
| COG | Clusters of Orthologous Genes |
| PGPTs | Plant Growth-Promoting Traits |
| P2RP | Predicted Prokaryotic Regulatory Proteins |
| COGs | Clusters of Orthologous Groups |
| GABA | γ-aminobutyric acid |
| EPS | Exopolysaccharides |
| TF | Transcription Factor |
| RP | Regulatory Protein |
| LB | Luria–Bertani |
References
- De Andrade, L.A.; Santos, C.H.B.; Frezarin, E.T.; Sales, L.R.; Rigobelo, E.C. Plant Growth-Promoting Rhizobacteria for Sustainable Agricultural Production. Microorganisms 2023, 11, 1088. [Google Scholar] [CrossRef]
- Kumar, M.; Poonam; Ahmad, S.; Singh, R.P. Plant growth promoting microbes: Diverse roles for sustainable and ecofriendly agriculture. Energy Nexus 2022, 7, 100133. [Google Scholar] [CrossRef]
- Abou Jaoudé, R.; Luziatelli, F.; Ficca, A.G.; Ruzzi, M. A plant ‘s perception of growth-promoting bacteria and their metabolites. Front. Plant Sci. 2024, 14, 1332864. [Google Scholar] [CrossRef] [PubMed]
- Soltani, H.; Hassani, A.; Baba Akbari Sari, M.; Hanifei, M. The phosphate solubilizing and N fixing Pantoea agglomerans bacteria affecting yield and biochemical properties including nutrient uptake of different tomato genotypes. J. Trace Elem. Min. 2025, 12, 100225. [Google Scholar] [CrossRef]
- Shariati, J.V.; Malboobi, M.A.; Tabrizi, Z.; Tavakol, E.; Owlia, P.; Safari, M. Author Correction: Comprehensive genomic analysis of a plant growth-promoting rhizobacterium Pantoea agglomerans strain P5. Sci. Rep. 2017, 7, 15610, Erratum in Sci Rep. 2020, 10, 12068. https://doi.org/10.1038/s41598-020-69169-7. [Google Scholar] [CrossRef]
- Lee, S.I.; Tran, T.D.; Hnasko, R.; McGarvey, J.A. Use of Pantoea agglomerans ASB05 as a biocontrol agent to inhibit the growth of Salmonella enterica on intact cantaloupe melons. J. Appl. Microbiol. 2023, 134, lxad235, Erratum in: J. Appl. Microbiol. 2023, 134, lxad254. [Google Scholar] [CrossRef] [PubMed]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Baldassarre Švecová, E.; Ruzzi, M. Foliar application of vegetal-derived bioactive compounds stimulates the growth of beneficial bacteria and enhances microbiome biodiversity in lettuce. Front. Plant Sci. 2019, 10, 60. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ahmad, I.; Pichtel, J.; Husain, F.M. Pantoea agglomerans FAP10: A novel biofilm-producing PGPR strain improves wheat growth and soil resilience under salinity stress. Environ. Exp. Bot. 2024, 222, 105759. [Google Scholar] [CrossRef]
- Elizondo-Reyna, E.; Martínez-Montoya, H.; Tamayo-Ordoñez, Y.; Cruz-Hernández, M.A.; Carrillo-Tripp, M.; Tamayo-Ordoñez, M.C.; de Jesús Sosa-Santillán, G.; Rodríguez-de la Garza, J.A.; Hernández-Guzmán, M.; Bocanegra-García, V.; et al. Insights from a Genome-Wide Study of Pantoea agglomerans UADEC20: A Promising Strain for Phosphate Solubilization and Exopolysaccharides Production. Curr. Issues Mol. Biol. 2025, 47, 56. [Google Scholar] [CrossRef]
- Manikandan, S.K.; Elnaggar, A.; Ahmady, I.M.; Tsombou, F.M.; El-Tarabily, K.A.; Sheteiwy, M.S.; El-Keblawy, A. Endophytic Pantoea agglomerans enhances lead phytoremediation and stress resilience of Calotropis procera in hydroponic system. Sci. Rep. 2025, 15, 26712. [Google Scholar] [CrossRef] [PubMed]
- Kirk, A.; Davidson, E.; Stavrinides, J. The expanding antimicrobial diversity of the genus Pantoea. Microbiol. Res. 2024, 289, 127923. [Google Scholar] [CrossRef]
- Kawuri, R. The antagonistic potential of Pantoea sp. against Ralstonia solanacearum, the root cause of bacterial wilt disease in potatoes (Solanum tuberosum L.). Malays. J. Microbiol. 2023, 19, 560–569. [Google Scholar]
- Bakhora, D.; Sokhibjon, A.; Dilfuza, J.; Bakhora, T.; Guzal, K.; Kakhramon, D. Bacteria associated with grapevine (Vitis vinifera L.) rhizosphere and their efficacy in plant growth promotion. Plant Sci. Today 2024, 11, 1739–1746. [Google Scholar] [CrossRef]
- Stockwell, V.O.; Johnson, K.B.; Sugar, D.; Loper, J.E. Antibiosis contributes to biological control of fire blight by Pantoea agglomerans strain Eh252 in orchards. Phytopathology 2002, 92, 1202–1209. [Google Scholar] [CrossRef] [PubMed]
- Dagher, F.; Olishevska, S.; Philion, V.; Zheng, J.; Déziel, E. Development of a novel biological control agent targeting the phytopathogen Erwinia amylovora. Heliyon 2020, 6, e05222. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Bonini, P.; Muleo, R.; Gatti, L.; Meneghini, M.; Tronati, M.; Melini, F.; Ruzzi, M. A genetic and metabolomic perspective on the production of indole-3-acetic acid by Pantoea agglomerans and use of their metabolites as biostimulants in plant nurseries. Front. Microbiol. 2020, 11, 1475. [Google Scholar] [CrossRef] [PubMed]
- Sallami, A.; Rabeh, K.; Idrissi Lahsini, A.; El Khedri, H.; Douira, A.; El Modafar, C.; Medraoui, L.; Filali-Maltouf, A. The ability of two indigenous bacteria isolated from Moroccan olive tree to promote the growth of olive seedlings in the presence of the pathogen Verticillium dahliae. Biocontrol Sci. Technol. 2023, 33, 963–984. [Google Scholar] [CrossRef]
- Sabagh, E.L.; Islam, M.S.; Hossain, A.; Iqbal, M.A.; Mubeen, M.; Waleed, M.; Reginato, M.; Battaglia, M.; Ahmed, S.; Rehman, A.; et al. Phytohormones as growth regulators during abiotic stress tolerance in plants. Front. Agron. 2022, 4, 765068. [Google Scholar] [CrossRef]
- Chhaya; Yadav, B.; Jogawat, A.; Gnanasekaran, P.; Kumari, P.; Lakra, N.; Lal, S.K.; Pawar, J.; Narayan, O.P. An overview of recent advancement in phytohormones-mediated stress management and drought tolerance in crop plants. Plant Gene 2021, 25, 100264. [Google Scholar] [CrossRef]
- Duchateau, S.; Crouzet, J.; Dorey, S.; Aziz, A. The plant-associated Pantoea spp. as biocontrol agents: Mechanisms and diversity of bacteria-produced metabolites as a prospective tool for plant protection. Biol. Control 2024, 188, 105441. [Google Scholar] [CrossRef]
- Patakova, P.; Vasylkivska, M.; Sedlar, K.; Jureckova, K.; Bezdicek, M.; Lovecka, P.; Branska, B.; Kastanek, P.; Krofta, K. Whole genome sequencing and characterization of Pantoea agglomerans DBM 3797, endophyte, isolated from fresh hop (Humulus lupulus L.). Front. Microbiol. 2024, 15, 1305338. [Google Scholar] [CrossRef]
- Luziatelli, F.; Gatti, L.; Ficca, A.G.; Medori, G.; Silvestri, C.; Melini, F.; Muleo, R.; Ruzzi, M. Metabolites secreted by a plant-growth-promoting Pantoea agglomerans strain improved rooting of Pyrus communis L. cv Dar Gazi cuttings. Front. Microbiol. 2020, 11, 539359. [Google Scholar] [CrossRef]
- Saia, S.; Aissa, E.; Luziatelli, F.; Ruzzi, M.; Colla, G.; Ficca, A.G.; Cardarelli, M.; Rouphael, Y. Growth-promoting bacteria and arbuscular mycorrhizal fungi differentially benefit tomato and corn depending upon the supplied form of phosphorus. Mycorrhiza 2020, 30, 133–147. [Google Scholar] [CrossRef]
- Melini, F.; Luziatelli, F.; Bonini, P.; Ficca, A.G.; Melini, V.; Ruzzi, M. Optimization of the growth conditions through response surface methodology and metabolomics for maximizing the auxin production by Pantoea agglomerans C1. Front. Microbiol. 2023, 14, 1022248. [Google Scholar] [CrossRef] [PubMed]
- Khatri, I.; Kaur, S.; Devi, U.; Kumar, N.; Sharma, D.; Subramanian, S.; Saini, A.K. Draft genome sequence of plant growth-promoting rhizobacterium Pantoea sp. strain AS-PWVM4. Genome Announc. 2013, 1, e00947-13. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Cardarelli, M.; Melini, F.; Cavalieri, A.; Ruzzi, M. Genome sequencing of Pantoea agglomerans C1 provides insights into molecular and genetic mechanisms of plant growth-promotion and tolerance to heavy metals. Microorganisms 2020, 8, 153. [Google Scholar] [CrossRef]
- Luziatelli, F.; Melini, F.; Bonini, P.; Melini, V.; Cirino, V.; Ruzzi, M. Production of indole auxins by Enterobacter sp. strain P-36 under submerged conditions. Fermentation 2021, 7, 138. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, 1–21. [Google Scholar] [CrossRef]
- Blom, J.; Albaum, S.P.; Doppmeier, D.; Pühler, A.; Vorhölter, F.J.; Zakrzewski, M.; Goesmann, A. EDGAR: A software framework for the comparative analysis of prokaryotic genomes. BMC Bioinform. 2009, 10, 154. [Google Scholar] [CrossRef]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386–2392. [Google Scholar] [CrossRef] [PubMed]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Durkin, A.S.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef]
- Willenbrock, H.; Hallin, P.F.; Wassenaar, T.M.; Ussery, D.W. Characterization of probiotic Escherichia coli isolates with a novel pan-genome microarray. Genome Biol. 2007, 8, R267. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8658. [Google Scholar] [CrossRef]
- Patz, S.; Gautam, A.; Becker, M.; Ruppel, S.; Rodríguez-Palenzuela, P.; Huson, D.H. PLaBAse: A comprehensive web resource for analyzing the plant growth-promoting potential of plant-associated bacteria. biorXiv 2021. biorXiv:12.13.472471. [Google Scholar]
- Barakat, M.; Ortet, P.; Whitworth, D.E. P2RP: A web-based framework for the identification and analysis of regulatory proteins in prokaryotic genomes. BMC Genom. 2013, 14, 269. [Google Scholar] [CrossRef]
- Ortet, P.; De Luca, G.; Whitworth, D.E.; Barakat, M. P2TF: A comprehensive resource for analysis of prokaryotic transcription factors. BMC Genom. 2012, 13, 628. [Google Scholar] [CrossRef]
- Ortet, P.; Whitworth, D.E.; Santaella, C.; Achouak, W.; Barakat, M. P2CS: Updates of the prokaryotic two-component systems database. Nucleic Acids Res. 2015, 43, D585–D589. [Google Scholar] [CrossRef]
- Patten, C.L.; Glick, B.R. Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 1996, 42, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.; Sorrentino, M.; Lucini, L.; Rouphael, Y.; Cardarelli, M.; Bonini, P.; Miras Moreno, M.B.; Reynaud, H.; Canaguier, R.; Trtílek, M.; et al. A combined phenotypic and metabolomic approach for elucidating the biostimulant action of a plant-derived protein hydrolysate on tomato grown under limited water availability. Front. Plant Sci. 2019, 10, 493. [Google Scholar] [CrossRef] [PubMed]
- Bonini, P.; Kind, T.; Tsugawa, H.; Barupal, D.K.; Fiehn, O. Retip: Retention time prediction for compound annotation in untargeted metabolomics. Anal. Chem. 2020, 92, 7515–7522. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Valerio, C.; Medori, G.; Luziatelli, F.; Melini, F.; Gatti, L.; Ruzzi, M.; Muleo, R.; Forgione, I. Bioregulation of adventitious root induction by metabolites secreted from plant growth promoting Pantoea agglomerans strains. Acta Hortic. 2023, 1359, 33–42. [Google Scholar] [CrossRef]
- Ralser, M.; Varma, S.J.; Notebaart, R.A. The evolution of the metabolic network over long timelines. Curr. Opin. Syst. Biol. 2021, 28, 100402. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Švecová, E.; Ruzzi, M. Effects of a protein hydrolysate-based biostimulant and two micronutrient-based fertilizers on plant growth and epiphytic bacterial population of lettuce. Acta Hortic. 2016, 1148, 43–48. [Google Scholar] [CrossRef]
- Gan, H.Y.; Gan, H.M.; Savka, M.A.; Triassi, A.J.; Wheatley, M.S.; Smart, L.B.; Fabio, E.S.; Hudson, A.O. Whole-genome sequences of 13 endophytic bacteria isolated from shrub willow (Salix) grown in Geneva, New York. Genome Announc. 2014, 2, e00288-14. [Google Scholar] [CrossRef]
- Hall, R.B.; Heybroek, H. Biology of Populus and its implications for management and conservation. For. Sci. 1997, 43, 457–458. [Google Scholar] [CrossRef]
- Malette, A.; Xu, R.; Gerdis, S.; Chi, S.I.; Daniels, G.C.; Harding, M.W.; Tambong, J.T. Analysis of draft genome sequences of two new Pantoea strains associated with wheat leaf necrotic tissues caused by Xanthomonas translucens reveals distinct species. Microbiol. Resour. Announc. 2020, 9, e00638-20. [Google Scholar] [CrossRef]
- Snoeijers, S.S.; Pérez-García, A.; Joosten, M.H.; De Wit, P.J.G.M. The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant Pathol. 2000, 106, 493–506. [Google Scholar] [CrossRef]
- Hausladen, A.; Gow, A.J.; Stamler, J.S. Nitrosative stress: Metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. USA 1998, 95, 14100–14105. [Google Scholar] [CrossRef]
- Tucker, N.P.; Le Brun, N.E.; Dixon, R.; Hutchings, M.I. There’s NO stopping NsrR, a global regulator of the bacterial NO stress response. Trends Microbiol. 2010, 18, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A. Modularity and distribution of sulfur metabolism genes in bacterial populations: Search and design. J. Comput. Sci. Syst. Biol. 2010, 3, 91–106. [Google Scholar] [CrossRef]
- Rabus, R.; Venceslau, S.S.; Wöhlbrand, L.; Voordouw, G.; Wall, J.D.; Pereira, I.A. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 2015, 66, 55–321. [Google Scholar] [CrossRef]
- Pal, A.; Bhattacharjee, S.; Saha, J.; Sarkar, M.; Mandal, P. Bacterial survival strategies and responses under heavy metal stress: A comprehensive overview. Crit. Rev. Microbiol. 2021, 48, 327–355. [Google Scholar] [CrossRef]
- Nnaji, N.D.; Anyanwu, C.U.; Miri, T.; Onyeaka, H. Mechanisms of Heavy Metal Tolerance in Bacteria: A Review. Sustainability 2024, 16, 11124. [Google Scholar] [CrossRef]
- Baquero, F. Environmental stress and evolvability in microbial systems. Clin. Microbiol. Infect. 2009, 15 (Suppl. 1), 5–10. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tang, H.; Xiang, G.; Xiao, W.; Yang, Z.; Zhao, B. Microbial mediated remediation of heavy metals toxicity: Mechanisms and future prospects. Front. Plant Sci. 2024, 15, 1420408. [Google Scholar] [CrossRef]
- Nies, D.H. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, S.; Deng, Z.; Dedon, P.C.; Chen, S. DNA phosphorothioate modification—A new multi-functional epigenetic system in bacteria. FEMS Microbiol. Rev. 2019, 43, 109–122. [Google Scholar] [CrossRef]
- Gan, R.; Wu, X.; He, W.; Liu, Z.; Wu, S.; Chen, C.; Chen, S.; Xiang, Q.; Deng, Z.; Liang, D.; et al. DNA phosphorothioate modifications influence the global transcriptional response and protect DNA from double-stranded breaks. Sci. Rep. 2014, 4, 6642. [Google Scholar] [CrossRef]
- Zhu, M.; Dai, X. Shaping of microbial phenotypes by trade-offs. Nat. Comm. 2024, 15, 4238. [Google Scholar] [CrossRef]
- Davidson, A.L.; Dassa, E.; Orelle, C.; Chen, J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 2008, 72, 317–364. [Google Scholar] [CrossRef]
- Monnet, V. Bacterial oligopeptide-binding proteins. Cell. Mol. Life Sci. 2003, 60, 2100–2114. [Google Scholar] [CrossRef]
- Detmers, F.J.; Lanfermeijer, F.C.; Poolman, B. Peptides and ATP binding cassette peptide transporters. Res. Microbiol. 2001, 152, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Olasz, F.; Szabó, M.; Veress, A.; Bibó, M.; Kiss, J. The dynamic network of IS30 transposition pathways. PLoS ONE 2022, 17, e0271414. [Google Scholar] [CrossRef]
- Russell, A.B.; Singh, P.; Brittnacher, M.; Bui, N.K.; Hood, R.D.; Carl, M.A.; Agnello, D.M.; Schwarz, S.; Goodlett, D.R.; Vollmer, W.; et al. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 2012, 11, 538–549. [Google Scholar] [CrossRef]
- Humbert, M.V.; Awanye, A.M.; Lian, L.Y.; Derrick, J.P.; Christodoulides, M. Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor. PLoS Pathog. 2017, 13, e1006448. [Google Scholar] [CrossRef]
- Alcoforado Diniz, J.; Liu, Y.C.; Coulthurst, S.J. Molecular weaponry: Diverse effectors delivered by the Type VI secretion system. Cell Microbiol. 2015, 17, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Cianfanelli, F.R.; Monlezun, L.; Coulthurst, S.J. Aim, load, fire: The type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 2016, 24, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Gavini, F.; Mergaert, J.; Beji, A.; Mielcarek, C.; Izard, D.; Kersters, K.; De Ley, J. Transfer of Enterobacter agglomerans (Beijerinck 1888) Ewing and Fife 1972 to Pantoea gen. nov. as Pantoea agglomerans comb. nov. and description of Pantoea dispersa sp. nov. Int. J. Syst. Evol. Microbiol. 1989, 39, 337–345. [Google Scholar] [CrossRef]
- Kowalska, J.; Tyburski, J.; Matysiak, K.; Tylkowski, B.; Malusà, E. Field exploitation of multiple functions of beneficial microorganisms for plant nutrition and protection: Real possibility or just a hope? Front. Microbiol. 2020, 11, 1904. [Google Scholar] [CrossRef] [PubMed]
- Prasad, J.K.; Dey, R.; Raghuwanshi, R. Exopolysaccharide-producing rhizospheric bacteria enhance yield via promoting wheat (Triticum aestivum L.) growth at early stages. Microbiology 2022, 91, 757–769. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Ali, S.; Babar, M.A. Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 2020, 90, 189–203. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth-promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef]
- Keswani, C.; Singh, S.P.; Cueto, L.; García-Estrada, C.; Mezaache-Aichour, S.; Glare, T.R.; Borriss, R.; Singh, S.P.; Blázquez, M.A.; Sansinenea, E. Auxins of microbial origin and their use in agriculture. Appl. Microbiol. Biotechnol. 2020, 104, 8549–8565. [Google Scholar] [CrossRef]
- Duca, D.R.; Glick, B.R. Indole-3-acetic acid biosynthesis and its regulation in plant-associated bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 8607–8619. [Google Scholar] [CrossRef]
- Levy, A.; Salas Gonzalez, I.; Mittelviefhaus, M.; Clingenpeel, S.; Paredes, S.H.; Miao, J.; Wang, K.; Devescovi, G.; Stillman, K.; Monteiro, F.; et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2018, 50, 138–150. [Google Scholar] [CrossRef]
- Su, F.; Zhao, B.; Dhondt-Cordelier, S.; Vaillant-Gaveau, N. Plant-Growth-Promoting Rhizobacteria modulate carbohydrate metabolism in connection with host plant defense mechanisms. Int. J. Mol. Sci. 2024, 25, 1465. [Google Scholar] [CrossRef]

| Compound_Name | p-Value | Class | Edirection | efs | FDR |
| Indole-3-ethanol (Tryptophol) | 1.50 × 10−7 | indoles | up | 38 | 2.00 × 10−5 |
| D-Fructose | 2.40 × 10−6 | hexoses | up | 5.7 | 3.10 × 10−4 |
| 3-Indoleacetic acid | 3.00 × 10−7 | indoleacetic acids | up | 3 | 4.00 × 10−5 |
| Tyr-Pro | 6.50 × 10−9 | dipeptides | up | 2.8 | 9.40 × 10.7 |
| 4-Methyl-5-thiazoleethanol | 3.40 × 10−5 | azoles | up | 2.6 | 4.30 × 10−3 |
| 2′,3′-Dimethoxy-3-hydroxyflavone | 3.70 × 10−8 | chromones | up | 2.4 | 5.30 × 10−6 |
| Quinoline | 2.00 × 10−9 | quinolines | down | −2 | 2.90 × 10−7 |
| Adenine | 3.10 × 10−10 | purines | down | −2.5 | 4.60 × 10−8 |
| Deisopropylatrazine | 1.60 × 10−10 | triazines | down | −2.8 | 2.40 × 10−8 |
| DL-Pyroglutamic acid (5-Oxoproline) | 9.00 × 10−12 | dipeptides | down | −3.1 | 1.40 × 10−9 |
| Quinoline-2,8-diol | 4.20 × 10−10 | quinolones | down | −3.2 | 6.20 × 10−8 |
| Adenosine | 3.50 × 10−9 | purines | down | −6.3 | 5.10 × 10−7 |
| Hydrocortisone 17-acetate | 4.60 × 10−7 | methylprednisolone | down | −6.6 | 6.40 × 10−6 |
| 6-Methoxytryptamine | 2.60 × 10−7 | indoles | down | −11 | 3.50 × 10−5 |
| Marrubiin | 2.90 × 10−9 | diterpenes | down | −19 | 4.20 × 10−7 |
| 1-Butylimidazole | 6.50 × 10−7 | imidazoles | down | −240 | 9.00 × 10−6 |
| 2-Methyl-4-nitroaniline | 3.10 × 10−3 | others | down | −440 | 3.80 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ficca, A.G.; Luziatelli, F.; Abou Jaoudé, R.; Ruzzi, M. Integrative Genomics and Metabolomics Analyses Provide New Insights into the Molecular Basis of Plant Growth Promotion by Pantoea agglomerans. Microorganisms 2025, 13, 2138. https://doi.org/10.3390/microorganisms13092138
Ficca AG, Luziatelli F, Abou Jaoudé R, Ruzzi M. Integrative Genomics and Metabolomics Analyses Provide New Insights into the Molecular Basis of Plant Growth Promotion by Pantoea agglomerans. Microorganisms. 2025; 13(9):2138. https://doi.org/10.3390/microorganisms13092138
Chicago/Turabian StyleFicca, Anna Grazia, Francesca Luziatelli, Renée Abou Jaoudé, and Maurizio Ruzzi. 2025. "Integrative Genomics and Metabolomics Analyses Provide New Insights into the Molecular Basis of Plant Growth Promotion by Pantoea agglomerans" Microorganisms 13, no. 9: 2138. https://doi.org/10.3390/microorganisms13092138
APA StyleFicca, A. G., Luziatelli, F., Abou Jaoudé, R., & Ruzzi, M. (2025). Integrative Genomics and Metabolomics Analyses Provide New Insights into the Molecular Basis of Plant Growth Promotion by Pantoea agglomerans. Microorganisms, 13(9), 2138. https://doi.org/10.3390/microorganisms13092138







