Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions
Abstract
:1. Introduction
2. Microbial Diversity in Representative Hypersaline Mats

3. Metagenomic Studies in Representative Hypersaline Mats
4. Emerging Issues
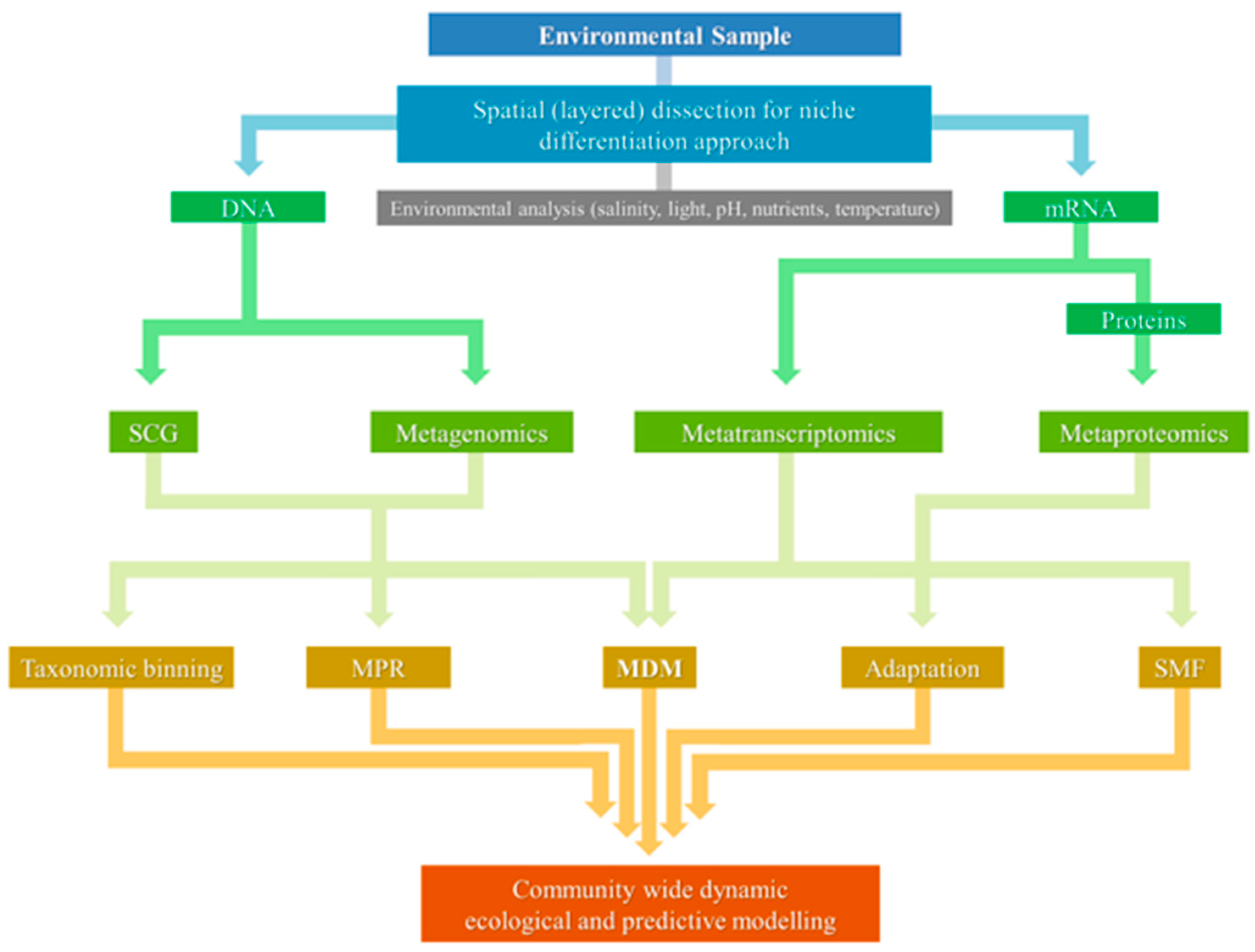
5. Combined Approaches for Molecular Ecology
6. Conclusions and Future Outlook
Author Contributions
Conflicts of Interest
References
- Walter, M.R.; Buick, R.; Dunlop, J.S.R. Stromatolites 3400–3500 Myr old from the North Pole area, Western Australia. Nature 1980, 284, 443–445. [Google Scholar] [CrossRef]
- Allen, M.A.; Goh, F.; Burns, B.P.; Neilan, B.A. Bacterial, archaeal and eukaryotic diversity of smooth and pustular microbial mat communities in the hypersaline lagoon of Shark Bay. Geobiology 2009, 7, 82–96. [Google Scholar] [CrossRef] [PubMed]
- Ruvindy, R.; White, R.A., III; Neilan, B.A.; Burns, B.P. Unravelling core microbial metabolisms in the hypersaline microbial mats of Shark Bay using high-throughput metagenomics. ISME J. 2015, 1, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Spring, S.; Brinkmann, N.; Murrja, M.; Spröer, C.; Reitner, J.; Klenk, H.P. High diversity of culturable prokaryotes in a lithifying hypersaline microbial mat. Geomicrobiol. J. 2015, 32, 332–346. [Google Scholar] [CrossRef]
- Paerl, H.W.; Pinckney, J.L.; Steppe, T.F. Cyanobacterial-bacterial mat consortia: Examining the functional unit of microbial survival and growth in extreme environments. Environ. Microbiol. 2000, 2, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Hoehler, T.M.; Bebout, B.M.; des Marais, D.J. The role of microbial mats in the production of reduced gases on the early Earth. Nature 2001, 412, 324–327. [Google Scholar] [CrossRef] [PubMed]
- DesMarais, D.J. Biogeochemistry of hypersaline microbial mats illustrates the dynamics of modern microbial ecosystems and the early evolution of the biosphere. Biol. Bull. 2003, 204, 160–167. [Google Scholar]
- Arp, G.; Reimer, A.; Reitner, J. Photosynthesis-Induced Biofilm Calcification and Calcium Concentrations in Phanerozoic Oceans. Science 2001, 292, 1701–1704. [Google Scholar] [CrossRef] [PubMed]
- Logan, B.W. Cryptozoon and associate stromatolites from the recent, Shark Bay, Western Australia. J. Geol. 1961, 69, 517–533. [Google Scholar] [CrossRef]
- Dravis, J.J. Hardened subtidal stromatolites, Bahamas. Science 1983, 219, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.T.; Prins, R.A.; van Gemerden, H. Rates of sulfate reduction and thiosulfate consumption in a marine microbial mat. FEMS Microbiol. Ecol. 1992, 86, 283–294. [Google Scholar] [CrossRef]
- Visscher, P.T.; Stolz, J.F. Microbial mats as bioreactors: Populations, processes and products. Palaeogeogr. Palaeocl. 2005, 219, 87–100. [Google Scholar] [CrossRef]
- Bauld, J.; Chambers, L.A.; Skyring, G.W. Primary productivity, sulfate reduction and sulfur isotope fractionation in algal mats and sediments of Hamelin Pool, Shark Bay, WA. Aust. J. Mar. Fresh. Res. 1979, 30, 753–764. [Google Scholar] [CrossRef]
- Moriarty, D.J.W. Bacterial biomass and productivity in sediments, stromatolites, and water of Hamelin Pool, Shark Bay, Western Australia. Geomicrobiol. J. 1983, 3, 121–133. [Google Scholar] [CrossRef]
- Krumbein, W.E. Stromatolites—The challenge of a term in space and time. Precambr. Res. 1983, 20, 493. [Google Scholar] [CrossRef]
- Fründ, C.; Cohen, Y. Diurnal cycles of sulfate reduction under oxic conditions in microbial mats. Appl. Environ. Microbiol. 1992, 58, 70–77. [Google Scholar] [PubMed]
- Revsbech, N.P.; Madsen, B.; Jørgensen, B.B. Oxygen production and consumption in sediments determined at high spatial resolution by computer simulation of oxygen microelectrode data. Limnol. Ocenogr. 1986, 31, 293–304. [Google Scholar] [CrossRef]
- Rinke, C.; Schwientek, P.; Sczyrba, A.; Ivanova, N.N.; Anderson, I.J.; Cheng, J.F.; Darling, A.; Malfatti, S.; Swan, B.K.; Gies, E.A.; et al. Insights into the Phylogeny and Coding Potential of Microbial Dark Matter. Nature 2013. [Google Scholar] [CrossRef] [PubMed]
- Berg, I.A.; Kockelkorn, D.; Buckel, W.; Fuchs, G. A 3-hydroxypropionate/4-hydroxybutyrate autotrophic carbon dioxide assimilation pathway in Archaea. Nature 2007, 318, 1782–1786. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.; Arp, G.; Reimer, A.; Reitner, J.; Daniel, R. Phylogenetic analysis of a microbialite-forming microbial mat from a hypersaline lake of the Kiritimati Atoll, Central Pacific. PLoS ONE 2013, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Yarza, P.; Richter, M.; Suárez-Suárez, A.; Antón, J.; Niemann, H.; Rosselló-Móra, R. Extremely halophilic microbial communities in anaerobic sediments from a solar saltern. Environ. Microb. Rep. 2010, 2, 258–271. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Harris, J.K.; Wilcox, J.; Spear, J.R.; Miller, S.R.; Bebout, B.M.; Maresca, J.A.; Bryant, D.A.; Sogin, M.L.; Pace, N.R. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 2006, 72, 3685–3695. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.K.; Caporaso, J.G.; Walker, J.J.; Spear, J.R.; Gold, N.J.; Robertson, C.E.; Hugenholtz, P.; Goodrich, J.; McDonald, D.; Knights, D.; et al. Phylogenetic Stratigraphy in the Guerrero Negro Hypersaline Microbial Mat. ISME J. 2013, 7, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.K.; Hutchison, J.R.; Renslow, R.S.; Kim, Y.M.; Chrisler, W.B.; Engelmann, H.E.; Dohnalkova, A.C.; Hu, D.; Metz, T.O.; Fredrickson, J.K.; et al. Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: Model systems for the study of autotroph-heterotroph interactions. Front. Microbiol. 2014, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Robertson, C.E.; Spear, J.R.; Harris, J.K.; Pace, N.R. Diversity and stratification of Archaea in a hypersaline microbial mat. Appl. Environ. Microbiol. 2009, 75, 1801–1810. [Google Scholar] [CrossRef] [PubMed]
- Minz, D.; Fishbain, S.; Green, S.J.; Gerard, M.; Cohen, Y.; Rittmann, B.E.; Stahl, D.A. Unexpected population distribution in a microbial mat community: Sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 1999, 65, 4659–4665. [Google Scholar] [PubMed]
- Minz, D.; Flax, J.L.; Green, S.J.; Gerard, M.; Cohen, Y.; Wagner, M.; Rittmann, B.E.; Stahl, D.A. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfate reductase genes. Appl. Environ. Microbiol. 1999, 65, 4666–4671. [Google Scholar] [PubMed]
- Risatti, J.B.; Capman, W.C.; Stahl, D.A. Community structure of a microbial mat: The phylogenetic dimension. Proc. Natl. Acad. Sci. USA 1994, 9117, 10173–10177. [Google Scholar] [CrossRef]
- Canfield, D.E.; des Marais, D.J. Biogeochemical cycles of carbon, sulphur, and free oxygen in a microbial mat. Geochim. Cosmochim. Acta 1993, 57, 3971–3984. [Google Scholar] [CrossRef]
- Visscher, P.T.; Baumgartner, L.K.; Buckley, D.H.; Rogers, D.R.; Hogan, M.E.; Raleigh, C.D.; Turk, K.A.; des Marais, D.J. Dimethyl sulphide and methanethiol formation in microbial mats: Potential pathways for biogenic signatures. Environ. Microbiol. 2003, 5, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Reimer, J.J.; Huerta-Diaz, M.A. Phosphorus speciation and sedimentary fluxes in hypersaline sediments of the Guerrero Negro salt evaporation area, Baja California Sur, Mexico. Estuar. Coasts 2011, 34, 514–528. [Google Scholar] [CrossRef]
- Spear, J.R.; Ley, R.E.; Berger, A.B.; Pace, N.R. Complexity in natural microbial ecosystems: The Guerrero Negro experience. Biol. Bull. 2003, 204, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Des Marais, D.J. The biogeochemistry of hypersaline microbial mats. Adv. Microb. Ecol. 1995, 14, 251–274. [Google Scholar] [PubMed]
- Jahnert, R.J.; Collins, L.B. Characteristics, Distribution and Morphogenesis of Subtidal Microbial Systems in Shark Bay, Australia. Mar. Geol. 2012, 303, 115–136. [Google Scholar] [CrossRef]
- Goh, F.; Allen, M.A.; Leuko, S.; Kawaguchi, T.; Decho, A.W.; Burns, B.P.; Neilan, B.A. Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 2009, 3, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Burns, B.P.; Goh, F.; Allen, M.; Neilan, B.A. Microbial diversity of extant stromatolites in the hypersaline marine environment of Shark Bay, Australia. Environ. Microbiol. 2004, 6, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.A.; Neilan, B.A.; Burns, B.P.; Jahnke, L.L.; Summons, R.E. Lipid biomarkers in Hamelin Pool microbial mats and stromatolites. Org. Geochem. 2010, 41, 1207–1218. [Google Scholar] [CrossRef]
- Goh, F.; Leuko, S.; Allen, M.A.; Bowman, J.P.; Kamekura, M.; Neilan, B.A.; Burns, B.P. Halococcus hamelinensis sp. nov., a novel halophilic archaeon isolated from stromatolites in Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2006, 56, 1323–1329. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.A.; Goh, F.; Leuko, S.; Echigo, A.; Mizuki, T.; Usami, R.; Kamekura, M.; Neilan, B.A.; Burns, B.P. Haloferax elongans sp. nov. and Haloferax mucosum sp. nov., isolated from microbial mats from Hamelin Pool, Shark Bay, Australia. Int. J. Syst. Evol. Microbiol. 2008, 58, 798–802. [Google Scholar] [CrossRef] [PubMed]
- Goh, F.; Jeon, Y.J.; Barrow, K.; Neilan, B.A.; Burns, B.P. Osmoadaptive strategies of the archaeon Halococcus hamelinensis isolated from a hypersaline stromatolite environment. Astrobiology 2011, 11, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Leuko, S.; Neilan, B.A.; Burns, B.P.; Walter, M.R.; Rothschild, L.J. Molecular Assessment of UVC Radiation-Induced DNA Damage Repair in the Stromatolitic halophilic archaeon, Halococcus hamelinensis. J. Photochem. Photobiol. B 2011, 102, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Leuko, S.; Goh, F.; Ibáñez-Peral, R.; Burns, B.P.; Walter, M.R.; Neilan, B.A. Lysis efficiency of standard DNA extraction methods for Halococcus sp. in an organic rich environment. Extremophiles 2008, 12, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Gudhka, R.K.; Neilan, B.A.; Burns, B.P. Adaptaion, ecology, and evolution of the halophilic stromatolite archaeon Halococcus hamelinensis inferred through genome analyses. Archaea 2015. [Google Scholar] [CrossRef] [PubMed]
- Kjeldsen, K.U.; Loy, A.; Jakobsen, T.F.; Thomsen, T.R.; Wagner, M.; Ingvorsen, K. Diversity of sulfate-reducing bacteria from an extreme hypersaline sediment, Great Salt Lake (Utah). FEMS Microbiol. Ecol. 2007, 60, 287–298. [Google Scholar] [CrossRef] [PubMed]
- López-López, A.; Richter, M.; Peña, A.; Tamames, J.; Rosselló-Móra, R. New insights into the archaeal diversity of a hypersaline microbial mat obtained by a metagenomics approach. Syst. Appl. Microbiol. 2013, 36, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Oremland, R.S.; Polcin, S. Methanogenesis and sulfate reduction: Competitive and non-competitive substrates in estuarine sediments. Appl. Environ. Microbiol. 1982, 44, 1270–1276. [Google Scholar] [PubMed]
- King, G.M.; Klug, M.J.; Lovley, D.R. Metabolism of acetate, methanol, and methylated amines in intertidal sediments of Lowes Cove, Maine. Appl. Environ. Microbiol. 1983, 45, 1848–1853. [Google Scholar] [PubMed]
- King, G.M. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine-sediments. Appl. Environ. Microbiol. 1984, 48, 719–725. [Google Scholar] [PubMed]
- Kiene, R.P.; Oremland, R.S.; Catena, A.; Miller, L.G.; Capone, D.G. Metabolism of reduced methylated sulphur-compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl. Environ. Microbiol. 1986, 52, 1037–1045. [Google Scholar] [PubMed]
- Buckley, D.H.; Baumgartner, L.K.; Visscher, P.T. Vertical distribution of methane metabolism in microbial mats of the Great Sippewissett Salt Marsh. Environ. Microbiol. 2008, 10, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Arp, G.; Helms, G.; Karlinska, K.; Schumann, G.; Reimer, A.; Reitner, J.; Trichet, J. Formation of microbialtes on the Atoll of Kiritimati, Republic of Kiribati, Central Pacific. Geomicrobiol. J. 2012, 29, 29–65. [Google Scholar] [CrossRef]
- Dröge, J.; McHardy, A.C. Taxonomic binning of metagenome samples generated by next-generation sequencing technologies. Brief. Bioinform. 2012, 13, 646–655. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Kjaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomics contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Saeed, I.; Tang, S.L.; Halgamuge, S.K. Unsupervised discovery of microbial population structure within metagenomes using nucleotide base composition. Nucleic Acid Res. 2012, 40, e34. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V.; Raes, J.; Harris, J.K.; Spear, J.R.; Walker, J.J.; Ivanova, N.; von Mering, C.; Bebout, B.M.; Pace, N.R.; Bork, P.; et al. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol. Syst. Biol. 2008, 4, 198–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soppa, J. From genomes to function: Haloarchaea as model organisms. Microbiology 2006, 152, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Bardavid, R.E.; Oren, A. Acid-shifted isoelectric point profiles of the proteins in a hypersaline microbial mat: An adaptation to life at high salt concentrations? Extremophiles 2012, 16, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Goh, F.; Barrow, K.D.; Neilan, B.A.; Burns, B.P. Identification and regulation of novel compatible solutes from hypersaline stromatolite-associated cyanobacteria. Arch. Microbiol. 2010, 192, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Pages, A.; Welsch, D.T.; Teasdale, P.R.; Grice, K.; Vacher, M.; Bennett, W.W.; Visscher, P.T. Diel fluctuations in solute distributions and biogeochemical cycling in a hypersaline microbial mat from Shark Bay, WA. Mar. Chem. 2014, 167, 102–112. [Google Scholar] [CrossRef]
- Jeffries, T.C.; Seymour, J.R.; Newton, K.; Smith, R.J.; Seuront, L.; Mitchell, J.G. Increases in the abundance of microbial genes encoding halotolerance and photosynthesis along a sediment salinity gradient. Biogeosciences 2012, 9, 815–825. [Google Scholar] [CrossRef] [Green Version]
- Hedlund, B.P.; Dodsworth, J.A.; Murugapiran, S.K.; Rinke, C.; Woyke, T. Impact of single-cell genomics and metagenomics on the emerging view of extremophile “microbial dark matter”. Extremophiles 2014. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.J.; Wrighton, K.C.; Thomas, B.C.; Hug, L.A.; Brown, C.T.; Wilkins, M.J.; Frischkorn, K.R.; Tringe, S.G.; Singh, A.; Markillie, L.M.; et al. Genomic expansion of domain Archaea highlights roles for organsims from new phyla in anaerobic carbon cycling. Curr. Biol. 2015, 25, 690–701. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Dick, G.J. Omic Approaches in Microbial Ecology: Charting the Unknown. Microbe 2013, 3, 353–360. [Google Scholar] [CrossRef]
- Dodsworth, J.A.; Blainey, P.C.; Murugapiran, S.K.; Swingley, W.D.; Ross, C.A.; Tringe, S.G.; Chain, P.S.G.; Scholz, M.B.; Lo, C.C.; Raymond, J.; et al. Single-cell and metagenomics analyses indicate a fermentative, saccharolytic lifestyle for members of the OP9 lineage. Nat. Commun. 2013, 4, 1854. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C. The future is now: Single-cell genomics of bacteria and archaea. Microbiol. Rev. 2013, 37, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Scholz, M.B.; Lo, C.C.; Chain, P.S.G. Next generation sequencing and bioinformatics bottlenecks: The current state of metagenomics data analysis. Curr. Opin. Biotechnol. 2012, 23, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Youssef, N.H.; Rinke, C.; Stepanauskas, R.; Farag, I.; Woyke, T.; Elshahed, M.S. Insights into the metabolism, lifestyle and putative evolutionary history of the novel archaeal phylum “Diapherotrites”. ISME J. 2015, 9, 447–460. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Xu, J.; Qin, D.; He, Y.; Xiao, X.; Wang, F. Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J. 2014, 8, 650–659. [Google Scholar] [CrossRef] [PubMed]
- Guy, L.; Ettema, T.J. The archaeal “TACK” superphylum and the origin of eukaryotes. Trends Micriobiol. 2011, 19, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.; Cahoon, N.; Trakhtenberg, E.M.; Pham, L.; Mehta, A.; Belanger, A.; Kanigan, T.; Lewis, K.; Epstein, S.S. Use of ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol. 2010, 76, 2445–2450. [Google Scholar] [CrossRef] [PubMed]
- Grünke, S.; Felden, J.; Lichtschlag, A.; Girnth, A.C.; de Beer, D.; Wenzhöfer, F.; Boetius, A. Niche differentiation among mat-forming, sulfide-oxidizing bacteria at cold seeps of the Nile Deep Sea Fan (Eastern Mediterranean Sea). Geobiology 2011, 9, 330–348. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Cruaud, P.; Roussel, E.G.; Pignet, P.; Caprais, J.C.; Callac, N.; Ciobanu, M.C.; Godfroy, A.; Cragg, B.A.; Parkes, J.R.; et al. Phylogenetic and functional diversity of microbial communities associated with subsurface sediments of the Sonora Margin, Guaymas Basin. PLoS ONE 2014, 9, e104427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Q.; Dutta, D.; Schwalbach, M.S.; Steele, J.A.; Fuhrman, J.A.; Sun, F. Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 2006, 22, 2532–2538. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A.; Steele, J.A. Community structure of marine bacterioplankton: Patterns, networks, and relationships to function. Aquat. Microb. Ecol. 2008, 59, 69–81. [Google Scholar] [CrossRef]
- Chaffron, S.; Rehrauer, H.; Pernthaler, J.; von Mering, C. A global network of coexisting microbes from environmental and whole—Genome sequence data. Genome Res. 2010, 20, 947–959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrei, A.Ş.; Robeson, M.S., II; Baricz, A.; Coman, C.; Muntean, V.; Ionescu, A.; Etiope, G.; Alexe, M.; Sicora, C.I.; Podar, M.; et al. Contrasting taxonomic stratification of microbial communities in two hypersaline meromictic lakes. ISME J. 2015, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ludemann, H.; Arth, I.; Liesack, W. Spatial changes in the bacterial community structure along a vertical oxygen gradient in flooded paddy soil cores. Appl. Environ. Microbiol. 2000, 66, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Stomp, M.; Huisman, J.; Stal, L.; Matthijs, H.C.P. Colorful niches of phototrophic microorganisms shaped by vibrations of the water molecule. ISME J. 2007, 1, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Long, R.A.; Eveillard, D.; Franco, S.L.M.; Reeves, E.; Pinckney, L.L. Antagonistic interactions between heterotrophic bacteria as a potential regulator of community structure of hypersaline microbial mats. FEMS Microbiol. Ecol. 2013, 83, 74–81. [Google Scholar] [CrossRef] [PubMed]
- De Jager, V.; Siezen, R.J. Single-cell genomics: Unravelling the genomes of unculturable microorganisms. Microb. Biotechnol. 2011, 4, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Stepanauskas, R. Single cell genomics: An individual look at microbes. Curr. Opin. Microbiol. 2012, 15, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Dean, F.B.; Hosono, S.; Fang, L.H.; Wu, X.H.; Faruqi, A.F.; Bray-Ward, P.; Sun, Z.Y.; Zong, Q.L.; Du, Y.F.; Du, J.; et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. USA 2002, 99, 5261–5266. [Google Scholar] [CrossRef] [PubMed]
- Walker, A. Adding genomic “foliage” to the tree of life. Nat. Rev. Microbiol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Kolinko, S.; Jogler, C.; Katzmann, E.; Wanner, G.; Peplies, J.; Schüler, D. Single-cell analysis reveals a novel uncultivated magnetotactic bacterium within the candidate division OP3. Environ. Microbiol. 2012, 14, 1709–1721. [Google Scholar] [CrossRef] [PubMed]
- Narasingarao, P.; Podell, S.; Ugalde, J.A.; Brochier-Armanet, C.; Emerson, J.B.; Brocks, J.J.; Heidelberg, K.B.; Banfield, J.F.; Allen, E.E. De novo metagenomics assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J. 2012, 6, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Ghai, R.; Pašic, L.; Fernández, A.B.; Martin-Cuadrado, A.B.; Mizuno, C.M.; McMahon, K.D.; Papke, R.T.; Stepanauskas, R.; Rodriguez-Brito, B.; Rohwer, F.; et al. New abundant microbial groups in aquatic hypersaline environments. Sci. Rep. 2011, 1, 135. [Google Scholar] [CrossRef] [PubMed]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Podar, M.; Abulencia, C.B.; Walcher, M.; Hutchison, D.; Zengler, K.; Garcia, J.A.; Holland, T.; Cotton, D.; Hauser, L.; Keller, M. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microb. 2007, 73, 3205–3214. [Google Scholar] [CrossRef] [PubMed]
- Dupraz, C.; Reid, R.P.; Braissant, O.; Decho, A.W.; Norman, R.S.; Visscher, P.T. Processes of carbonate precipitation in modern microbial mats. Earth Sci. Rev. 2008, 96, 141–162. [Google Scholar] [CrossRef]
- Sorek, R.; Cossart, P. Prokaryotic transcriptomics: A new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2010, 11, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Bhaya, B.; Grossman, A.R.; Steunou, A.S.; Khuri, N.; Cohan, F.M.; Hamamura, N.; Melendrez, M.C.; Bateson, M.M.; Ward, D.M.; Heidelberg, J.F. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomics analyses. ISME J. 2007, 1, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tyson, G.W.; Eppley, J.M.; DeLong, E.F. Integrated metatranscriptomic and metagenomics analyses of stratified microbial assemblages in the open ocean. ISME J. 2011, 5, 999–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.X.; Hu, M.; Huang, L.N.; Hua, Z.S.; Kuang, J.L.; Li, S.J.; Shu, W.S. Comparative metagenomics and metatranscriptomic analyses of microbial communities in acid mine drainage. ISME J. 2015, 9, 1579–1592. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Klatt, C.G.; Wood, J.M.; Rusch, D.B.; Ludwig, M.; Wittekindt, N.; Tomsho, L.P.; Schuster, S.C.; Ward, D.M.; Bryant, D.A. Metatranscriptomic analyses of chlorophototrophs of a hot-spring microbial mat. ISME J. 2011, 5, 1279–1290. [Google Scholar] [CrossRef] [PubMed]
- Mobberley, J.M.; Khodadad, C.L.M.; Visccher, P.T.; Reid, R.P.; Hagan, P.; Foster, J.S. Inner workings of thrombolites: Spatial gradients of metabolic activity as revealed by metatranscriptome profiling. Sci. Rep. 2015, 5, 12601. [Google Scholar] [CrossRef] [PubMed]
- Schaffert, C.S.; Ward, D.M.; Klatt, C.G.; Pauley, M.; Steinke, L.A. Identification and distribution of high-abundance proteins in the Octopus Spring microbial mat community. Appl. Environ. Microbiol. 2012, 78, 8481–8484. [Google Scholar] [CrossRef] [PubMed]
- Jungblut, A.D.J.; Allen, M.A.; Burns, B.P.; Neilan, B.A. Lipid biomarker analysis of cyanobacterial dominated microbial mats in melt water ponds on the McMurdo Ice Shelf, Antarctica. Org. Geochem. 2009, 40, 258–269. [Google Scholar] [CrossRef]
- Rubakhin, S.S.; Romanova, E.V.; Nemes, P.; Sweedler, J.V. Profiling metabolites and peptides in single cells. Nat. Methods 2011, 8, 20–29. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wong, H.L.; Ahmed-Cox, A.; Burns, B.P. Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions. Microorganisms 2016, 4, 6. https://doi.org/10.3390/microorganisms4010006
Wong HL, Ahmed-Cox A, Burns BP. Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions. Microorganisms. 2016; 4(1):6. https://doi.org/10.3390/microorganisms4010006
Chicago/Turabian StyleWong, Hon Lun, Aria Ahmed-Cox, and Brendan Paul Burns. 2016. "Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions" Microorganisms 4, no. 1: 6. https://doi.org/10.3390/microorganisms4010006
APA StyleWong, H. L., Ahmed-Cox, A., & Burns, B. P. (2016). Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions. Microorganisms, 4(1), 6. https://doi.org/10.3390/microorganisms4010006





