Cellulases from Thermophiles Found by Metagenomics
Abstract
1. Introduction
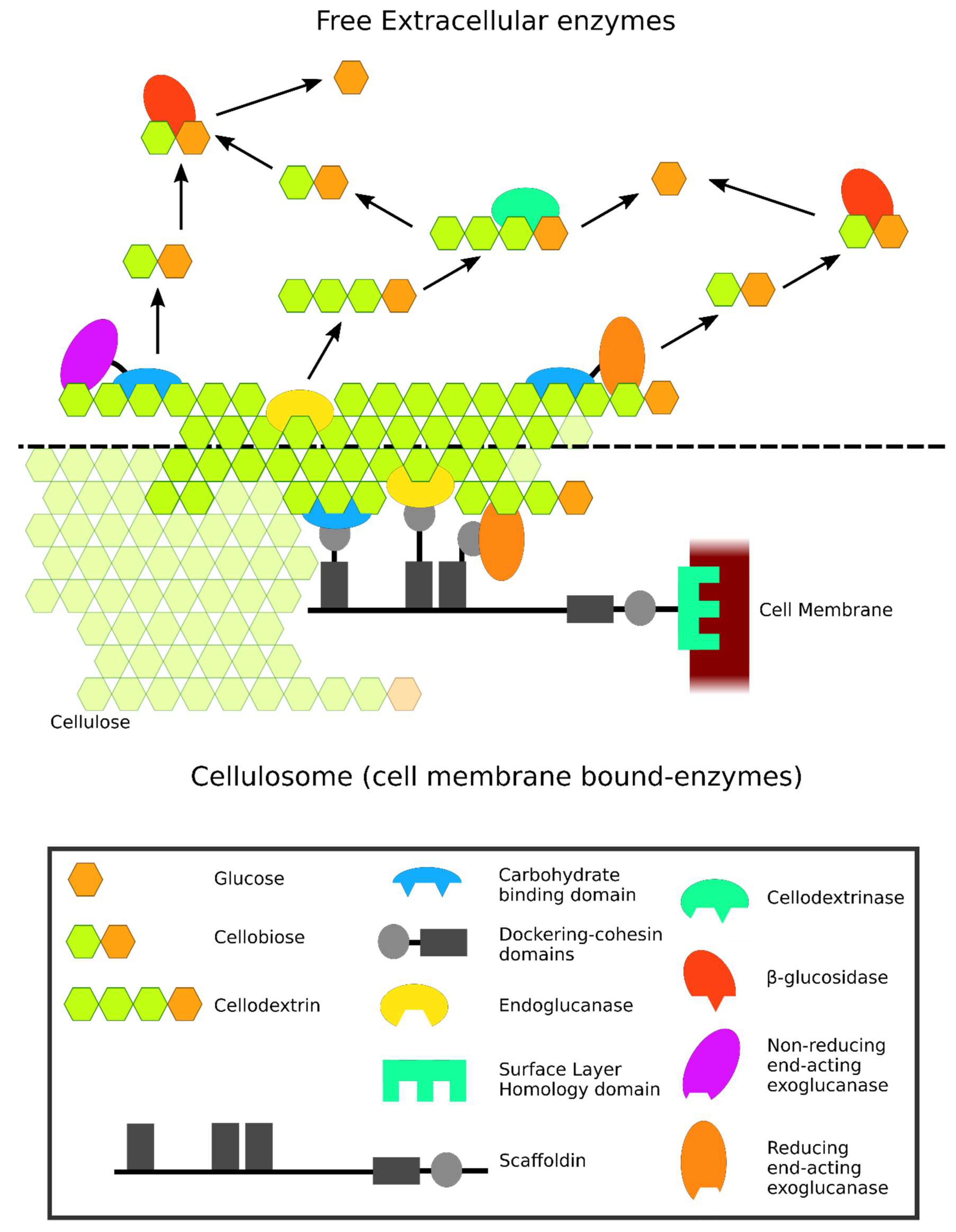
2. Modular Structure of Cellulases and their Classification
3. Factors Influencing Thermostability of Thermophile Cellulases
4. Biotechnological Applications by Thermophile Cellulases
4.1. Endoglucanase-Specific Industrial Applications
4.2. Exoglucanase-Specific Industrial Applications
4.3. β-glucosidase-Specific Industrial Applications
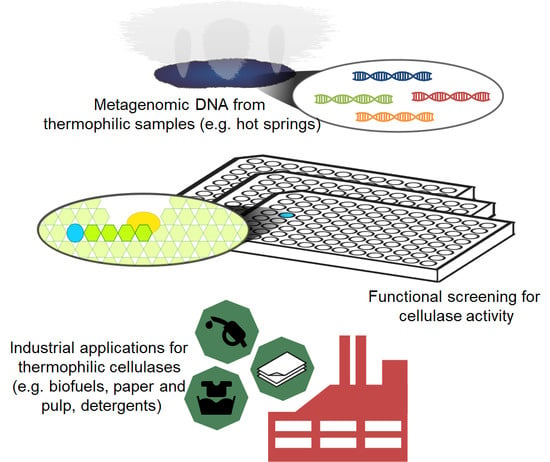
5. Metagenomics for the Search of Novel Cellulases
6. Thermophile Cellulases Characterized
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sandgren, M.; Ståhlberg, J.; Mitchinson, C. Structural and biochemical studies of GH family 12 cellulases: Improved thermal stability, and ligand complexes. Prog. Biophys. Mol. Biol. 2005, 89, 246–291. [Google Scholar] [CrossRef] [PubMed]
- Blumer-Schuette, S.E.; Kataeva, I.; Westpheling, J.; Adams, M.W.; Kelly, R.M. Extremely thermophilic microorganisms for biomass conversion: Status and prospects. Curr. Opin. Biotechnol. 2008, 19, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.-J.; Feng, J.-X. Mining metagenomes for novel cellulase genes. Biotechnol. Lett. 2010, 32, 1765–1775. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Tewari, R.; Rana, S.S.; Soni, R.; Soni, S.K. Cellulases: Classification, Methods of Determination and Industrial Applications. Appl. Biochem. Biotechnol. 2016, 179, 1346–1380. [Google Scholar] [CrossRef] [PubMed]
- Couturier, M.; Feliu, J.; Haon, M.; Navarro, D.; Lesage-Meessen, L.; Coutinho, P.M.; Berrin, J.-G. A thermostable GH45 endoglucanase from yeast: Impact of its atypical multimodularity on activity. Microb. Cell Fact. 2011, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- López-Mondéjar, R.; Zühlke, D.; Becher, D.; Riedel, K.; Baldrian, P. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci. Rep. 2016, 6, 25279. [Google Scholar] [CrossRef] [PubMed]
- Kaur, B.; Chadha, B.S. Approaches for Bioprospecting Cellulases. In Extremophilic Enzymatic Processing of Lignocellulosic Feedstocks to Bioenergy; Sani, R.K., Krishnaraj, R.N., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 53–71. ISBN 978-3-319-54683-4. [Google Scholar]
- Bok, J.D.; Yernool, D.A.; Eveleigh, D.E. Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitana. Appl. Environ. Microbiol. 1998, 64, 4774–4781. [Google Scholar] [PubMed]
- Elleuche, S.; Schäfers, C.; Blank, S.; Schröder, C.; Antranikian, G. Exploration of extremophiles for high temperature biotechnological processes. Curr. Opin. Microbiol. 2015, 25, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, R.; Nain, L.; Labrou, N.E.; Shukla, P. Bioprospecting of functional cellulases from metagenome for second generation biofuel production: A review. Crit. Rev. Microbiol. 2018, 44, 244–257. [Google Scholar] [CrossRef] [PubMed]
- DeCastro, M.-E.; Rodríguez-Belmonte, E.; González-Siso, M.-I. Metagenomics of Thermophiles with a Focus on Discovery of Novel Thermozymes. Front. Microbiol. 2016, 7, 1521. [Google Scholar] [CrossRef] [PubMed]
- Wagner, I.D.; Wiegel, J. Diversity of thermophilic anaerobes. Ann. N. Y. Acad. Sci. 2008, 1125, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J.; Liu, Q.; Zhang, C.; Ma, Q. Molecular cloning of novel cellulase genes cel9A and cel12A from Bacillus licheniformis GXN151 and synergism of their encoded polypeptides. Curr. Microbiol. 2004, 49, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Crennell, S.J.; Hreggvidsson, G.O.; Nordberg Karlsson, E. The structure of Rhodothermus marinus Cel12A, a highly thermostable family 12 endoglucanase, at 1.8 A resolution. J. Mol. Biol. 2002, 320, 883–897. [Google Scholar] [CrossRef]
- Chhabra, S.R.; Kelly, R.M. Biochemical characterization of Thermotoga maritima endoglucanase Cel74 with and without a carbohydrate binding module (CBM). FEBS Lett. 2002, 531, 375–380. [Google Scholar] [CrossRef]
- Peer, A.; Smith, S.P.; Bayer, E.A.; Lamed, R.; Borovok, I. Noncellulosomal cohesin- and dockerin-like modules in the three domains of life. FEMS Microbiol. Lett. 2009, 291, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bayer, E.A.; Belaich, J.-P.; Shoham, Y.; Lamed, R. The cellulosomes: Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 2004, 58, 521–554. [Google Scholar] [CrossRef] [PubMed]
- Sathya, T.A.; Khan, M. Diversity of glycosyl hydrolase enzymes from metagenome and their application in food industry. J. Food Sci. 2014, 79, R2149–R2156. [Google Scholar] [CrossRef] [PubMed]
- Poidevin, L.; Feliu, J.; Doan, A.; Berrin, J.-G.; Bey, M.; Coutinho, P.M.; Henrissat, B.; Record, E.; Heiss-Blanquet, S. Insights into exo- and endoglucanase activities of family 6 glycoside hydrolases from Podospora anserina. Appl. Environ. Microbiol. 2013, 79, 4220–4229. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Lee, M.J.; Cho, H.-Y.; Lee, J.S.; Lee, M.-H.; Chung, C.W.; Shin, D.-H.; Rhee, Y.H.; Son, K.-H.; Park, H.-Y. Genetic and functional characterization of an extracellular modular GH6 endo-β-1,4-glucanase from an earthworm symbiont, Cellulosimicrobium funkei HY-13. Antonie Van Leeuwenhoek 2016, 109, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Halldórsdóttir, S.; Thórólfsdóttir, E.T.; Spilliaert, R.; Johansson, M.; Thorbjarnardóttir, S.H.; Palsdottir, A.; Hreggvidsson, G.O.; Kristjánsson, J.K.; Holst, O.; Eggertsson, G. Cloning, sequencing and overexpression of a Rhodothermus marinus gene encoding a thermostable cellulase of glycosyl hydrolase family 12. Appl. Microbiol. Biotechnol. 1998, 49, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Ausili, A.; Cobucci-Ponzano, B.; Di Lauro, B.; D’Avino, R.; Perugino, G.; Bertoli, E.; Scirè, A.; Rossi, M.; Tanfani, F.; Moracci, M. A comparative infrared spectroscopic study of glycoside hydrolases from extremophilic archaea revealed different molecular mechanisms of adaptation to high temperatures. Proteins 2007, 67, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, C.F.; Sanderson, I.; Moracci, M.; Ciaramella, M.; Nucci, R.; Rossi, M.; Pearl, L.H. Crystal structure of the β-glycosidase from the hyperthermophilic archeon Sulfolobus solfataricus: Resilience as a key factor in thermostability. J. Mol. Biol. 1997, 271, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Wu, I.; Arnold, F.H. Engineered thermostable fungal Cel6A and Cel7A cellobiohydrolases hydrolyze cellulose efficiently at elevated temperatures. Biotechnol. Bioeng. 2013, 110, 1874–1883. [Google Scholar] [CrossRef] [PubMed]
- Rawat, R.; Kumar, S.; Chadha, B.S.; Kumar, D.; Oberoi, H.S. An acidothermophilic functionally active novel GH12 family endoglucanase from Aspergillus niger HO: Purification, characterization and molecular interaction studies. Antonie Van Leeuwenhoek 2015, 107, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Voutilainen, S.P.; Puranen, T.; Siika-Aho, M.; Lappalainen, A.; Alapuranen, M.; Kallio, J.; Hooman, S.; Viikari, L.; Vehmaanperä, J.; Koivula, A. Cloning, expression, and characterization of novel thermostable family 7 cellobiohydrolases. Biotechnol. Bioeng. 2008, 101, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Voutilainen, S.P.; Murray, P.G.; Tuohy, M.G.; Koivula, A. Expression of Talaromyces emersonii cellobiohydrolase Cel7A in Saccharomyces cerevisiae and rational mutagenesis to improve its thermostability and activity. Protein Eng. Des. Sel. 2010, 23, 69–79. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; Li, H.; Li, A.-N.; Li, D.-C. Cloning of a gene encoding thermostable cellobiohydrolase from the thermophilic fungus Chaetomium thermophilum and its expression in Pichia pastoris. J. Appl. Microbiol. 2009, 106, 1867–1875. [Google Scholar] [CrossRef] [PubMed]
- Karnaouri, A.C.; Topakas, E.; Christakopoulos, P. Cloning, expression, and characterization of a thermostable GH7 endoglucanase from Myceliophthora thermophila capable of high-consistency enzymatic liquefaction. Appl. Microbiol. Biotechnol. 2014, 98, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Hua, C.; Yang, S.; Li, Y.; Jiang, Z. High level expression of extracellular secretion of a β-glucosidase gene (PtBglu3) from Paecilomyces thermophila in Pichia pastoris. Protein Expr. Purif. 2012, 84, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Dun, B.; Shi, P.; Ma, R.; Luo, H.; Bai, Y.; Xie, X.; Yao, B. A Novel GH7 Endo-β-1,4-Glucanase from Neosartorya fischeri P1 with Good Thermostability, Broad Substrate Specificity and Potential Application in the Brewing Industry. PLoS ONE 2015, 10, e0137485. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, M.J.; D’haeseleer, P.; Hazen, T.C.; Simmons, B.A.; Adams, P.D.; Hadi, M.Z. Glycoside hydrolases from a targeted compost metagenome, activity-screening and functional characterization. BMC Biotechnol. 2012, 12, 38. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Gao, G.; Li, Y.; Yang, L.; Liang, Y.; Jin, H.; Han, W.; Feng, Y.; Zhang, Z. Cloning, Expression, and Characterization of a Thermophilic Endoglucanase, AcCel12B from Acidothermus cellulolyticus 11B. Int. J. Mol. Sci. 2015, 16, 25080–25095. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bao, L.; Chang, L.; Liu, Z.; You, C.; Lu, H. Beta-xylosidase activity of a GH3 glucosidase/xylosidase from yak rumen metagenome promotes the enzymatic degradation of hemicellulosic xylans. Lett. Appl. Microbiol. 2012, 54, 79–87. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, R.P.; Park, J.I.; Heins, R.A.; Reindl, W.; Friedland, G.D.; D’haeseleer, P.; Northen, T.; Sale, K.L.; Simmons, B.A.; Adams, P.D. From soil to structure, a novel dimeric β-glucosidase belonging to glycoside hydrolase family 3 isolated from compost using metagenomic analysis. J. Biol. Chem. 2013, 288, 14985–14992. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-M.; Lee, Y.-S.; Seo, S.-H.; Yoon, S.-H.; Kim, S.-J.; Hahn, B.-S.; Sim, J.-S.; Koo, B.-S. Screening and Characterization of a Novel Cellulase Gene from the Gut Microflora of Hermetia illucens Using Metagenomic Library. J. Microbiol. Biotechnol. 2014, 24, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Girfoglio, M.; Rossi, M.; Cannio, R. Cellulose degradation by Sulfolobus solfataricus requires a cell-anchored endo-β-1-4-glucanase. J. Bacteriol. 2012, 194, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Biver, S.; Stroobants, A.; Portetelle, D.; Vandenbol, M. Two promising alkaline β-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose. J. Ind. Microbiol. Biotechnol. 2014, 41, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Konishi, T.; Takeda, T. Biochemical characterization of Magnaporthe oryzae β-glucosidases for efficient β-glucan hydrolysis. Appl. Microbiol. Biotechnol. 2011, 91, 1073–1082. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Bansal, N.; Kumar, S.; Bischoff, K.M.; Sani, R.K. Improved lignocellulose conversion to biofuels with thermophilic bacteria and thermostable enzymes. Bioresour. Technol. 2013, 128, 751–759. [Google Scholar] [CrossRef] [PubMed]
- Harnpicharnchai, P.; Champreda, V.; Sornlake, W.; Eurwilaichitr, L. A thermotolerant beta-glucosidase isolated from an endophytic fungi, Periconia sp., with a possible use for biomass conversion to sugars. Protein Expr. Purif. 2009, 67, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Fusco, F.A.; Fiorentino, G.; Pedone, E.; Contursi, P.; Bartolucci, S.; Limauro, D. Biochemical characterization of a novel thermostable β-glucosidase from Dictyoglomus turgidum. Int. J. Biol. Macromol. 2018, 113, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Boyce, A.; Walsh, G. Characterisation of a novel thermostable endoglucanase from Alicyclobacillus vulcanalis of potential application in bioethanol production. Appl. Microbiol. Biotechnol. 2015, 99, 7515–7525. [Google Scholar] [CrossRef] [PubMed]
- Ando, S.; Ishida, H.; Kosugi, Y.; Ishikawa, K. Hyperthermostable Endoglucanase from Pyrococcus horikoshii. Appl. Environ. Microbiol. 2002, 68, 430–433. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Wang, J.; Zhang, Z.; Shi, P.; Luo, H.; Huang, H.; Feng, Y.; Yao, B. Extremely acidic beta-1,4-glucanase, CelA4, from thermoacidophilic Alicyclobacillus sp. A4 with high protease resistance and potential as a pig feed additive. J. Agric. Food Chem. 2010, 58, 1970–1975. [Google Scholar] [CrossRef] [PubMed]
- Vuong, T.V.; Wilson, D.B. Processivity, synergism, and substrate specificity of Thermobifida fusca Cel6B. Appl. Environ. Microbiol. 2009, 75, 6655–6661. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Liu, S.; Jing, H.; Sun, R.; Liu, M.; Chen, H.; Wu, Q.; Han, X. Cloning and expression of A. oryzae β-glucosidase in Pichia pastoris. Mol. Biol. Rep. 2014, 41, 7567–7573. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Yan, Q.; Yang, Y.; Yang, S.; Liu, Y.; Jiang, Z. Expression and characterization of a novel β-glucosidase, with transglycosylation and exo-β-1,3-glucanase activities, from Rhizomucor miehei. Food Chem. 2015, 175, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Villanueva, A.; Ramón, D.; Vallés, S.; Lluch, M.A.; MacCabe, A.P. Heterologous Expression in Aspergillus nidulans of a Trichoderma longibrachiatum Endoglucanase of Enological Relevance. J. Agric. Food Chem. 2000, 48, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Lee, C.-M.; Kim, M.-Y.; Yeo, Y.-S.; Yoon, S.-H.; Kang, H.-C.; Koo, B.-S. Screening and characterization of an enzyme with beta-glucosidase activity from environmental DNA. J. Microbiol. Biotechnol. 2007, 17, 905–912. [Google Scholar] [PubMed]
- Lee, G.-W.; Yoo, M.-H.; Shin, K.-C.; Kim, K.-R.; Kim, Y.-S.; Lee, K.-W.; Oh, D.-K. β-glucosidase from Penicillium aculeatum hydrolyzes exo-, 3-O-, and 6-O-β-glucosides but not 20-O-β-glucoside and other glycosides of ginsenosides. Appl. Microbiol. Biotechnol. 2013, 97, 6315–6324. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-S.; Yeom, S.-J.; Oh, D.-K. Characterization of a GH3 family β-glucosidase from Dictyoglomus turgidum and its application to the hydrolysis of isoflavone glycosides in spent coffee grounds. J. Agric. Food Chem. 2011, 59, 11812–11818. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Ryan, B.; Henehan, G.T.M. β-Glucosidase from Streptomyces griseus: Nanoparticle immobilisation and application to alkyl glucoside synthesis. Protein Expr. Purif. 2017, 132, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Placido, A.; Hai, T.; Ferrer, M.; Chernikova, T.N.; Distaso, M.; Armstrong, D.; Yakunin, A.F.; Toshchakov, S.V.; Yakimov, M.M.; Kublanov, I.V.; et al. Diversity of hydrolases from hydrothermal vent sediments of the Levante Bay, Vulcano Island (Aeolian archipelago) identified by activity-based metagenomics and biochemical characterization of new esterases and an arabinopyranosidase. Appl. Microbiol. Biotechnol. 2015, 99, 10031–10046. [Google Scholar] [CrossRef] [PubMed]
- Leis, B.; Heinze, S.; Angelov, A.; Pham, V.T.T.; Thürmer, A.; Jebbar, M.; Golyshin, P.N.; Streit, W.R.; Daniel, R.; Liebl, W. Functional Screening of Hydrolytic Activities Reveals an Extremely Thermostable Cellulase from a Deep-Sea Archaeon. Front. Bioeng. Biotechnol. 2015, 3, 95. [Google Scholar] [CrossRef] [PubMed]
- Graham, J.E.; Clark, M.E.; Nadler, D.C.; Huffer, S.; Chokhawala, H.A.; Rowland, S.E.; Blanch, H.W.; Clark, D.S.; Robb, F.T. Identification and characterization of a multidomain hyperthermophilic cellulase from an archaeal enrichment. Nat. Commun. 2011, 2, 375. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Elleuche, S.; Blank, S.; Antranikian, G. Characterization of a heat-active archaeal β-glucosidase from a hydrothermal spring metagenome. Enzyme Microb. Technol. 2014, 57, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Yasir, M.; Khan, H.; Azam, S.S.; Telke, A.; Kim, S.W.; Chung, Y.R. Cloning and functional characterization of endo-β-1,4-glucanase gene from metagenomic library of vermicompost. J. Microbiol. 2013, 51, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kwon, E.J.; Jeong, Y.S.; Kim, Y.H.; Kim, S.K.; Na, H.B.; Kim, J.; Yun, H.D.; Kim, H. Construction of a Metagenomic Library from Compost and Screening of Cellulase- and Xylanase-positive Clones. J. Korean Soc. Appl. Biol. Chem. 2010, 53, 702–708. [Google Scholar] [CrossRef]
- Okano, H.; Ozaki, M.; Kanaya, E.; Kim, J.-J.; Angkawidjaja, C.; Koga, Y.; Kanaya, S. Structure and stability of metagenome-derived glycoside hydrolase family 12 cellulase (LC-CelA) a homolog of Cel12A from Rhodothermus marinus. FEBS Open Bio 2014, 4, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Geng, A.; Zhang, J.; Wei, Y.; Zhang, L.; Qian, C.; Wang, Q.; Wang, S.; Zhou, Z. Discovery of (hemi-)cellulase genes in a metagenomic library from a biogas digester using 454 pyrosequencing. Appl. Microbiol. Biotechnol. 2013, 97, 8173–8182. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, T.M.; Paiva, J.H.; Ruiz, D.M.; Cairo, J.P.L.F.; Pereira, I.O.; Paixão, D.A.A.; de Almeida, R.F.; Tonoli, C.C.C.; Ruller, R.; Santos, C.R.; et al. Structure and function of a novel cellulase 5 from sugarcane soil metagenome. PLoS ONE 2013, 8, e83635. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, W.-D.; Zhao, X.-L.; Shen, W.-J.; Cao, H.; Cui, Z.-L. Cloning and functional characterization of a novel endo-β-1,4-glucanase gene from a soil-derived metagenomic library. Appl. Microbiol. Biotechnol. 2011, 89, 1083–1092. [Google Scholar] [CrossRef] [PubMed]
- Pottkämper, J.; Barthen, P.; Ilmberger, N.; Schwaneberg, U.; Schenk, A.; Schulte, M.; Ignatiev, N.; Streit, W.R. Applying metagenomics for the identification of bacterial cellulases that are stable in ionic liquids. Green Chem. 2009, 11, 957. [Google Scholar] [CrossRef]
- Feng, Y.; Duan, C.-J.; Pang, H.; Mo, X.-C.; Wu, C.-F.; Yu, Y.; Hu, Y.-L.; Wei, J.; Tang, J.-L.; Feng, J.-X. Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl. Microbiol. Biotechnol. 2007, 75, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, M.V.; Fernández-Arrojo, L.; Gil-Martínez, J.; Montesinos, A.; Chernikova, T.N.; Nechitaylo, T.Y.; Waliszek, A.; Tortajada, M.; Rojas, A.; Huws, S.A.; et al. Microbial β-glucosidases from cow rumen metagenome enhance the saccharification of lignocellulose in combination with commercial cellulase cocktail. Biotechnol. Biofuels 2012, 5, 73. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Escudero, M.; Del Pozo, M.V.; Marín-Navarro, J.; González, B.; Golyshin, P.N.; Polaina, J.; Ferrer, M.; Sanz-Aparicio, J. Structural and Functional Characterization of a Ruminal β-Glycosidase Defines a Novel Subfamily of Glycoside Hydrolase Family 3 with Permuted Domain Topology. J. Biol. Chem. 2016, 291, 24200–24214. [Google Scholar] [CrossRef] [PubMed]
- Beloqui, A.; Nechitaylo, T.Y.; López-Cortés, N.; Ghazi, A.; Guazzaroni, M.-E.; Polaina, J.; Strittmatter, A.W.; Reva, O.; Waliczek, A.; Yakimov, M.M.; et al. Diversity of glycosyl hydrolases from cellulose-depleting communities enriched from casts of two earthworm species. Appl. Environ. Microbiol. 2010, 76, 5934–5946. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Qian, C.; Zhang, X.-Z.; Liu, N.; Liu, N.; Yan, X.; Zhou, Z. Characterization of a novel thermostable β-glucosidase from a metagenomic library of termite gut. Enzyme Microb. Technol. 2012, 51, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Liu, N.; Qian, C.; Wang, Q.; Wang, Q.; Long, Y.; Huang, Y.; Zhou, Z.; Yan, X. Phylogenetic and functional analysis of gut microbiota of a fungus-growing higher termite: Bacteroidetes from higher termites are a rich source of β-glucosidase genes. Microb. Ecol. 2014, 68, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Himmel, M.E.; Adney, W.S.; Tucker, M.P.; Grohmann, K. Thermostable Purified Endoglucanas from Acidothermus cellulolyticus ATCC 43068. U.S. Patent 5,275,944, 4 January 1994. [Google Scholar]
- Nurachman, Z.; Kurniasih, S.D.; Puspitawati, F.; Hadi, S.; Radjasa, O.K.; Natalia, D. Cloning of the Endoglucanase Gene from a Bacillus amyloliquefaciens PSM 3.1 in Escherichia coli Revealed Catalytic Triad Residues Thr-His-Glu. Am. J. Biochem. Biotechnol. 2010, 6, 268–274. [Google Scholar] [CrossRef]
- Bischoff, K.M.; Rooney, A.P.; Li, X.-L.; Liu, S.; Hughes, S.R. Purification and characterization of a family 5 endoglucanase from a moderately thermophilic strain of Bacillus licheniformis. Biotechnol. Lett. 2006, 28, 1761–1765. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.-J.; Lee, Y.-S.; Park, I.-H.; Chandra, M.S.; Kim, K.-K.; Choi, Y.-L. Molecular cloning, purification and characterization of thermostable beta-1,3-1,4 glucanase from Bacillus subtilis A8-8. Indian J. Biochem. Biophys. 2010, 47, 203–210. [Google Scholar] [PubMed]
- Chhabra, S.R.; Shockley, K.R.; Ward, D.E.; Kelly, R.M. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl. Environ. Microbiol. 2002, 68, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Bauer, S.; Vasu, P.; Persson, S.; Mort, A.J.; Somerville, C.R. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. USA 2006, 103, 11417–11422. [Google Scholar] [CrossRef] [PubMed]
- Calza, R.E.; Irwin, D.C.; Wilson, D.B. Purification and characterization of two β-1,4-endoglucanases from Thermomonospora fusca. Biochemistry 1985, 24, 7797–7804. [Google Scholar] [CrossRef]
- Yin, Y.-R.; Zhang, F.; Hu, Q.-W.; Xian, W.-D.; Hozzein, W.N.; Zhou, E.-M.; Ming, H.; Nie, G.-X.; Li, W.-J. Heterologous expression and characterization of a novel halotolerant, thermostable, and alkali-stable GH6 endoglucanase from Thermobifida halotolerans. Biotechnol. Lett. 2015, 37, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Cazemier, A.E.; Verdoes, J.C.; Op den Camp, H.J.M.; Hackstein, J.H.P.; van Ooyen, A.J. A beta-1,4-endoglucanase-encoding gene from Cellulomonas pachnodae. Appl. Microbiol. Biotechnol. 1999, 52, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, J.; Zhang, W.; Huang, H.; Shi, P.; Luo, H.; Liu, B.; Zhang, Y.; Zhang, Z.; Fan, Y.; et al. A Neutral Thermostable β-1,4-Glucanase from Humicola insolens Y1 with Potential for Applications in Various Industries. PLoS ONE 2015, 10, e0124925. [Google Scholar] [CrossRef] [PubMed]
- Li, X.L.; Chen, H.; Ljungdahl, L.G. Two cellulases, CelA and CelC, from the polycentric anaerobic fungus Orpinomyces strain PC-2 contain N-terminal docking domains for a cellulase-hemicellulase complex. Appl. Environ. Microbiol. 1997, 63, 4721–4728. [Google Scholar] [PubMed]
- Takashima, S.; Nakamura, A.; Hidaka, M.; Masaki, H.; Uozumi, T. Cloning, sequencing, and expression of the cellulase genes of Humicola grisea var. thermoidea. J. Biotechnol. 1996, 50, 137–147. [Google Scholar] [CrossRef]
- Wei, X.-M.; Qin, Y.; Qu, Y. Molecular Cloning and Characterization of Two Major Endoglucanases from Penicillium decumbens. J. Microbiol. Biotechnol. 2010, 20, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Miettinen-Oinonen, A.; Londesborough, J.; Joutsjoki, V.; Lantto, R.; Vehmaanperä, J. Three cellulases from Melanocarpus albomyces for textile treatment at neutral pH. Enzyme Microb. Technol. 2004, 34, 332–341. [Google Scholar] [CrossRef]
- Yoo, J.-S.; Jung, Y.-J.; Chung, S.-Y.; Lee, Y.-C.; Choi, Y.-L. Molecular cloning and characterization of CMCase gene (celC) from Salmonella typhimurium UR. J. Microbiol. 2004, 42, 205–210. [Google Scholar] [PubMed]
- Kim, J.O.; Park, S.R.; Lim, W.J.; Ryu, S.K.; Kim, M.K.; An, C.L.; Cho, S.J.; Park, Y.W.; Kim, J.H.; Yun, H.D. Cloning and characterization of thermostable endoglucanase (Cel8Y) from the hyperthermophilic Aquifex aeolicus VF5. Biochem. Biophys. Res. Commun. 2000, 279, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Hakamada, Y.; Endo, K.; Takizawa, S.; Kobayashi, T.; Shirai, T.; Yamane, T.; Ito, S. Enzymatic properties, crystallization, and deduced amino acid sequence of an alkaline endoglucanase from Bacillus circulans. Biochim. Biophys. Acta 2002, 1570, 174–180. [Google Scholar] [CrossRef]
- Ul Haq, I.; Akram, F.; Khan, M.A.; Hussain, Z.; Nawaz, A.; Iqbal, K.; Shah, A.J. CenC, a multidomain thermostable GH9 processive endoglucanase from Clostridium thermocellum: Cloning, characterization and saccharification studies. World J. Microbiol. Biotechnol. 2015, 31, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Zverlov, V.; Mahr, S.; Riedel, K.; Bronnenmeier, K. Properties and gene structure of a bifunctional cellulolytic enzyme (CelA) from the extreme thermophile “Anaerocellum thermophilum” with separate glycosyl hydrolase family 9 and 48 catalytic domains. Microbiology 1998, 144, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Sathitsuksanoh, N.; Zhang, Y.-H.P. Glycoside hydrolase family 9 processive endoglucanase from Clostridium phytofermentans: Heterologous expression, characterization, and synergy with family 48 cellobiohydrolase. Bioresour. Technol. 2010, 101, 5534–5538. [Google Scholar] [CrossRef] [PubMed]
- Liebl, W.; Ruile, P.; Bronnenmeier, K.; Riedel, K.; Lottspeich, F.; Greif, I. Analysis of a Thermotoga maritima DNA fragment encoding two similar thermostable cellulases, CelA and CelB, and characterization of the recombinant enzymes. Microbiology 1996, 142, 2533–2542. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.W.; Driskill, L.E.; Callen, W.; Snead, M.A.; Mathur, E.J.; Kelly, R.M. An Endoglucanase, EglA, from the Hyperthermophilic Archaeon Pyrococcus Furiosus Hydrolyzes β-1,4 Bonds in Mixed-Linkage (1→3),(1→4)-β-D-Glucans and Cellulose. J. Bacteriol. 1999, 181, 284–290. [Google Scholar] [PubMed]
- Huang, Y.; Krauss, G.; Cottaz, S.; Driguez, H.; Lipps, G. A highly acid-stable and thermostable endo-beta-glucanase from the thermoacidophilic archaeon Sulfolobus solfataricus. Biochem. J. 2005, 385, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Irdani, T.; Perito, B.; Mastromei, G. Characterization of a Streptomyces rochei endoglucanase. Ann. N. Y. Acad. Sci. 1996, 782, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, J.; Siika-aho, M.; Tenkanen, M.; Tjerneld, F. Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 2002, 99, 63–78. [Google Scholar] [CrossRef]
- Warner, C.D.; Hoy, J.A.; Shilling, T.C.; Linnen, M.J.; Ginder, N.D.; Ford, C.F.; Honzatko, R.B.; Reilly, P.J. Tertiary structure and characterization of a glycoside hydrolase family 44 endoglucanase from Clostridium acetobutylicum. Appl. Environ. Microbiol. 2010, 76, 338–346. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.K.; Diderichsen, B.; Jørgensen, P.L. celA from Bacillus lautus PL236 encodes a novel cellulose-binding endo-beta-1,4-glucanase. J. Bacteriol. 1992, 174, 3522–3531. [Google Scholar] [CrossRef] [PubMed]
- Ahsan, M.M.; Matsumoto, M.; Karita, S.; Kimura, T.; Sakka, K.; Ohmiya, K. Purification and Characterization of the Family J Catalytic Domain Derived from the Clostridium thermocellum Endoglucanase CelJ. Biosci. Biotechnol. Biochem. 1997, 61, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Koga, J.; Baba, Y.; Shimonaka, A.; Nishimura, T.; Hanamura, S.; Kono, T. Purification and characterization of a new family 45 endoglucanase, STCE1, from Staphylotrichum coccosporum and its overproduction in Humicola insolens. Appl. Environ. Microbiol. 2008, 74, 4210–4217. [Google Scholar] [CrossRef] [PubMed]
- Wonganu, B.; Pootanakit, K.; Boonyapakron, K.; Champreda, V.; Tanapongpipat, S.; Eurwilaichitr, L. Cloning, expression and characterization of a thermotolerant endoglucanase from Syncephalastrum racemosum (BCC18080) in Pichia pastoris. Protein Expr. Purif. 2008, 58, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Baba, Y.; Shimonaka, A.; Koga, J.; Murashima, K.; Kubota, H.; Kono, T. Purification and characterization of a new endo-1,4-beta-D-glucanase from Beltraniella portoricensis. Biosci. Biotechnol. Biochem. 2005, 69, 1198–1201. [Google Scholar] [CrossRef] [PubMed]
- Eckert, K.; Schneider, E. A thermoacidophilic endoglucanase (CelB) from Alicyclobacillus acidocaldarius displays high sequence similarity to arabinofuranosidases belonging to family 51 of glycoside hydrolases. Eur. J. Biochem. 2003, 270, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- Brás, J.L.A.; Cartmell, A.; Carvalho, A.L.M.; Verzé, G.; Bayer, E.A.; Vazana, Y.; Correia, M.A.S.; Prates, J.A.M.; Ratnaparkhe, S.; Boraston, A.B.; et al. Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis. Proc. Natl. Acad. Sci. USA 2011, 108, 5237–5242. [Google Scholar] [CrossRef] [PubMed]
- Park, C.-S.; Kawaguchi, T.; Sumitani, J.-I.; Takada, G.; Izumori, K.; Arai, M. Cloning and sequencing of an exoglucanase gene from Streptomyces sp. M 23, and its expression in Streptomyces lividans TK-24. J. Biosci. Bioeng. 2005, 99, 434–436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Lao, G.; Wilson, D.B. Characterization of a Thermomonospora fusca exocellulase. Biochemistry 1995, 34, 3386–3395. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-A.; Li, D.-C.; Er, S.-J.; Zhang, Y. Cloning and expressing of cellulase gene (cbh2) from thermophilic fungi Chaetomium thermophilum CT2. Sheng Wu Gong Cheng Xue Bao 2005, 21, 892–899. [Google Scholar] [PubMed]
- Bukhtojarov, F.E.; Ustinov, B.B.; Salanovich, T.N.; Antonov, A.I.; Gusakov, A.V.; Okunev, O.N.; Sinitsyn, A.P. Cellulase complex of the fungus Chrysosporium lucknowense: Isolation and characterization of endoglucanases and cellobiohydrolases. Biochemistry. 2004, 69, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Toda, H.; Nagahata, N.; Amano, Y.; Nozaki, K.; Kanda, T.; Okazaki, M.; Shimosaka, M. Gene cloning of cellobiohydrolase II from the white rot fungus Irpex lacteus MC-2 and its expression in Pichia pastoris. Biosci. Biotechnol. Biochem. 2008, 72, 3142–3147. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, F.; Gao, F.; Wang, L.; Zhao, J.; Qu, Y. Purification and characterization of a novel cellobiohydrolase (PdCel6A) from Penicillium decumbens JU-A10 for bioethanol production. Bioresour. Technol. 2011, 102, 8339–8342. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Liu, B.; Liu, Z.; Yang, Q. Cloning of two cellobiohydrolase genes from Trichoderma viride and heterogenous expression in yeast Saccharomyces cerevisiae. Mol. Biol. Rep. 2010, 37, 2135–2140. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.N.; Akimenko, V.K. Isolation of a Cellobiohydrolase of Clostridium thermocellum Capable of Degrading Natural Crystalline Substrates. Biochem. Biophys. Res. Commun. 1993, 192, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Kataeva, I.; Li, X.-L.; Chen, H.; Choi, S.-K.; Ljungdahl, L.G. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: Evidence for gene duplication and recombination. J. Bacteriol. 1999, 181, 5288–5295. [Google Scholar] [PubMed]
- Zverlov, V.V.; Velikodvorskaya, G.A.; Schwarz, W.H. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: Investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology 2002, 148, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Takada, G.; Kawaguchi, T.; Sumitani, J.; Arai, M. Expression of Aspergillus aculeatus No. F-50 cellobiohydrolase I (cbhI) and beta-glucosidase 1 (bgl1) genes by Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 1998, 62, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Moroz, O.V.; Maranta, M.; Shaghasi, T.; Harris, P.V.; Wilson, K.S.; Davies, G.J. The three-dimensional structure of the cellobiohydrolase Cel7A from Aspergillus fumigatus at 1.5 Å resolution. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 114–120. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, D.; Teng, F. Purification and characterization of a cellobiohydrolase from the thermophilic fungus Chaetomium thermophilus CT2. Wei Sheng Wu Xue Bao 2006, 46, 143–146. [Google Scholar] [PubMed]
- Hobdey, S.E.; Knott, B.C.; Haddad Momeni, M.; Taylor, L.E.; Borisova, A.S.; Podkaminer, K.K.; VanderWall, T.A.; Himmel, M.E.; Decker, S.R.; Beckham, G.T.; et al. Biochemical and Structural Characterizations of Two Dictyostelium Cellobiohydrolases from the Amoebozoa Kingdom Reveal a High Level of Conservation between Distant Phylogenetic Trees of Life. Appl. Environ. Microbiol. 2016, 82, 3395–3409. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Iikura, H.; Nakamura, A.; Hidaka, M.; Masaki, H.; Uozumi, T. Isolation of the gene and characterization of the enzymatic properties of a major exoglucanase of Humicola grisea without a cellulose-binding domain. J. Biochem. 1998, 124, 717–725. [Google Scholar] [CrossRef] [PubMed]
- Voutilainen, S.P.; Boer, H.; Linder, M.B.; Puranen, T.; Rouvinen, J.; Vehmaanperä, J.; Koivula, A. Heterologous expression of Melanocarpus albomyces cellobiohydrolase Cel7B, and random mutagenesis to improve its thermostability. Enzyme Microb. Technol. 2007, 41, 234–243. [Google Scholar] [CrossRef]
- Texier, H.; Dumon, C.; Neugnot-Roux, V.; Maestracci, M.; O’Donohue, M.J. Redefining XynA from Penicillium funiculosum IMI 378536 as a GH7 cellobiohydrolase. J. Ind. Microbiol. Biotechnol. 2012, 39, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Colussi, F.; Serpa, V.; Delabona, P.D.S.; Manzine, L.R.; Voltatodio, M.L.; Alves, R.; Mello, B.L.; Pereira, N.; Farinas, C.S.; Golubev, A.M.; et al. Purification, and biochemical and biophysical characterization of cellobiohydrolase I from Trichoderma harzianum IOC 3844. J. Microbiol. Biotechnol. 2011, 21, 808–817. [Google Scholar] [CrossRef] [PubMed]
- Bronnenmeier, K.; Rücknagel, K.P.; Staudenbauer, W.L. Purification and properties of a novel type of exo-1,4-beta-glucanase (avicelase II) from the cellulolytic thermophile Clostridium stercorarium. Eur. J. Biochem. 1991, 200, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-Z.; Zhang, Z.; Zhu, Z.; Sathitsuksanoh, N.; Yang, Y.; Zhang, Y.-H.P. The noncellulosomal family 48 cellobiohydrolase from Clostridium phytofermentans ISDg: Heterologous expression, characterization, and processivity. Appl. Microbiol. Biotechnol. 2010, 86, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Kruus, K.; Wang, W.K.; Ching, J.; Wu, J.H.D. Exoglucanase activities of the recombinant Clostridium thermocellum CelS, a major cellulosome component. J. Bacteriol. 1995, 177, 1641–1644. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Bevan, D.R.; Zhang, Y.-H.P. The family 1 glycoside hydrolase from Clostridium cellulolyticum H10 is a cellodextrin glucohydrolase. Appl. Biochem. Biotechnol. 2010, 161, 264–273. [Google Scholar] [CrossRef] [PubMed]
- Yernool, D.A.; McCarthy, J.K.; Eveleigh, D.E.; Bok, J.D. Cloning and characterization of the glucooligosaccharide catabolic pathway β-glucan glucohydrolase and cellobiose phosphorylase in the marine hyperthermophile Thermotoga neapolitana. J. Bacteriol. 2000, 182, 5172–5179. [Google Scholar] [CrossRef] [PubMed]
- Kengen, S.W.M.; Luesink, E.J.; Stams, A.J.M.; Zehnder, A.J.B. Purification and characterization of an extremely thermostable β-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur. J. Biochem. 1993, 213, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Lee, J.E.; Lee, H.S.; Kwon, K.K.; Kang, S.G.; Lee, J. Novel substrate specificity of a thermostable β-glucosidase from the hyperthermophilic archaeon, Thermococcus pacificus P-4. Korean J. Microbiol. 2015, 51, 68–74. [Google Scholar] [CrossRef]
- Matsui, I.; Sakai, Y.; Matsui, E.; Kikuchi, H.; Kawarabayasi, Y.; Honda, K. Novel substrate specificity of a membrane-bound beta-glycosidase from the hyperthermophilic archaeon Pyrococcus horikoshii. FEBS Lett. 2000, 467, 195–200. [Google Scholar] [CrossRef]
- Wu, Y.; Yuan, S.; Chen, S.; Wu, D.; Chen, J.; Wu, J. Enhancing the production of galacto-oligosaccharides by mutagenesis of Sulfolobus solfataricus β-galactosidase. Food Chem. 2013, 138, 1588–1595. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.K.; Datta, S. β-Glucosidase from the hyperthermophilic archaeon Thermococcus sp. is a salt-tolerant enzyme that is stabilized by its reaction product glucose. Appl. Microbiol. Biotechnol. 2016, 100, 8399–8409. [Google Scholar] [CrossRef] [PubMed]
- Di Lauro, B.; Rossi, M.; Moracci, M. Characterization of a beta-glycosidase from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius. Extremophiles 2006, 10, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, R.; Wang, J.; Zhang, X.; Jia, R.; Gao, Y.; Peng, H. Increased enzymatic hydrolysis of sugarcane bagasse by a novel glucose- and xylose-stimulated β-glucosidase from Anoxybacillus flavithermus subsp. yunnanensis E13T. BMC Biochem. 2017, 18, 4. [Google Scholar] [CrossRef] [PubMed]
- Paavilainen, S.; Hellman, J.; Korpela, T. Purification, characterization, gene cloning, and sequencing of a new β-glucosidase from Bacillus circulans subsp. alkalophilus. Appl. Environ. Microbiol. 1993, 59, 927–932. [Google Scholar] [PubMed]
- Xu, H.; Xiong, A.-S.; Zhao, W.; Tian, Y.-S.; Peng, R.-H.; Chen, J.-M.; Yao, Q.-H. Characterization of a glucose-, xylose-, sucrose-, and D-galactose-stimulated β-glucosidase from the alkalophilic bacterium Bacillus halodurans C-125. Curr. Microbiol. 2011, 62, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Love, D.R.; Fisher, R.; Bergquist, P.L. Sequence structure and expression of a cloned β-glucosidase gene from an extreme thermophile. MGG Mol. Gen. Genet. 1988, 213, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, A.; Arai, T.; Doi, R.H. Degradation of cellulosome-produced cello-oligosaccharides by an extracellular non-cellulosomal beta-glucan glucohydrolase, BglA, from Clostridium cellulovorans. Biochem. Biophys. Res. Commun. 2006, 349, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.-Z.; Yu, H.-L.; Li, C.-X.; Zhou, X.-W.; Hayashi, C.; Sun, J.; Liu, B.-H.; Imanaka, T.; Xu, J.-H. A new thermostable β-glucosidase mined from Dictyoglomus thermophilum: Properties and performance in octyl glucoside synthesis at high temperatures. Bioresour. Technol. 2012, 118, 425–430. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, D.; Klippel, B.; Antranikian, G. A novel thermostable and glucose-tolerant β-glucosidase from Fervidobacterium islandicum. Appl. Microbiol. Biotechnol. 2012, 93, 1947–1956. [Google Scholar] [CrossRef] [PubMed]
- Gefen, G.; Anbar, M.; Morag, E.; Lamed, R.; Bayer, E.A. Enhanced cellulose degradation by targeted integration of a cohesin-fused β-glucosidase into the Clostridium thermocellum cellulosome. Proc. Natl. Acad. Sci. USA 2012, 109, 10298–10303. [Google Scholar] [CrossRef] [PubMed]
- Brognaro, H.; Almeida, V.M.; de Araujo, E.A.; Piyadov, V.; Santos, M.A.M.; Marana, S.R.; Polikarpov, I. Biochemical Characterization and Low-Resolution SAXS Molecular Envelope of GH1 β-Glycosidase from Saccharophagus degradans. Mol. Biotechnol. 2016, 58, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.R.; Coutinho, P.M.; Videira, P.; Fialho, A.M.; Sá-Correia, I. Sphingomonas paucimobilis β-glucosidase Bgl1: A member of a new bacterial subfamily in glycoside hydrolase family 1. Biochem. J. 2003, 370, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Perez-Pons, J.A.; Cayetano, A.; Rebordosa, X.; Lloberas, J.; Guasch, A.; Querol, E. A beta-glucosidase gene (bgl3) from Streptomyces sp. strain QM-B814. Molecular cloning, nucleotide sequence, purification and characterization of the encoded enzyme, a new member of family 1 glycosyl hydrolases. Eur. J. Biochem. 1994, 223, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Breves, R.; Bronnenmeier, K.; Wild, N.; Lottspeich, F.; Staudenbauer, W.L.; Hofemeister, J. Genes encoding two different β-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl. Environ. Microbiol. 1997, 63, 3902–3910. [Google Scholar] [PubMed]
- Song, X.; Xue, Y.; Wang, Q.; Wu, X. Comparison of three thermostable β-glucosidases for application in the hydrolysis of soybean isoflavone glycosides. J. Agric. Food Chem. 2011, 59, 1954–1961. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Pang, Q.; Zhao, L.; Fan, S.; Shi, H. Thermoanaerobacterium thermosaccharolyticum β-glucosidase: A glucose-tolerant enzyme with high specific activity for cellobiose. Biotechnol. Biofuels 2012, 5, 31. [Google Scholar] [CrossRef] [PubMed]
- Spiridonov, N.A.; Wilson, D.B. Cloning and biochemical characterization of BglC, a beta-glucosidase from the cellulolytic actinomycete Thermobifida fusca. Curr. Microbiol. 2001, 42, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Wright, R.M.; Yablonsky, M.D.; Shalita, Z.P.; Goyal, A.K.; Eveleigh, D.E. Cloning, characterization, and nucleotide sequence of a gene encoding Microbispora bispora BglB, a thermostable beta-glucosidase expressed in Escherichia coli. Appl. Environ. Microbiol. 1992, 58, 3455–3465. [Google Scholar] [PubMed]
- Haq, I.U.; Khan, M.A.; Muneer, B.; Hussain, Z.; Afzal, S.; Majeed, S.; Rashid, N.; Javed, M.M.; Ahmad, I. Cloning, characterization and molecular docking of a highly thermostable β-1,4-glucosidase from Thermotoga petrophila. Biotechnol. Lett. 2012, 34, 1703–1709. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.-J.; Lee, Y.-J.; Chol, J.J.; Seo, M.S.; Lee, M.S.; Kim, G.A.; Kwon, S.-T. Mutational analysis of Thermus caldophilus GK24 beta-glycosidase: Role of His119 in substrate binding and enzyme activity. J. Microbiol. Biotechnol. 2008, 18, 287–294. [Google Scholar] [PubMed]
- Xiangyuan, H.; Shuzheng, Z.; Shoujun, Y. Cloning and expression of thermostable beta-glycosidase gene from Thermus nonproteolyticus HG102 and characterization of recombinant enzyme. Appl. Biochem. Biotechnol. 2001, 94, 243–255. [Google Scholar] [CrossRef]
- Kang, S.K.; Cho, K.K.; Ahn, J.K.; Bok, J.D.; Kang, S.H.; Woo, J.H.; Lee, H.G.; You, S.K.; Choi, Y.J. Three forms of thermostable lactose-hydrolase from Thermus sp. IB-21: Cloning, expression, and enzyme characterization. J. Biotechnol. 2005, 116, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Nam, E.S.; Kim, M.S.; Lee, H.B.; Ahn, J.K. β-Glycosidase of Thermus thermophilus KNOUC202: Gene and biochemical properties of the enzyme expressed in Escherichia coli. Appl. Biochem. Microbiol. 2010, 46, 515–524. [Google Scholar] [CrossRef]
- Zhao, Z.; Ramachandran, P.; Kim, T.-S.; Chen, Z.; Jeya, M.; Lee, J.-K. Characterization of an acid-tolerant β-1,4-glucosidase from Fusarium oxysporum and its potential as an animal feed additive. Appl. Microbiol. Biotechnol. 2013, 97, 10003–10011. [Google Scholar] [CrossRef] [PubMed]
- Takashima, S.; Nakamura, A.; Hidaka, M.; Masaki, H.; Uozumi, T. Molecular Cloning and Expression of the Novel Fungal -Glucosidase Genes from Humicola grisea and Trichoderma reesei. J. Biochem. 1999, 125, 728–736. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Ljungdahl, L.G.; Ximenes, E.A.; Chen, H.; Felix, C.R.; Cotta, M.A.; Dien, B.S. Properties of a recombinant beta-glucosidase from polycentric anaerobic fungus Orpinomyces PC-2 and its application for cellulose hydrolysis. Appl. Biochem. Biotechnol. 2004, 113, 233–250. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Shi, P.; Yang, P.; Wang, Y.; Luo, H.; Yao, B. Molecular cloning and expression of a novel β-glucosidase gene from Phialophora sp. G5. Appl. Biochem. Biotechnol. 2013, 169, 941–949. [Google Scholar] [CrossRef] [PubMed]
- Schröder, C.; Blank, S.; Antranikian, G. First Glycoside Hydrolase Family 2 Enzymes from Thermus antranikianii and Thermus brockianus with β-Glucosidase Activity. Front. Bioeng. Biotechnol. 2015, 3, 76. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, X.; Dang, W.; Tran, P.L.; Park, S.-H.; Oh, B.-C.; Hong, W.-S.; Lee, J.-S.; Park, K.-H. Characterization and application of an acidophilic and thermostable β-glucosidase from Thermofilum pendens. J. Biosci. Bioeng. 2013, 115, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Wei, P.; Chen, Q.; Chen, X.; Wang, S.; Li, J.; Gao, C. Functional and structural characterization of a β-glucosidase involved in saponin metabolism from intestinal bacteria. Biochem. Biophys. Res. Commun. 2018, 496, 1349–1356. [Google Scholar] [CrossRef] [PubMed]
- Lau, A.T.Y.; Wong, W.K.R. Purification and characterization of a major secretory cellobiase, Cba2, from Cellulomonas biazotea. Protein Expr. Purif. 2001, 23, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wakarchuk, W. Characterization of five β-glycoside hydrolases from Cellulomonas fimi ATCC 484. J. Bacteriol. 2014, 196, 4103–4110. [Google Scholar] [CrossRef] [PubMed]
- Bronnenmeier, K.; Staudenbauer, W.L. Purification and properties of an extracellular β-glucosidase from the cellulolytic thermophile Clostridium stercorarium. Appl. Microbiol. Biotechnol. 1988, 28, 380–386. [Google Scholar] [CrossRef]
- Li, Y.-K.; Lee, J.-A. Cloning and expression of β-glucosidase from Flavobacterium meningosepticum: A new member of family B β-glucosidase. Enzyme Microb. Technol. 1999, 24, 144–150. [Google Scholar] [CrossRef]
- Xie, J.; Zhao, D.; Zhao, L.; Pei, J.; Xiao, W.; Ding, G.; Wang, Z. Overexpression and characterization of a Ca2+ activated thermostable β-glucosidase with high ginsenoside Rb1 to ginsenoside 20(S)-Rg3 bioconversion productivity. J. Ind. Microbiol. Biotechnol. 2015, 42, 839–850. [Google Scholar] [CrossRef] [PubMed]
- Colabardini, A.C.; Valkonen, M.; Huuskonen, A.; Siika-aho, M.; Koivula, A.; Goldman, G.H.; Saloheimo, M. Expression of Two Novel β-Glucosidases from Chaetomium atrobrunneum in Trichoderma reesei and Characterization of the Heterologous Protein Products. Mol. Biotechnol. 2016, 58, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhang, R.; Yang, X.; Zhang, Z.; Song, S.; Miao, Y.; Shen, Q. Characterization of a thermostable β-glucosidase from Aspergillus fumigatus Z5, and its functional expression in Pichia pastoris X33. Microb. Cell Fact. 2012, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Teng, F.; Zhang, C.; Li, D. Cloning of a Gene Encoding β-Glucosidase from Chaetomium thermophilum CT2 and Its Expression in Pichia pastoris. J. Mol. Microbiol. Biotechnol. 2011, 20, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Dotsenko, G.S.; Sinitsyna, O.A.; Hinz, S.W.A.; Wery, J.; Sinitsyn, A.P. Characterization of a GH family 3 β-glycoside hydrolase from Chrysosporium lucknowense and its application to the hydrolysis of β-glucan and xylan. Bioresour. Technol. 2012, 112, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.; Zhao, J.; Cai, P.; Sun, W.; Ren, J.; Wu, Q.; Zhang, S.; Tian, C. Heterologous expression of a GH3 β-glucosidase from Neurospora crassa in Pichia pastoris with high purity and its application in the hydrolysis of soybean isoflavone glycosides. Protein Expr. Purif. 2016, 119, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Kato, Y.; Nomura, T.; Ogita, S.; Takano, M.; Hoshino, K. Two new β-glucosidases from ethanol-fermenting fungus Mucor circinelloides NBRC 4572: Enzyme purification, functional characterization, and molecular cloning of the gene. Appl. Microbiol. Biotechnol. 2013, 97, 10045–10056. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ma, R.; Shi, P.; Huang, H.; Bai, Y.; Wang, Y.; Yang, P.; Fan, Y.; Yao, B. Molecular characterization of a highly-active thermophilic β-glucosidase from Neosartorya fischeri P1 and its application in the hydrolysis of soybean isoflavone glycosides. PLoS ONE 2014, 9, e106785. [Google Scholar] [CrossRef] [PubMed]
- Krogh, K.B.R.M.; Harris, P.V.; Olsen, C.L.; Johansen, K.S.; Hojer-Pedersen, J.; Borjesson, J.; Olsson, L. Characterization and kinetic analysis of a thermostable GH3 β-glucosidase from Penicillium brasilianum. Appl. Microbiol. Biotechnol. 2010, 86, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Qin, Y.; Liu, Z.; Liu, K.; Wang, F.; Qu, Y. Isolation and characterization of a β-glucosidase from Penicillium decumbens and improving hydrolysis of corncob residue by using it as cellulase supplementation. Enzyme Microb. Technol. 2010, 46, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Machida, M.; Ohtsuki, I.; Fukui, S.; Yamashita, I. Nucleotide sequences of Saccharomycopsis fibuligera genes for extracellular β-glucosidases as expressed in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1988, 54, 3147–3155. [Google Scholar] [PubMed]
- Murray, P.; Aro, N.; Collins, C.; Grassick, A.; Penttilä, M.; Saloheimo, M.; Tuohy, M. Expression in Trichoderma reesei and characterisation of a thermostable family 3 beta-glucosidase from the moderately thermophilic fungus Talaromyces emersonii. Protein Expr. Purif. 2004, 38, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Xu, X.; Qian, L.; Shi, P.; Bai, Y.; Luo, H.; Ma, R.; Yao, B. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol. Biofuels 2016, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Tamaki, H.; Kumagai, H. Cloning and functional expression of thermostable beta-glucosidase gene from Thermoascus aurantiacus. Appl. Microbiol. Biotechnol. 2007, 73, 1331–1339. [Google Scholar] [CrossRef] [PubMed]
- Karnaouri, A.; Topakas, E.; Paschos, T.; Taouki, I.; Christakopoulos, P. Cloning, expression and characterization of an ethanol tolerant GH3 β-glucosidase from Myceliophthora thermophila. PeerJ 2013, 1, e46. [Google Scholar] [CrossRef] [PubMed]
- Brunner, F.; Wirtz, W.; Rose, J.K.C.; Darvill, A.G.; Govers, F.; Scheel, D.; Nürnberger, T. A β-glucosidase/xylosidase from the phytopathogenic oomycete, Phytophthora infestans. Phytochemistry 2002, 59, 689–696. [Google Scholar] [CrossRef]
- Ferrara, M.C.; Cobucci-Ponzano, B.; Carpentieri, A.; Henrissat, B.; Rossi, M.; Amoresano, A.; Moracci, M. The identification and molecular characterization of the first archaeal bifunctional exo-β-glucosidase/N-acetyl-β-glucosaminidase demonstrate that family GH116 is made of three functionally distinct subfamilies. Biochim. Biophys. Acta 2014, 1840, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Sansenya, S.; Mutoh, R.; Charoenwattanasatien, R.; Kurisu, G.; Ketudat Cairns, J.R. Expression and crystallization of a bacterial glycoside hydrolase family 116 β-glucosidase from Thermoanaerobacterium xylanolyticum. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015, 71, 41–44. [Google Scholar] [CrossRef] [PubMed]
| Enzyme | GH Family Domains | Optimum Temperature | Optimum pH | Temperature Stability 1 | Source | Reference |
|---|---|---|---|---|---|---|
| EGPh | 5 | >97 °C | 5.4–6.0 | 80%; 97 °C; 3 h | Archaea (Pyrococcus horikoshii) | [46] |
| EG1 | 5 | 83 °C | 5.0 | 20%; 90 °C; 2 h | Bacteria (Acidothermus cellolyticus) | [73] |
| EglII | 5 | 50 °C | 6.0 | NM | Bacteria (Bacillus amyloliquefaciens) | [74] |
| EG | 5 | 65 °C | 6.0 | 72%; 55 °C; 42 h 50%; 65 °C; 12 min | Bacteria (Bacillus licheniformis) | [75] |
| CelA | 5 | 60 °C | 8.0 | 30%; 70 °C; 1 h | Bacteria (Bacillus subtilis) | [76] |
| TmCel5A | 5 | 80 °C | 6.0 | 50%; 80 °C; 18 h | Bacteria (Thermotoga maritima) | [77] |
| EglA | 5 | 57 °C | 4.0 | NM | Fungi (Aspergillus nidulans) | [78] |
| EglB | 5 | 52 °C | 4.0 | NM | Fungi (Aspergillus nidulans) | [78] |
| EBI-244 | 5 | 109 °C | 5.5 | 50%; 100 °C; 4.5 h 50%; 105 °C; 0.57 h 108 °C; 50%; 0.17 h | Uncultured Archaea (Continental geothermal pool enrichment) | [58] |
| CelE1 | 5 | 50 °C | 7.0 | NM | Uncultured organism (Sugarcane field soil metagenome) | [64] |
| CelA10 | 5 | 55 °C | 7.5 | NM | Uncultured organism (Aquatic community and soil sample) | [66] |
| CelA24 | 5 | 55 °C | 7.0 | NM | Uncultured organism (Aquatic community and soil sample) | [66] |
| cMGL504 | 5 | 50 °C | 5.5 | NM | Uncultured organism (Vermicompost sample) | [60] |
| Cel5G | 5 | 50 °C | 4.8 | >90%; 50 °C; 30 min | Uncultured organism (Soil metagenome) | [65] |
| En1 | 5 | 55 °C | 5.5 | 87%; 45 °C; 16 h 67%; 50 °C; 6 h 42%; 55 °C; 30 min | Uncultured organism (Biogas digester metagenome) | [63] |
| RC1 | 5 | 55 °C | 6.0–6.5 | >90%; 50 °C; 30 min | Uncultured organism (Rabbit cecum metagenome) | [67] |
| RC3 | 5 | 50 °C | 6.0–7.0 | NM | Uncultured organism (Rabbit cecum metagenome) | [67] |
| RC5 | 5 | 50 °C | 6.5–7.0 | NM | Uncultured organism (Rabbit cecum metagenome) | [67] |
| CelL | 6 | 50 °C | 5.0 | 50%; 50 °C; 12 min | Bacteria (Cellulosimicrobium funkei) | [22] |
| Cel6A | 6 | 58 °C | 6.5 | >80%; 56 °C; 18 h | Bacteria (Thermobifida fusca) | [79] |
| ThCel6A | 6 | 55 °C | 8.5 | 58%; 90 °C; 1 h | Bacteria (Thermobifida halotolerans) | [80] |
| Cel6A | 6 | 50–55 °C | 5.5 | NM | Bacteria (Xylanimicrobium pachnodae) | [81] |
| HiCel6C | 6 | 70 °C | 6.5 | >90%; 60 °C; 1 h | Fungi (Humicola insolens) | [82] |
| Cel6A | 6 | 50 °C | 4.8 | >90%; 45 °C; 24 h 92%; 50 °C; 5 h | Fungi (Orpinomyces sp.) | [83] |
| C1 | 6 | 50 °C | 6.0 | 100%; 60 °C; 30 min | Uncultured organism (Compost metagenome) | [61] |
| pre-LC-CelB | 6 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| pre-LC-CelJ | 6 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| EGI | 7 | 55–60 °C | 5.0 | >80%; 60 °C; 10 min | Fungi (Humicola grisea var. thermoidea) | [84] |
| Cel7B | 7 | 60 °C | 4.0 | >90%; 60 °C; 1 h | Fungi (Penicillium decumbens) | [85] |
| Cel7A | 7 | 60 °C | 5.0 | 100%; 60 °C; 1 h 16.1%; 70 °C; 1 h | Fungi (Neosartorya fischeri) | [33] |
| MtEG7 | 7 | 60 °C | 5.0 | 50%; 70 °C; 9.96 h 50%; 80 °C; 6.5 h | Fungi (Myceliophthora thermophila) | [31] |
| EGL1 | 7 | 62 °C | 4.8 | NM | Fungi (Trichoderma longibrachiatum) | [51] |
| MaCel7A | 7 | 65–70 °C | 6.0 | NM | Fungi (Melanocarpus albomyces) | [86] |
| CelC | 8 | 50 °C | 6.5 | NM | Bacteria (Salmonella typhimurium) | [87] |
| Cel8Y | 8 | 80 °C | 7.0 | 50%; 90 °C; 4 h 50%; 100 °C; 2 h | Bacteria (Aquifex geolicus) | [88] |
| Egl-257 | 8 | 55 °C | 8.5 | 100%; 50 °C; 15 min >50%; 60 °C; 15 min | Bacteria (Bacillus circulans) | [89] |
| CenC | 9 | 70 °C | 6.0 | 100%; 60 °C; 2 h 60%; 70 °C; 1 h | Bacteria (Clostridium thermocellum) | [90] |
| CelA | 9 (endoglucanase) and 48 (cellobiohydrolase) | 95 °C (endoglucanase) and 85 °C (cellobiohydrolase) | 5.0–6.0 | 50%; 95 °C; 40 min (endoglucanase) 100%; 85 °C; 4 h (cellobiohydrolase) | Bacteria (Caldicellulosiruptor bescii) | [91] |
| Cel9A | 9 | 65 °C | 6.5 | NM | Bacteria (Lachnoclostridium phytofermentans) | [92] |
| CelA20 | 9 | 55 °C | 5.0 | NM | Uncultured organism (Aquatic community and soil metagenome) | [66] |
| AcCel12B | 12 | 75 °C | 4.5 | 50%; 60 °C; 90 h 50%; 65 °C; 55 h 50%; 70 °C; 2 h | Bacteria (Acidothermus cellulolyticus) | [35] |
| CelA | 12 | 95 °C | 6.0 | NM | Bacteria (Thermotoga neapolitana) | [8] |
| CelB | 12 | 106 °C | 6.0–6.6 | 50%; 106 °C; 130 min 50%; 110 °C; 26 min 73%; 100 °C; 4 h | Bacteria (Thermotoga neapolitana) | [8] |
| TmCel12A | 12 | 90 °C | 7.0 | >40%; 85 °C; 48 h 50%; 90 °C; 3 h | Bacteria (Thermotoga maritima) | [93] |
| TmCel12B | 12 | 85 °C | 6.0 | 50%; 90 °C; 9 h | Bacteria (Thermotoga maritima) | [93] |
| CelA | 12 | >100 °C | 6.0–7.0 | 45%; 90 °C; 8 h | Bacteria (Rhodothermus marinus) | [23] |
| EglA | 12 | 100 °C | 6.0 | 50%; 95 °C; 40 h | Archaea (Pyrococcus furiosus) | [94] |
| SSO1949 | 12 | 80 °C | 1.8 | 50%; 80 °C; 8 h | Archeaea (Sulfolobus solfataricus) | [95] |
| SSO1354 | 12 | 90 °C | 4.0 | 50%; 90 °C; 180 min | Archaea (Sulfolobus solfataricus) | [39] |
| EglS | 12 | 65 °C | 6.0 | >40%; 60 °C; 30 min | Bacteria (Streptomyces rochei) | [96] |
| Cel12A | 12 | 50 °C | 5.0 | NM | Fungi (Trichoderma reseei) | [97] |
| EG | 12 | 70 °C | 3.5 | 50%; 70 °C; 3 h 50%; 80 °C; 1 h | Fungi (Aspergillus niger) | [27] |
| Pre-LC-CelA | 12 | 90 °C | 5.0–9.0 | 100%; 90 °C; 30 min | Uncultured organism (Compost metagenome) | [62] |
| Pre-LC-CelD | 12 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| Pre-LC-CelE | 12 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| Cel12E | 12 | 92 °C | 5.5 | >80%; 80 °C; 4.5 h | Uncharacterized archeon (deep sea vents metagenome enrichment) | [57] |
| GH44EG | 44 | 55 °C | 5.0 | NM | Bacteria (Clostridium acetobutylicum) | [98] |
| CelA | 44 | 60 °C | 5.0–8.5 | 50%; 60 °C; 70 min | Bacteria (Paenibacillus lautus) | [99] |
| CelJ | 44 | 70 °C | 6.5 | >90%; 80 °C; 10 min | Bacteria (Ruminiclostridium thermocellum) | [100] |
| pre-LC-CelH | 44 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| Cel45A | 45 | 60 °C | 5.0 | NM | Fungi (Trichoderma reseei) | [97] |
| PpCel45A | 45 | 65 °C | 4.8 | 70%; 65 °C; 48 h 60%; 80 °C; 4 h | Fungi (Picchia pastoris) | [5] |
| STCE1 | 45 | 60 °C | 6.0 | NM | Fungi (Staphylotrichum coccosporum) | [101] |
| BCC18080 | 45 | 70 °C | 6.0 | >70%; 70 °C; 2 h >50%; 70 °C; 4 h | Fungi (Syncephalastrum racemosum) | [102] |
| BCE1 | 45 | 55 °C | 4.5 | NM | Fungi (Beltraniella portoricensis) | [103] |
| MaCel45A | 45 | 70 °C | 7.0 | NM | Fungi (Melanocarpus albomyces) | [86] |
| CelB | 51 | 80 °C | 4.0 | 60%; 80 °C; 1 h | Bacteria (Alicyclobacillus acidocaldarius) | [104] |
| CelA4 | 51 | 65 °C | 2.6 | >85%; 60 °C; 1 h | Bacteria (Alicyclobacillus sp. A4) | [47] |
| CelVA | 51 | 80 °C | 3.6–4.5 | 70%; 70 °C; 2 h | Bacteria (Alicyclobacillus vulcanalis) | [45] |
| pre-LC-CelC | 51 | NM | NM | NM | Uncultured organism (Compost metagenome) | [62] |
| TmCel74 | 74 | 90 °C | 6.0 | 50%; 90 °C; 5 h | Bacteria (Thermotoga maritima) | [15] |
| CtCel124 | 124 | NM | NM | NM | Bacteria (Ruminiclostridium thermocellum) | [105] |
| Enzyme | GH Family Domains | Optimum Temperature | Optimum pH | Temperature Stability 1 | Source | Reference |
|---|---|---|---|---|---|---|
| CBHII | 6 | 60 °C | 4.0 | 30%; 100 °C; 10 min | Bacteria (Streptomyces sp. M23) | [106] |
| Cel6B | 6 | NM | 7.0–8.0 | 100%; 55 °C; 16 h | Bacteria (Thermobifida fusca) | [107] |
| CBHII | 6 | 57 °C | 5.5 | NM | Fungi (Aspergillus nidulans) | [78] |
| Cel6A | 6 | 50 °C | 4.0 | 50%; 70 °C; 30 min | Fungi (Chaetomium thermophilum) | [108] |
| CBHII (Cel6A) | 6 | 60 °C | 5.0–5.5 | >90%; 50 °C; 5 h | Fungi (Chrysosporium lucknowense) | [109] |
| HiCel6A | 6 | 60–65 °C | NM | 50%; 75 °C; <25 min | Fungi (Humicola insolens) | [26] |
| Ex-4 | 6 | 50 °C | 5.0 | 80%; 60 °C; 60 min | Fungi (Irpex Lacteus) | [110] |
| PoCel6A | 6 | 50 °C | 5.0 | 90%; 50 °C; 2 h 80%; 60 °C; 4 h | Fungi (Penicillium oxalicum) | [111] |
| PaCel6A | 6 | 55 °C | 5.0–9.0 | 100%; 35 °C; 24 h >20%; 45 °C; 24 h | Fungi (Podospora anserina) | [19] |
| CBHII | 6 | 70 °C | 5.0 | NM | Fungi (Trichoderma viride) | [112] |
| G10-6 | 6 | 55 °C | 9.5 | NM | Uncultured organism (Eathworm casts metagenome) | [70] |
| Cbh9A | 9 | 60 °C | 6.5 | NM | Bacteria (Ruminiclostridium thermocellum) | [113] |
| Cel9K | 9 | 65 °C | 6.0 | 97%; 60 °C; 200 h | Bacteria (Ruminiclostridium thermocellum) | [114] |
| Enzyme | GH Family Domains | Optimum Temperature | Optimum pH | Temperature Stability 1 | Source | Reference |
|---|---|---|---|---|---|---|
| CelO | 5 | 65 °C | 6.6 | NM | Bacteria (Ruminiclostridium thermocellum) | [115] |
| AtCel7A | 7 | 60 °C | 5.0 | NM | Fungi (Acremonium thermophilum) | [28] |
| CBHI | 7 | 60 °C | 3.0 | NM | Fungi (Aspergillus aculeatus) | [116] |
| CBHI | 7 | 55 °C | NM | NM | Fungi (Aspergillus fumigatus) | [117] |
| CtCel7A | 7 | 65 °C | 4.0 | NM | Fungi (Chaetomium thermophilum) | [28] |
| CBH3 | 7 | 65 °C | 5.0 | 50%; 70 °C; 1 h 20%; 80 °C; 20 min | Fungi (Chaetomium thermophilum) | [118] |
| DpuCel7A | 7 | 55 °C | 5.0 | NM | Metazoa (Dictyostelium purpureum) | [119] |
| CBHI | 7 | 60 °C | 5.0 | >90%; 55 °C; 10 min | Fungi (Humicola grisea var. thermoidea) | [84] |
| EXO1 | 7 | 65 °C | 5.0 | >80%; 65 °C; 10 min | Fungi (Humicola grisea var. thermoidea) | [120] |
| MaCel7B | 7 | 55 °C | NM | NM | Fungi (Melanocarpus albomyces) | [121] |
| TeCel7A | 7 | 65 °C | 4.0–5.0 | 50%; 70 °C; 30 min | Fungi (Talaromyces emersonii) | [29] |
| Cel7A | 7 | 55 °C | 3.7–5.2 | 50%; 50 °C; 2.5 h | Fungi (Penicillium funiculosum) | [122] |
| TaCel7A | 7 | 65 °C | 5.0 | NM | Fungi (Thermoascus aurantiacus) | [28] |
| ThCBHI | 7 | 50 °C | 5.0 | NM | Fungi (Trichoderma harzianum) | [123] |
| CBHI | 7 | 60 °C | 5.8 | NM | Fungi (Trichoderma viride) | [112] |
| CelA | 9 (endoglucanase) and 48 (cellobiohydrolase) | 95 °C (endoglucanase) and 85 °C (cellobiohydrolase) | 5.0–6.0 | 50%; 95 °C; 40 min (endoglucanase) 100%; 85 °C; 4 h (cellobiohydrolase) | Bacteria (Caldicellulosiruptor bescii) | [91] |
| CelY | 48 | 70 °C | 5.0–6.0 | NM | Bacteria (Clostridium stercorarium) | [124] |
| CpCel48 | 48 | 55 °C | 5.0–6.0 | >70%; 50 °C; 30 min>20%; 55 °C; 30 min | Bacteria (Lachnoclostridium phytofermentans) | [125] |
| CelS | 48 | 70 °C | 5.5 | NM | Bacteria (Ruminiclostridium thermocellum) | [126] |
| Enzyme | GH family Domains | Optimum Temperature | Optimum pH | Temperature Stability 1 | Source | Reference |
|---|---|---|---|---|---|---|
| CcGH1 | 1 | 60 °C | 6.5 | 61%; 50 °C; 30 min | Bacteria (Clostridium Cellulolyticum) | [127] |
| GghA | 1 | 95 °C | 6.5 | 85%; 90 °C; 9 h 88%; 95 °C; 1 h | Bacteria (Thermotoga neapolitana) | [128] |
| Enzyme | GH family Domains | Optimum Temperature | Optimum pH | Temperature Stability 1 | Source | Reference |
|---|---|---|---|---|---|---|
| CelB | 1 | 102–105 °C | 5.0 | 50%; 100 °C; 85 h 50%; 110 °C; 13 h | Arquea (Pyrococcus furiosus) | [129] |
| Tpa-glu | 1 | 75 °C | 7.5 | 50%; 90 °C; 6 h | Arquea (Thermococcus pacificus) | [130] |
| BGPh | 1 | >100 °C | 6.0 | 50%; 90 °C; 15 h | Arquea (Pyrococcus horikoshii) | [131] |
| LacS | 1 (β-glucosidase and β-galactosidase) | 90 °C | 6.0 | 90%; 75 °C; 80 h | Arquea (Sulfolobus solfataricus) | [132] |
| O08324 | 1 | 78 °C | 5.0–6.8 | 50%; 78 °C; 860 min | Arquea (Thermococcus sp.) | [133] |
| Bgl1 | 1 | 90 °C | 6.5 | 67%; 90 °C; 1.5 h 78%; 50 °C; 24 h 68%; 60 °C; 24 h | Uncultured Arquea (hot spring metagenome) | [59] |
| GlyB | 1 (multiple substrates) | 85 °C | 5.5 | 8%; 80 °C; 10 min >70%; 65 °C; 3 h | Bacteria (Alicyclobacillus acidocaldarius) | [134] |
| Bglp | 1 | 60 °C | 7.0 | 50%; 60 °C; 10 h | Bacteria (Anoxybacillus flavithermus) | [135] |
| BglA | 1 | 55 °C | 6.0–9.0 | 80%; 50 °C; 15 min 1%; 60 °C; 15 min | Bacteria (Bacillus circulans subsp. Alkalophilus) | [136] |
| BhbglA | 1 | 50 °C | 7.0 | 50%; 50 °C; 30 min | Bacteria (Bacillus halodurans) | [137] |
| BglA | 1 | 85 °C | 6.25 | 50%; 70 °C; 2280 min | Bacteria (Caldicellulosiruptor saccharolyticus) | [138] |
| BglA | 1 | 50 °C | 6.0 | NM | Bacteria (Clostridium cellulovorans) | [139] |
| DtGH | 1 | 90 °C | 7.0 | 50%; 70 °C; 533 h 50%; 80 °C; 44 h 50%; 90 °C; 5 h | Bacteria (Dictyoglomus thermophilum) | [140] |
| DturβGlu | 1 | 80 °C | 5.4 | 70%; 70 °C; 2 h | Bacteria (Dictyoglomus turgidum) | [44] |
| FiBgl1A | 1 | 90 °C | 6.0–7.0 | 50%; 90 °C 25 min 50%; 100 °C; 15 min | Bacteria (Fervidobacterium islandicum) | [141] |
| BglA | 1 | 60 °C | 6.5 | 91%; 60 °C; 3 h 34%; 60 °C; 43 h | Bacteria (Ruminiclostridium thermocellum) | [142] |
| SdBgl1B | 1 | 50 °C | 6.0–7.5 | NM | Bacteria (Saccharophagus degradans) | [143] |
| Bgl1 | 1 | 50 °C | 5.1–5.7 | 60%; 40 °C; 4 h | Bacteria (Sphingomonas paucimobilis) | [144] |
| SGR_2426 | 1 | 69 °C | 6.9 | 50%; 69 °C; 1.5 h | Bacteria (Streptomyces griseus) | [55] |
| Bgl3 | 1 | 50 °C | 6.5 | NM | Bacteria (Streptomyces sp. strain QM-B814) | [145] |
| CglT | 1 | 75 °C | 5.5 | 100%; 60 °C; 24 h | Bacteria (Thermoanaerobacter brockii) | [146] |
| TeBglA | 1 | 80 °C | 7.0 | 10%; 65 °C; 5 h | Bacteria (Thermoanaerobacter ethanolicus) | [147] |
| TmBglA | 1 | 90 °C | 6.2 | >80%; 65 °C; 5 h | Bacteria (Thermotoga maritima) | [147] |
| Bgl | 1 | 70 °C | 6.4 | 50%; 68 °C; 1 h >80%; 60 °C; 2 h | Bacteria (Thermoanaerobacterium thermosaccharolyticum) | [148] |
| BglC | 1 | 50 °C | 7.0 | NM | Bacteria (Thermobifida fusca) | [149] |
| BglB | 1 | 60 °C | 6.2 | 70%; 60 °C; 48 h | Bacteria (Thermobispora bispora) | [150] |
| BglA | 1 | 80–90 °C | 7.0–8.0 | 100%; 70 °C; 6 h | Bacteria (Thermotoga petrophila) | [151] |
| TcaBglA | 1 | 90 °C | 5.5–6.5 | >40%; 80 °C; 30 min >20%; 80 °C; 30 min | Bacteria (Thermus caldophilus) | [152] |
| TnGly | 1 | 90 °C | 5.6 | 50%; 90 °C; 2.5 h | Bacteria (Thermus nonproteolyticus) | [153] |
| BglA | 1 | 70 °C | 5.0–6.0 | 50%; 70 °C; 38 h 50%; 80 °C; <0.4 h 50%; 90 °C; <0.3 h | Bacteria (Thermus sp. IB-21) | [154] |
| BglB | 1 | 80 °C | 5.0–6.0 | 50%; 70 °C; 38 h 50%; 80 °C; 2.7 h 50%; 90 °C; 24 min | Bacteria (Thermus sp. IB-21) | [154] |
| Bgly | 1 | 90 °C | 5.4 | 100%; 80 °C; 2 h 50%; 90 °C; 1.5 h 50%; 95 °C; 20 min | Bacteria (Thermus thermophilus) | [155] |
| BglA | 1 | 55 °C | 6.5 | 82%; 50 °C; 60 min 20%; 55 °C; 60 min | Uncultured organism (soil metagenome) | [52] |
| AS-Esc10 | 1 | 60 °C | 8.0 | 100%; 50 °C; 1 h | Uncultured organism (agricultural soil metagenome) | [40] |
| Bgl-gs1 | 1 | 90 °C | 6.0 | 50%; 90 °C; 5 min 50%; 85 °C; 15 min 50%; 80 °C; 45 min | Uncultured organism (termite gut metagenome) | [71] |
| Bgl | 1 | 60 °C | 5.0 | 50%; 60 °C; 540 min | Fungi (Fusarium oxysporum) | [156] |
| Bgl4 | 1 | 55 °C | 6.0 | 80%; 50 °C; 10 min | Fungi (Humicola grisea var. thermoidea IFO9854) | [157] |
| Bgl1 | 1 | 55 °C | 5.5–7.5 | 100%; 50 °C; 8 h 50%; 55 °C; 8 h | Fungi (Orpinomyces sp. PC-2) | [158] |
| Bgl1G5 | 1 | 50 °C | 6.0 | 50%; 50 °C; 6 h | Fungi (Phialophora sp. G5) | [159] |
| TaGH2 | 2 | 95 °C | 6.5 | 100%; 90 °C; 3 h 50%; 70 °C; 22 h | Bacteria (Thermus antranikianii) | [160] |
| TbGH2 | 2 | 90 °C | 6.5 | 17%; 80 °C; 3 h 50%; 70 °C; 12 h | Bacteria (Thermus brockianus) | [160] |
| TbBgl | 3 | 90 °C | 3.5 | 50%; 95 °C; 60 min | Arquea (Thermofilum pendens) | [161] |
| BlBG3 | 3 | 50 °C | 6.0 | NM | Bacteria (Bifidobacterium longum) | [162] |
| Cba2 | 3 | 70 °C | 4.8 | NM | Bacteria (Cellulomonas biazotea) | [163] |
| CfBgl3A | 3 | 55 °C | 7.5 | NM | Bacteria (Cellulomonas fimi) | [164] |
| Bgl3Z | 3 | 65 °C | 5.5 | 50%; 60 °C; 5 h | Bacteria (Clostridium stercorarium) | [165] |
| Dtur_0219 | 3 | 85 °C | 5.0 | 50%; 70 °C; 1575 min 50%; 75 °C; 854 min 50%; 80 °C; 524 min 50%; 85 °C; 334 min 50%; 90 °C; 20 min | Bacteria (Dictyoglomus turgidum) | [54] |
| Bgl | 3 | 50 °C | 4.2–5.0 | NM | Bacteria (Elizabethkingia meningoseptica) | [166] |
| TmBglB | 3 | 80 °C | 4.2 | >80%; 65 °C; 5 h | Bacteria (Thermotoga maritima) | [147] |
| Tpebgl3 | 3 | 90 °C | 5.0 | >90%; 70 °C; 3 h >50%; 90 °C; 3 h | Bacteria (Thermotoga petrophila) | [167] |
| Cel3A | 3 | 50–60 °C | 5.0 | 98%; 60 °C; 6 h >50%; 60 °C; 24 h >50%; 70 °C; 24 h | Fungi (Amesia atrobrunnea) | [168] |
| Cel3B | 3 | 50–60 °C | 5.0 | 88%; 60 °C; 6 h >50%; 60 °C; 24 h >50%; 70 °C; 24 h | Fungi (Amesia atrobrunnea) | [168] |
| Bgl3 | 3 | 60 °C | 6.0 | >50%; 70 °C; 1 h | Fungi (Aspergillus fumigatus) | [169] |
| BglB | 3 | 52 °C | 5.5 | NM | Fungi (Aspergillus nidulans) | [78] |
| BglC | 3 | 52 °C | 6.0 | NM | Fungi (Aspergillus nidulans) | [78] |
| Bgl | 3 | 50 °C | 5.0 | 100%; 50 °C; 30 min 60%; 60 °C; 30 min | Fungi (Aspergillus oryzae) | [49] |
| Bgl | 3 | 60 °C | 5.0 | 67.7%; 60 °C; 1 h 50%; 65 °C; 55 min 29.7%; 70 °C; 10 min | Fungi (Chaetomium thermophilum) | [170] |
| Bxl5 | 3 | 75 °C | 4.6 | 50%; 65 °C; 5 h 50%; 70 °C; 20 min 50%; 75 °C; 5 min | Fungi (Chrysosporium lucknowense) | [171] |
| MoCel3A | 3 | 50 °C | 5.0–5.5 | NM | Fungi (Magnaporthe oryzae) | [41] |
| MoCel3B | 3 | 50 °C | 5.0–5.5 | NM | Fungi (Magnaporthe oryzae) | [41] |
| Bgl2 | 3 | 60 °C | 5.4 | >50%; 40 °C; 2 h >45%; 50 °C; 2 h 25%; 55 °C; 1 h | Fungi (Neurospora crassa) | [172] |
| Bgl1 | 3 | 50 °C | 3.5–5.0 | 100%; 45 °C; 30 min | Fungi (Mucor circinelloides) | [173] |
| Bgl2 | 3 | 55 °C | 3.5–5.5 | 100%; 55 °C; 30 min | Fungi (Mucor circinelloides) | [173] |
| NfBGL1 | 3 | 80 °C | 5.0 | >80%; 70 °C; 2 h | Fungi (Neosartorya fischeri) | [174] |
| PtBglu3 | 3 | 65 °C | 6.0 | >85%; 60 °C; 30 min | Fungi (Paecilomyces thermophila) | [32] |
| Bgl1 | 3 | 70 °C | 4.8 | 50%; 65 °C; 24 h | Fungi (Penicillium brasilianum) | [175] |
| pBGL1 | 3 | 65–70 °C | 4.5–5.50 | 96.3%; 50 °C; 12 h 50%; 70 °C; 4 h | Fungi (Penicillium decumbens) | [176] |
| Bgl1 | 3 | 70 °C | 5.0–6.0 | 60%; 70 °C; 1.5 h | Fungi (Periconia sp.) | [43] |
| RmBglu3B | 3 | 50 °C | 5.0 | 50%; 50 °C; 30 min | Fungi (Rhizomucor miehei) | [50] |
| Bgl1 | 3 | 50 °C | 5.0 | >70%; 50 °C; 30 min <10%; 60 °C; 30 min | Fungi (Saccharomycopsis fibuligera) | [177] |
| Bgl2 | 3 | 50 °C | 5.0 | >70%; 50 °C; 30 min <10%; 60 °C; 30 min | Fungi (Saccharomycopsis fibuligera) | [177] |
| β-glucosidase | 3 | 75 °C | 4.5 | 50%; 60 °C; 136 h 50%; 65 °C; 55 h 50%; 70 °C; 10 h 50%; 75 °C; 1 h | Fungi (Talaromyces aculeatus) | [53] |
| Cel3a | 3 | 71.5 °C | 4.02 | 50%; 65 °C; 62 min 50%; 75 °C; 18 min | Fungi (Talaromyces emersonii) | [178] |
| Bgl3A | 3 | 75 °C | 4.5 | >65%; 60 °C; 1 h | Fungi (Talaromyces leycettanus) | [179] |
| Bgl1 | 3 | 70 °C | 5.0 | >70%; 60 °C; 1 h | Fungi (Thermoascus auranticus) | [180] |
| Bgl3a | 3 | 70 °C | 5.0 | 50%; 60 °C; 143 min | Fungi (Myceliophthora thermophila) | [181] |
| RG3 | 3 | 50–55 °C | 5.5–6.0 | NM | Uncultured organism (Rabbit cecum metagenome) | [67] |
| RG14 | 3 | 50–55 °C | 5.5–7.0 | NM | Uncultured organism (Rabbit cecum metagenome) | [67] |
| BGL7 | 3 | 50 °C | 6.5 | NM | Uncultured organism (Termite gut metagenome) | [72] |
| LAB25g2 | 3 | 55 °C | 4.5 | 82%; 50 °C; 5 d | Uncultured organism (Cow rumen metagenome) | [68] |
| SRF2g14 | 3 | 55 °C | 5.0 | 50%; 50 °C; 18.06 h | Uncultured organism (Cow rumen metagenome) | [68] |
| SRF2g18 | 3 | 50 °C | 4.0 | 50%; 50 °C; 37.5 h | Uncultured organism (Cow rumen metagenome) | [68] |
| RuBGX1 | 3 | 50 °C | 6.0 | 62%; 50 °C; 10 min | Uncultured organism (Yak rumen metagenome) | [36] |
| JMB19063 | 3 | 50–55 °C | 6.5 | NM | Uncultured organism (Compost metagenome) | [37] |
| GlyA1 | 3 | 55 °C | 6.5 | NM | Uncultured organism (Cow rumen metagenome) | [69] |
| Bgx1 | 30 | 50 °C | 4.0–6.0 | NM | Oomycota (Phytophthora infestans) | [182] |
| SSO3039 | 116 | >70 °C | 4.0 | >70%; 65 °C; 48 h >50%; 85 °C; 8 h | Arquea (Sulfolobus solfataricus) | [183] |
| TxGH116 | 116 | 85 °C | 6.0 | NM | Bacteria (Thermoanaerobacterium xylanolyticum) | [184] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escuder-Rodríguez, J.-J.; DeCastro, M.-E.; Cerdán, M.-E.; Rodríguez-Belmonte, E.; Becerra, M.; González-Siso, M.-I. Cellulases from Thermophiles Found by Metagenomics. Microorganisms 2018, 6, 66. https://doi.org/10.3390/microorganisms6030066
Escuder-Rodríguez J-J, DeCastro M-E, Cerdán M-E, Rodríguez-Belmonte E, Becerra M, González-Siso M-I. Cellulases from Thermophiles Found by Metagenomics. Microorganisms. 2018; 6(3):66. https://doi.org/10.3390/microorganisms6030066
Chicago/Turabian StyleEscuder-Rodríguez, Juan-José, María-Eugenia DeCastro, María-Esperanza Cerdán, Esther Rodríguez-Belmonte, Manuel Becerra, and María-Isabel González-Siso. 2018. "Cellulases from Thermophiles Found by Metagenomics" Microorganisms 6, no. 3: 66. https://doi.org/10.3390/microorganisms6030066
APA StyleEscuder-Rodríguez, J.-J., DeCastro, M.-E., Cerdán, M.-E., Rodríguez-Belmonte, E., Becerra, M., & González-Siso, M.-I. (2018). Cellulases from Thermophiles Found by Metagenomics. Microorganisms, 6(3), 66. https://doi.org/10.3390/microorganisms6030066