Investigating Potential Chromosomal Rearrangements during Laboratory Culture of Neisseria gonorrhoeae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth Conditions
2.2. Pulsed Field Gel Electrophoresis
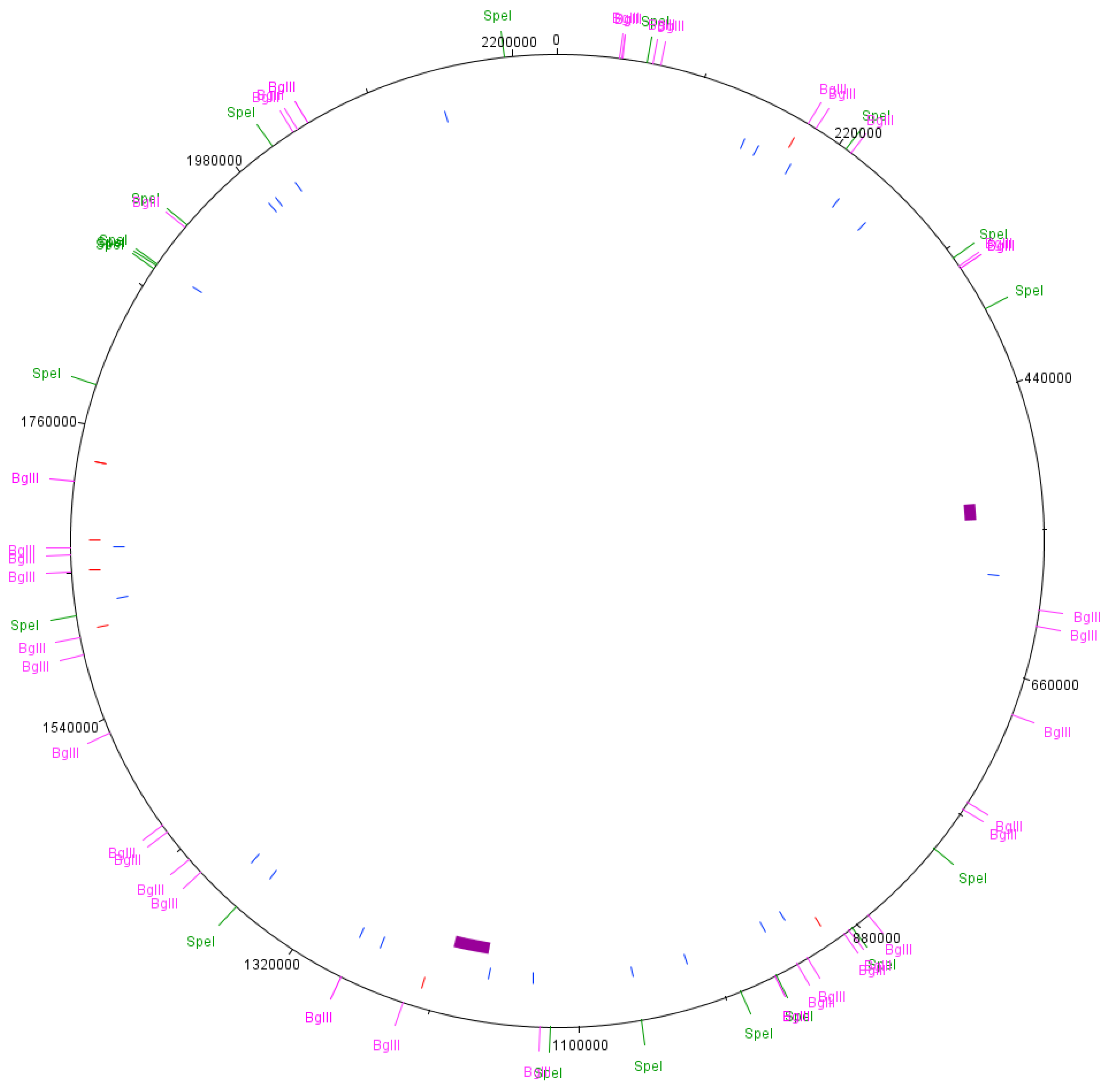
2.3. Predicted PFGE Band Patterns
2.4. Next Generation Genome Sequencing
3. Results
3.1. Growth of Bacteria in the Laboratory under Different Conditions
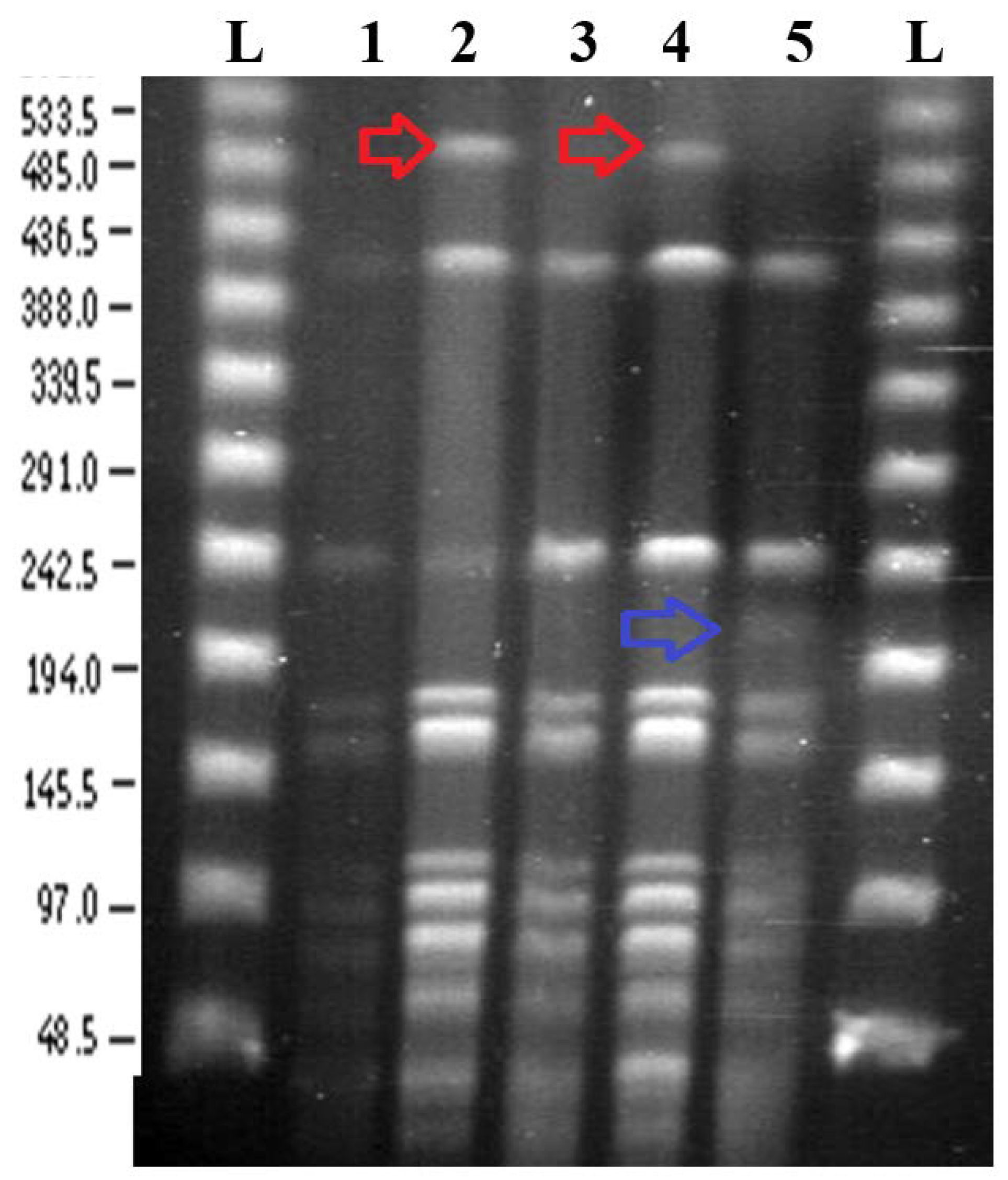
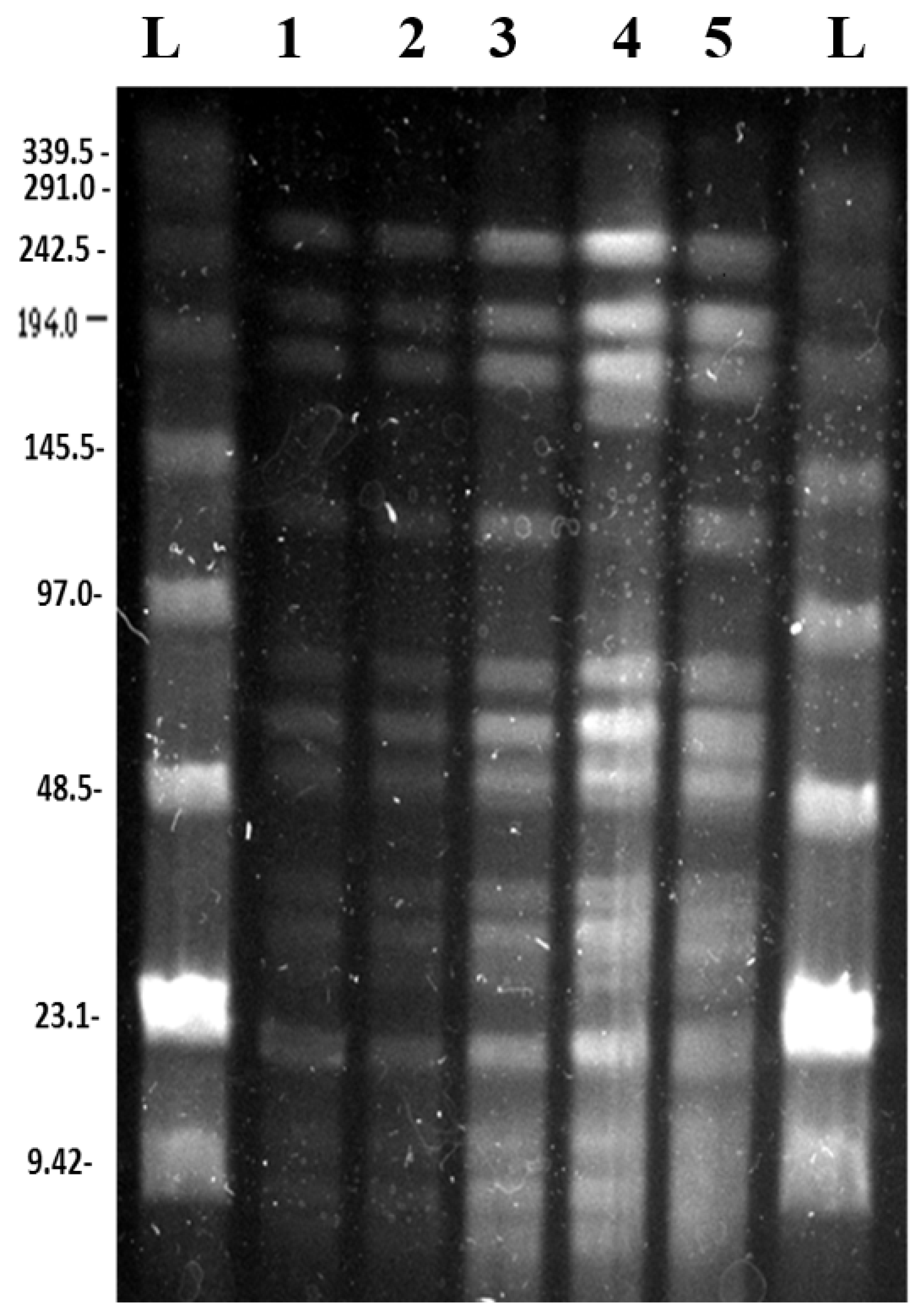
3.2. Pulsed Field Gel Electrophoresis
3.2.1. SpeI Digestion and PFGE
3.2.2. BglII Digestion and PFGE
3.3. Predicted PFGE Band Patterns Based on Genome Sequence Data
3.4. Comparison of PFGE Results with Genome Sequence Data from the Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chung, G.T.; Yoo, J.S.; Oh, H.B.; Lee, Y.S.; Cha, S.H.; Kim, S.J.; Yoo, C.K. Complete genome sequence of Neisseria gonorrhoeae NCCP11945. J. Bacteriol. 2008, 190, 6035–6036. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L.A.; Cole, J.A.; Pallen, M.J. Comparative analysis of two Neisseria gonorrhoeae genome sequences reveals evidence of mobilization of Correia Repeat Enclosed Elements and their role in regulation. BMC Genomics 2009, 10, 70. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.S.; Bentley, S.D.; Vernikos, G.S.; Quail, M.A.; Cherevach, I.; White, B.; Parkhill, J.; Maiden, M.C. Independent evolution of the core and accessory gene sets in the genus Neisseria: Insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics 2010, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, M.; Watanabe, Y.; Ono, E.; Takahashi, C.; Oya, H.; Kuroki, T.; Shimuta, K.; Okazaki, N.; Nakayama, S.; Watanabe, H. Spread of a chromosomal cefixime-resistant penA gene among different Neisseria gonorrhoeae lineages. Antimicrob. Agents Chemother. 2010, 54, 1060–1067. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.T.; Seifert, H.S. Opportunity and means: Horizontal gene transfer from the human host to a bacterial pathogen. MBio 2011, 2, e00005-11. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Hsia, K.C.; Huang, C.T.; Wong, W.W.; Yen, M.Y.; Li, L.H.; Lin, K.Y.; Chen, K.W.; Li, S.Y. Draft genome sequence of a dominant, multidrug-resistant Neisseria gonorrhoeae strain, TCDC-NG08107, from a sexual group at high risk of acquiring human immunodeficiency virus infection and syphilis. J. Bacteriol. 2011, 193, 1788–1789. [Google Scholar] [CrossRef] [PubMed]
- De Curraize, C.; Kumanski, S.; Micaëlo, M.; Fournet, N.; La Ruche, G.; Meunier, F.; Amarsy, R.; Jacquier, H.; Cambau, E.; Goubard, A.; et al. Ceftriaxone-resistant Neisseria gonorrhoeae isolates (2010 to 2014) in France characterized by using whole-genome sequencing. Antimicrob. Agents Chemother. 2016, 60, 6962–6964. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Ryan, C.S.; Davies, J.K. Neisserial Correia repeat-enclosed elements do not influence the transcription of pil genes in Neisseria gonorrhoeae and Neisseria meningitidis. J. Bacteriol. 2011, 193, 5728–5736. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.S.; Jolley, K.A.; Earle, S.G.; Corton, C.; Bentley, S.D.; Parkhill, J.; Maiden, M.C. A genomic approach to bacterial taxonomy: An examination and proposed reclassification of species within the genus Neisseria. Microbiology 2012, 158, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Spencer-Smith, R.; Varkey, E.M.; Fielder, M.D.; Snyder, L.A. Sequence features contributing to chromosomal rearrangements in Neisseria gonorrhoeae. PLoS ONE 2012, 7, e46023. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.B.; Xiong, L.; Teng, J.L.; Yuen, K.Y.; Lau, S.K.; Woo, P.C. Re-annotation of protein-coding genes in 10 complete genomes of Neisseriaceae family by combining similarity-based and composition-based methods. DNA Res. 2013, 20, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Putonti, C.; Nowicki, B.; Shaffer, M.; Fofanov, Y.; Nowicki, S. Where does Neisseria acquire foreign DNA from: An examination of the source of genomic and pathogenic islands and the evolution of the Neisseria genus. BMC Evol. Biol. 2013, 13, 184. [Google Scholar] [CrossRef] [PubMed]
- Jamet, A.; Jousset, A.B.; Euphrasie, D.; Mukorako, P.; Boucharlat, A.; Ducousso, A.; Charbit, A.; Nassif, X. A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog. 2015, 11, e1004592. [Google Scholar] [CrossRef] [PubMed]
- McClure, R.; Nudel, K.; Massari, P.; Tjaden, B.; Su, X.; Rice, P.A.; Genco, C.A. The Gonococcal Transcriptome during Infection of the Lower Genital Tract in Women. PLoS ONE 2015, 10, e0133982. [Google Scholar] [CrossRef] [PubMed]
- Spencer-Smith, R.; Roberts, S.; Gurung, N.; Snyder, L.A.S. DNA uptake sequences in Neisseria gonorrhoeae as intrinsic transcriptional terminators and markers of horizontal gene transfer. Microb. Genomics 2016, 2, e000069. [Google Scholar] [CrossRef] [PubMed]
- Zelewska, M.A.; Pulijala, M.; Spencer-Smith, R.; Mahmood, H.A.; Norman, B.; Churchward, C.P.; Calder, A.; Snyder, L.A. Phase variable DNA repeats in Neisseria gonorrhoeae influence transcription, translation, and protein sequence variation. Microb. Genomics 2016, 2, e000078. [Google Scholar] [CrossRef] [PubMed]
- Dempsey, J.A.; Wallace, A.B.; Cannon, J.G. The physical map of the chromosome of a serogroup A strain of Neisseria meningitidis shows complex rearrangements relative to the chromosomes of the two mapped strains of the closely related species N. gonorrhoeae. J. Bacteriol. 1995, 177, 6390–6400. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, C.P.; Meyer, T.F. Genome plasticity in Neisseria gonorrhoeae. FEMS Microbiol. Lett. 1996, 145, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Uchiyama, I.; Kobayashi, I. Genome comparison in silico in Neisseria suggests integration of filamentous bacteriophages by their own transposase. DNA Res. 2005, 12, 389–401. [Google Scholar] [CrossRef] [PubMed]
- Piekarowicz, A.; Klyz, A.; Majchrzak, M.; Adamczyk-Poplawska, M.; Maugel, T.K.; Stein, D.C. Characterization of the dsDNA prophage sequences in the genome of Neisseria gonorrhoeae and visualization of productive bacteriophage. BMC Microbiol. 2007, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Skaar, E.P.; LeCuyer, B.; Lenich, A.G.; Lazio, M.P.; Perkins-Balding, D.; Seifert, H.S.; Karls, A.C. Analysis of the Piv recombinase-related gene family of Neisseria gonorrhoeae. J. Bacteriol. 2005, 187, 1276–1286. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Nakao, K.; Uchiyama, I.; Kobayashi, I. How genomes rearrange: Genome comparison within bacteria Neisseria suggests roles for mobile elements in formation of complex genome polymorphisms. Gene 2006, 383, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Hebeler, B.H.; Young, F.E. Autolysis of Neisseria gonorrhoeae. J. Bacteriol. 1976, 122, 385–392. [Google Scholar]
- Cehovin, A.; Lewis, S.B. Mobile genetic elements in Neisseria gonorrhoeae: Movement for change. Pathog. Dis. 2017, 75, ftx071. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, D.S.; Peacock, W.L.; Deacon, W.E.; Brown, L.; Pirkle, C.I. Neisseria gonorrhoeae I: Virulence genetically linked to clonal variation. J. Bacteriol. 1963, 85, 1274–1279. [Google Scholar] [PubMed]
- Cowlishaw, J.; Ginoza, W. Induction of lambda prophage by nalidixic acid. Virology 1970, 41, 244–255. [Google Scholar] [CrossRef]
- Vihanová, D.; Vondrejs, V. Induction of bacteriophage lambda with oxolinic and nalidixic acid. Folia Microbiol. (Praha) 1974, 19, 390–393. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.-D.; Harwig, S.S.L.; Oren, A.; Shafer, W.M.; Lehrer, R.I. Susceptibility of Neisseria gonorrhoeae to protegrins. Infect. Immun. 1996, 64, 1240–1245. [Google Scholar] [PubMed]
- Stothard, P. The Sequence Manipulation Suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 2000, 28, 1102–1104. [Google Scholar] [PubMed]
- Bihlmaier, A.; Römlin, U.; Meyer, T.F.; Tümmler, B.; Gibbs, C.P. Physical and genetic map of the Neisseria gonorrhoeae strain MS11-N198 chromosome. Mol. Microbiol. 1991, 5, 2529–2539. [Google Scholar] [CrossRef]
- Dempsey, J.A.; Litaker, W.; Madhure, A.; Snodgrass, T.L.; Cannon, J.G. Physical map of the chromosome of Neisseria gonorrhoeae FA1090 with locations of genetic markers, including opa and pil genes. J. Bacteriol. 1991, 173, 5476–5486. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.; Thomson, N.; Bleasby, A.; Berriman, M.; Parkhill, J. DNAPlotter: Circular and linear interactive genome visualization. Bioinformatics 2009, 25, 119–120. [Google Scholar] [CrossRef] [PubMed]
- Barnett, D.W.; Garrison, E.K.; Quinlan, A.R.; Strömberg, M.P.; Marth, G.T. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 2011, 27, 1691–1692. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Galaxy Team. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.A.; Hirsh, H.L.; Zeller, W.W.; Dowling, H.F. Gonococcal arthritis; a study of 202 patients treated with penicillin, sulfonamides or fever therapy. Ann. Intern. Med. 1949, 30, 1212–1223. [Google Scholar] [PubMed]
- Dillard, J.P.; Seifert, H.S. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 2001, 41, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L.A.; Jarvis, S.A.; Saunders, N.J. Complete and variant forms of the ‘gonococcal genetic island’ in Neisseria meningitidis. Microbiology 2005, 151, 4005–4013. [Google Scholar] [CrossRef] [PubMed]
- Elbeyioglu, F.; Roberts, S.B.; Spencer-Smith, R.; Pulijala, M.; Zelewska, M.A.; Nebel, J.C.; Snyder, L.A. Inversion of Correia Repeat Enclosed Elements in Neisseria gonorrhoeae. Microbiology 2016, 163, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Zöllner, R.; Oldewurtel, E.R.; Kouzel, N.; Maier, B. Phase and antigenic variation govern competition dynamics through positioning in bacterial colonies. Sci. Rep. 2017, 7, 12151. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, J.; Dittrich, M.; Blom, J.; Mitesser, V.; Vogel, U.; Frosch, M.; Goesmann, A.; Müller, T.; Schoen, C. Comparative genome sequencing reveals within-host genetic changes in Neisseria meningitidis during invasive disease. PLoS ONE 2017, 12, e0169892. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.G.; Fulcher, N.B.; Jerse, A.E. Opacity proteins increase Neisseria gonorrhoeae fitness in the female genital tract due to a factor under ovarian control. Infect. Immun. 2010, 78, 1629–1641. [Google Scholar] [CrossRef] [PubMed]
- Snyder, L.A.; Butcher, S.A.; Saunders, N.J. Comparative whole-genome analyses reveal over 100 putative phase-variable genes in the pathogenic Neisseria spp. Microbiology 2001, 147, 2321–2332. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spencer-Smith, R.; Gould, S.W.; Pulijala, M.; Snyder, L.A.S. Investigating Potential Chromosomal Rearrangements during Laboratory Culture of Neisseria gonorrhoeae. Microorganisms 2018, 6, 10. https://doi.org/10.3390/microorganisms6010010
Spencer-Smith R, Gould SW, Pulijala M, Snyder LAS. Investigating Potential Chromosomal Rearrangements during Laboratory Culture of Neisseria gonorrhoeae. Microorganisms. 2018; 6(1):10. https://doi.org/10.3390/microorganisms6010010
Chicago/Turabian StyleSpencer-Smith, Russell, Simon W. Gould, Madhuri Pulijala, and Lori A. S. Snyder. 2018. "Investigating Potential Chromosomal Rearrangements during Laboratory Culture of Neisseria gonorrhoeae" Microorganisms 6, no. 1: 10. https://doi.org/10.3390/microorganisms6010010
APA StyleSpencer-Smith, R., Gould, S. W., Pulijala, M., & Snyder, L. A. S. (2018). Investigating Potential Chromosomal Rearrangements during Laboratory Culture of Neisseria gonorrhoeae. Microorganisms, 6(1), 10. https://doi.org/10.3390/microorganisms6010010





