Probiotic Bifunctionality of Bacillus subtilis—Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
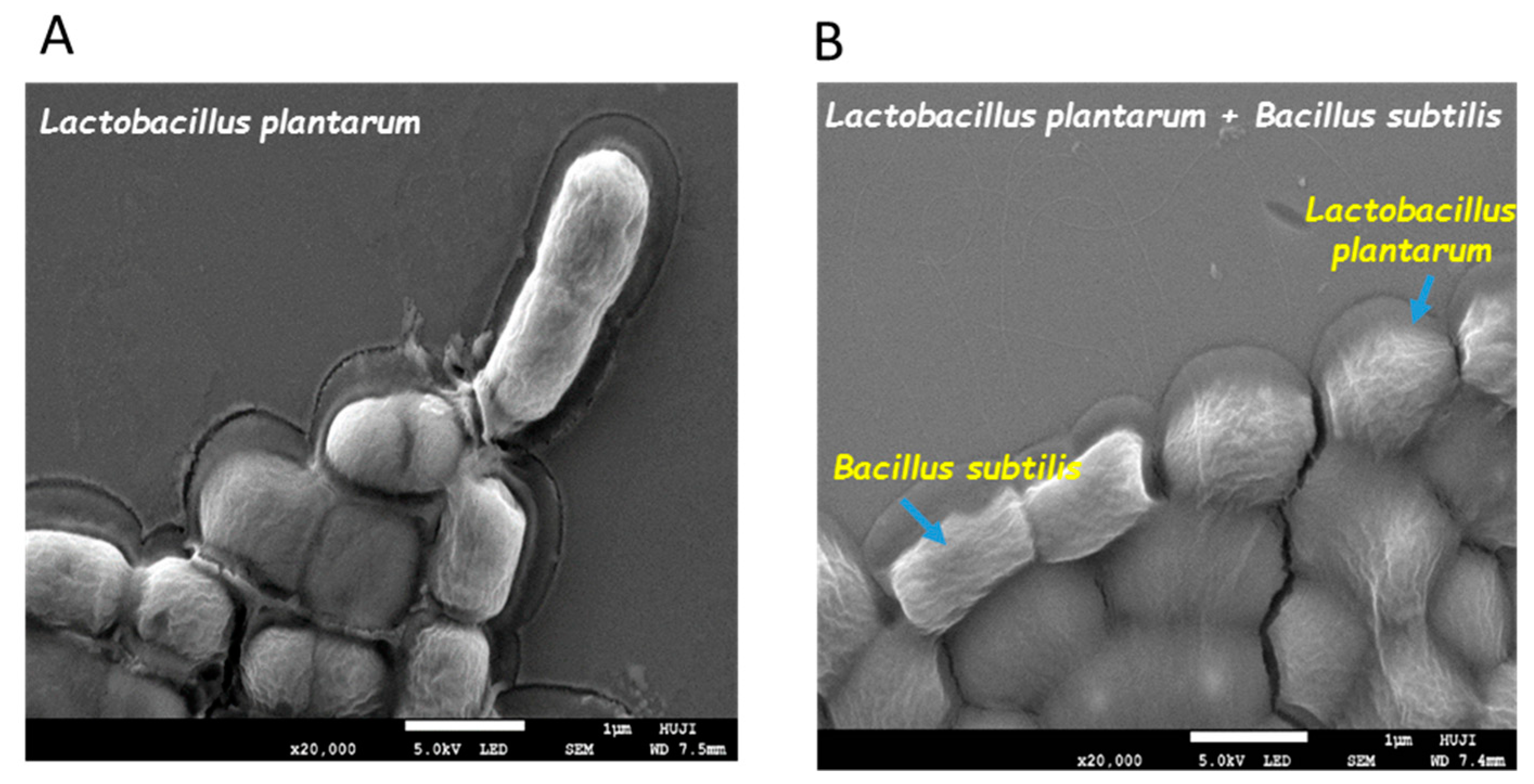
2.2. Visualizing Biofilm-Forming Cells Using Confocal Laser Scanning Microscopy (CLSM)
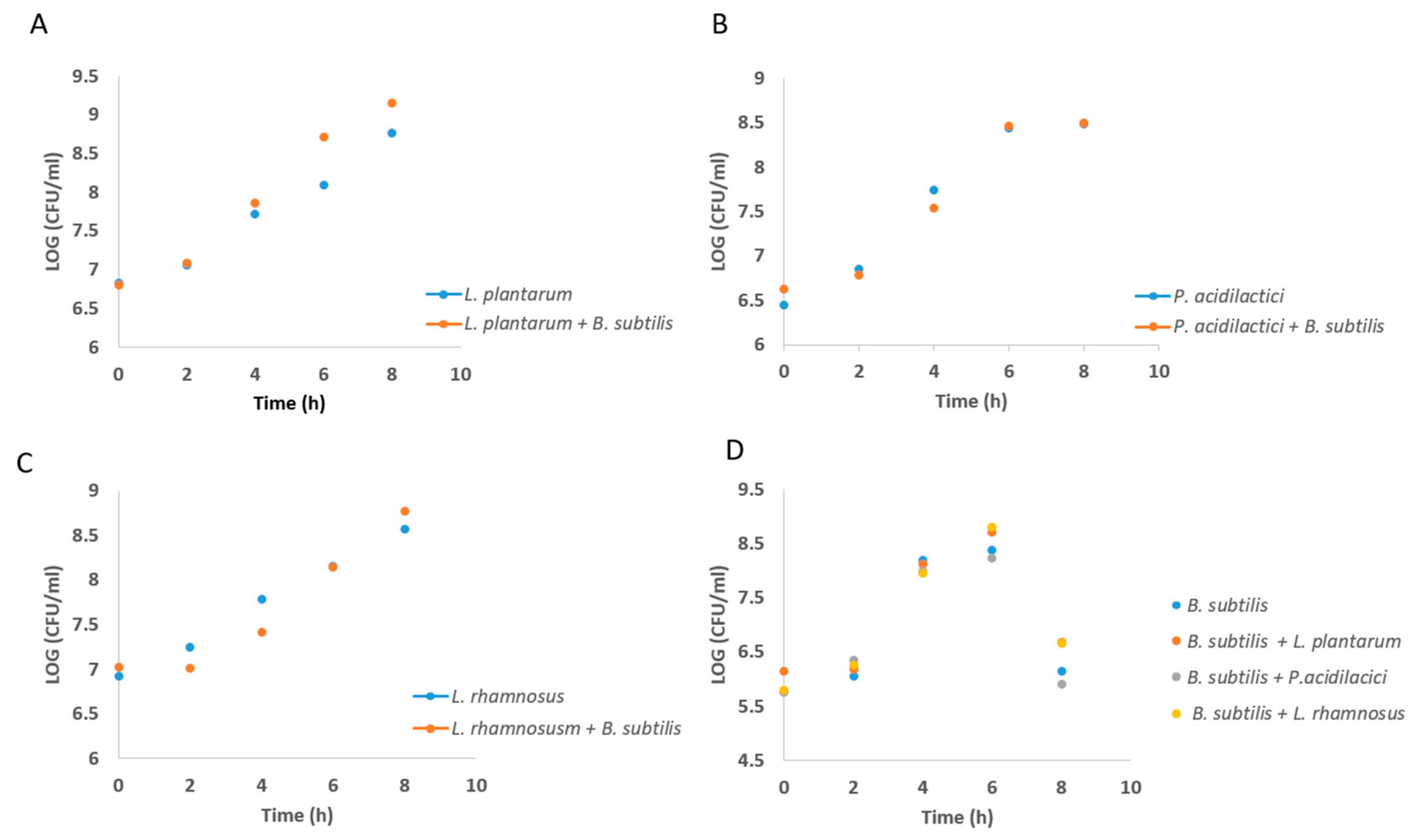
2.3. Growth Curve Analysis of Lab During Growth in Co-Culture
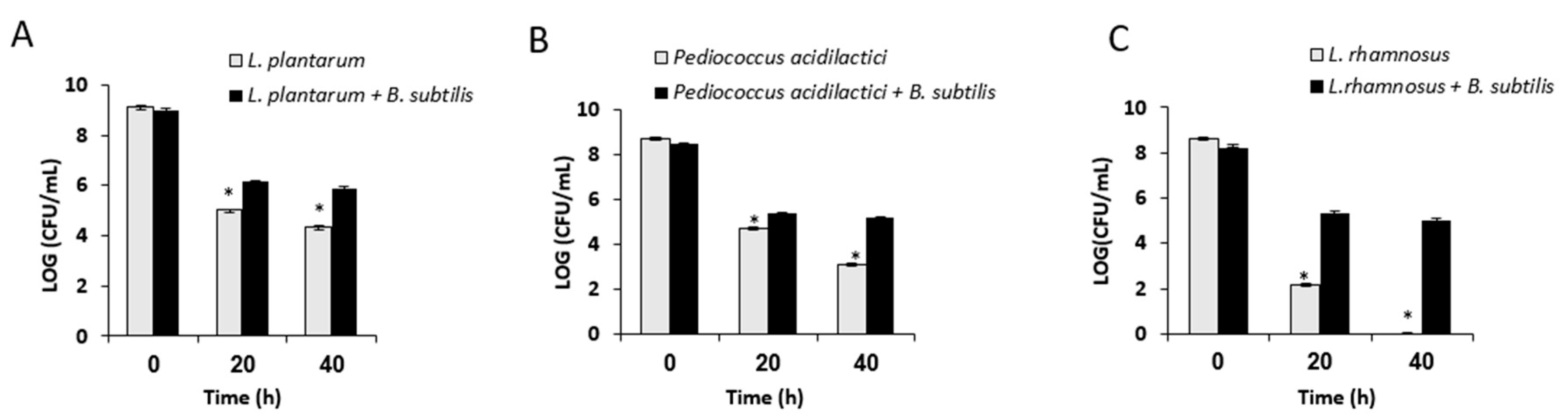
2.4. Analysis of Survival Rates Following Desiccation Treatment
2.5. Visualizing Co-Culture Biofilm Following Desiccation Treatment Using Scanning Electron Microscopy (SEM)
2.6. Analysis of Survival Rates Following Freeze-Drying
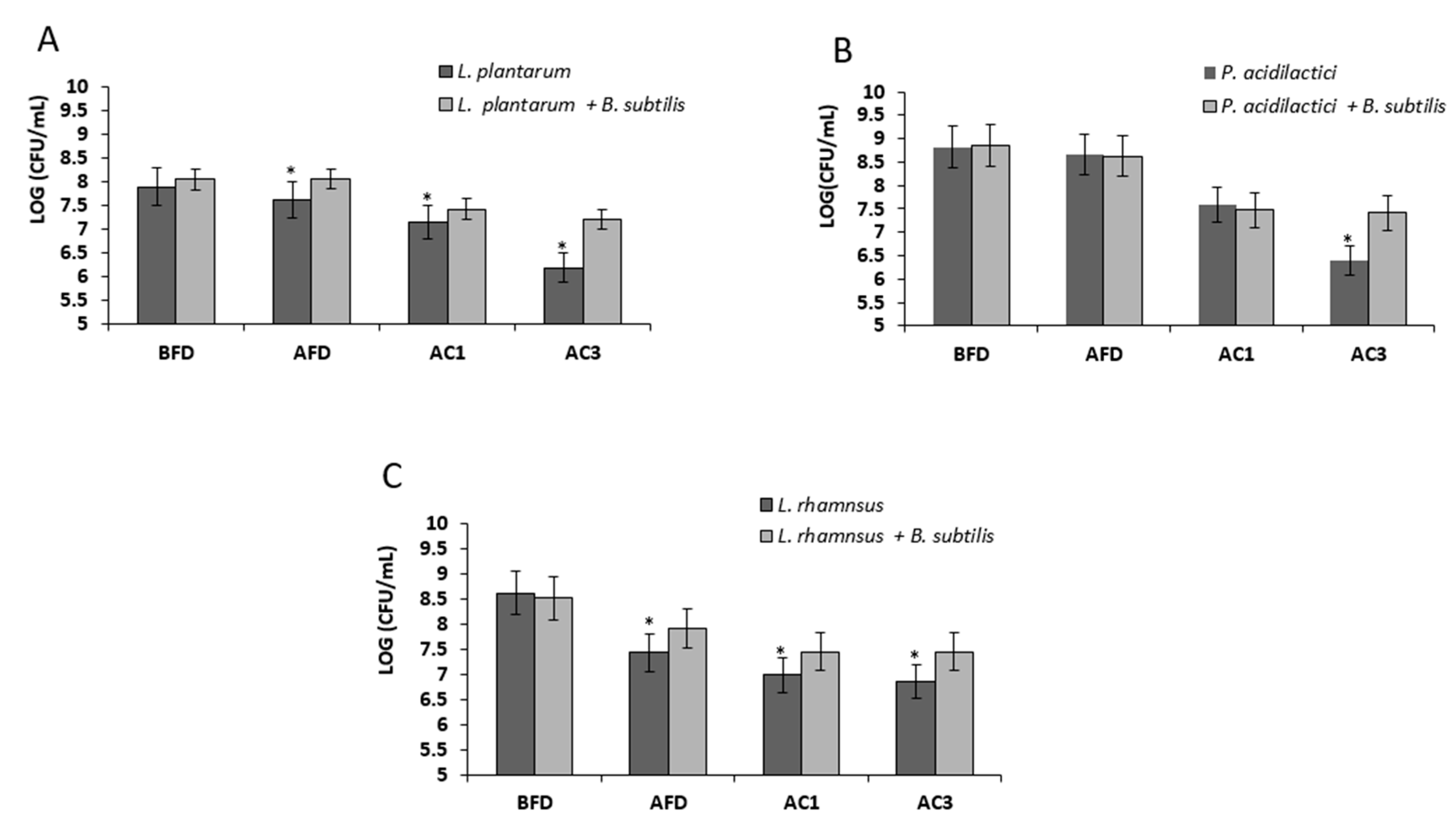
2.7. Analysis of Survival Rates Following Transition within In Vitro Digestion System
2.8. Determining the Effect of Conditioning Supernatant (CSN) on S. aureus Biofilm Formation
2.9. Biofilm Quantitation Assay
2.10. Effect of Cell-Free Culture Supernatant on S. aureus Growth
2.11. Confocal Laser Scan Microscopy (CLSM) Analysis
2.12. Real-Time PCR
2.13. Statistical Analysis
3. Results
3.1. Formation of Mutual Probiotic Biofilm of B. subtilis with Lactic Acid Bacteria (LAB)
3.2. Growth in Mutual Biofilm Increases the Survivability of the LAB during Desiccation
3.3. Growth in Mutual Biofilm Increases the Survivability of the LAB during Acid Stress Following Freeze-Drying
3.4. Bio-Coating Retains the LAB Survivability during In Vitro Gastrointestinal Digestion Following Freeze-Drying
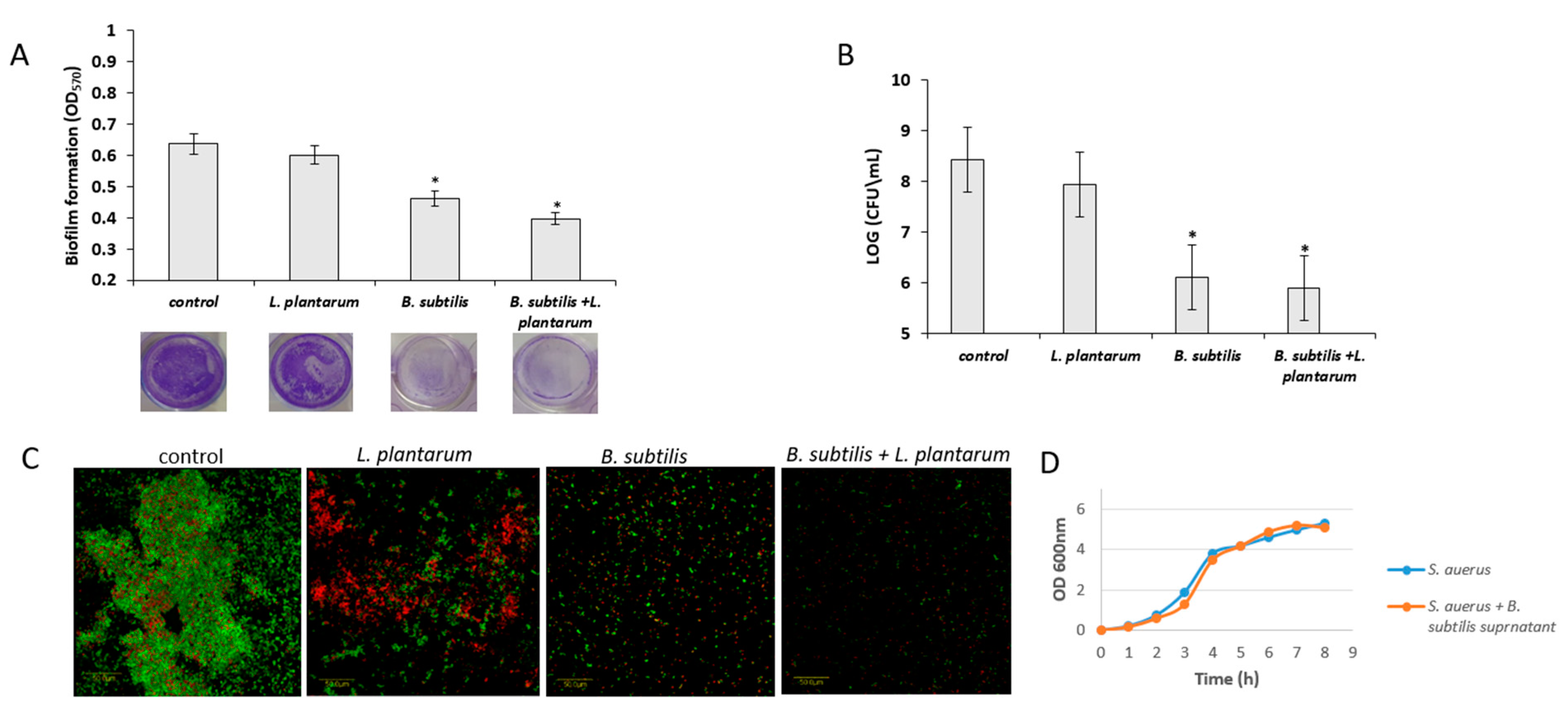
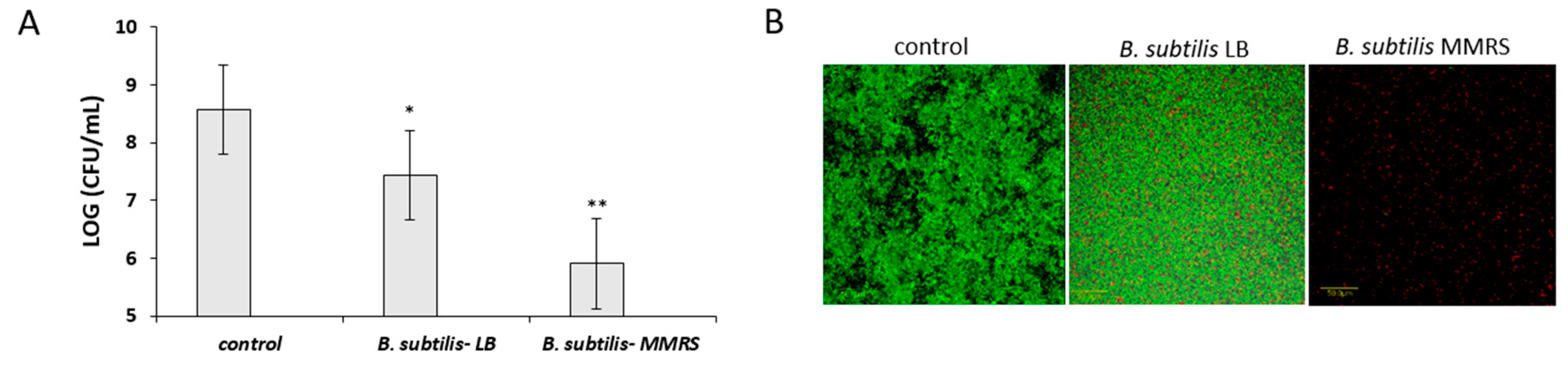
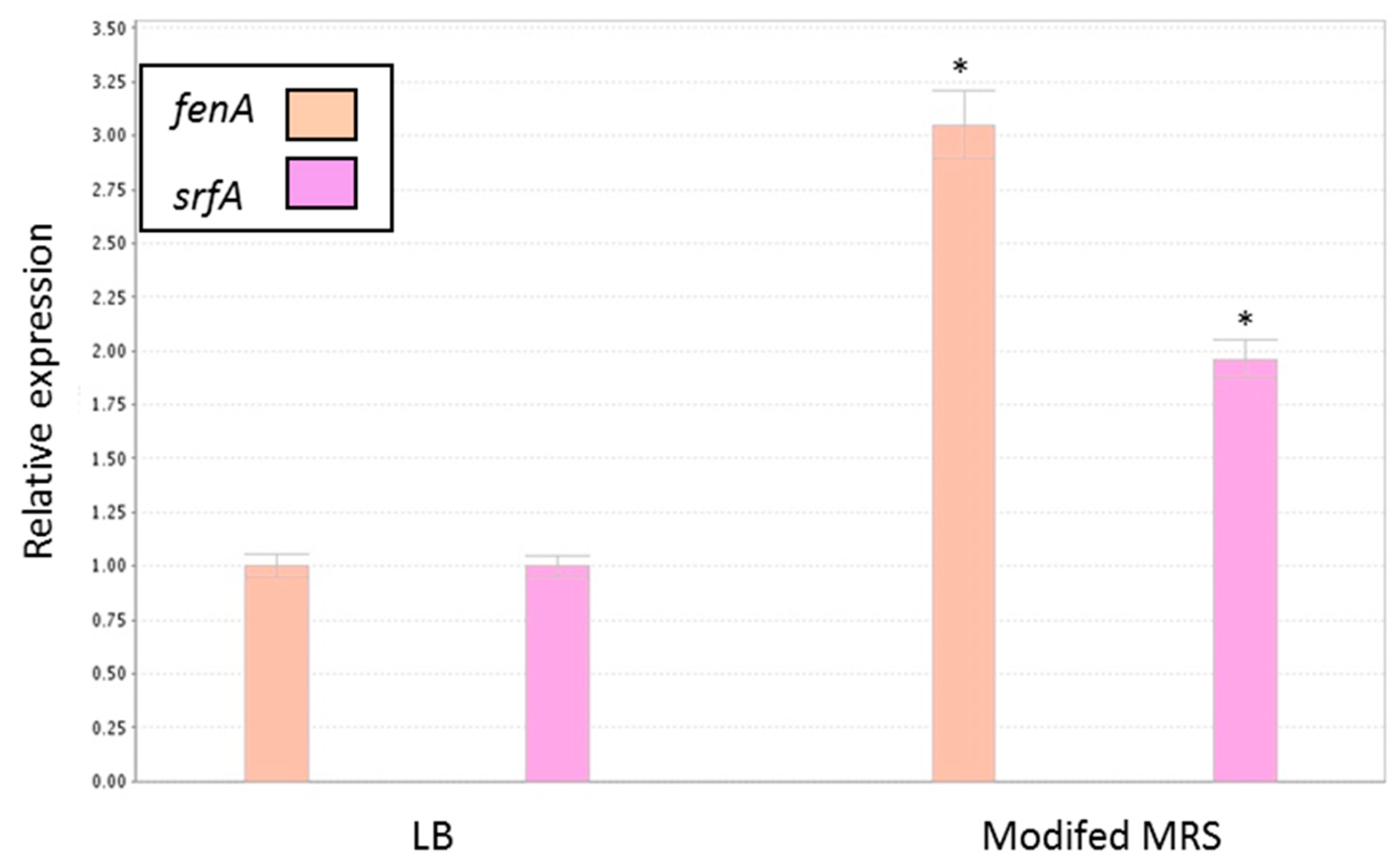
3.5. Antagonistic Effect of Probiotic Cells against the Biofilm-Forming, S. aureus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reid, G.; Jass, J.; Sebulsky, M.T.; McCormick, J.K. Potential uses of probiotics in clinical practice. Clin. Microbiol. Rev. 2003, 16, 658–672. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Haberer, P.; Geisen, R.; Björkroth, J.; Schillinger, U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 2001, 73, 365S–373S. [Google Scholar] [CrossRef]
- Anukam, K.C.; Reid, G. Probiotics: 100 years (1907–2007) after Elie Metchnikoff’s Observation. Commun. Curr. Res. Educ. Top. Trends Appl. Microbiol. 2007, 1, 466–474. [Google Scholar]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.S.; Villaruz, A.E.; Chiou, J.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; et al. Pathogen elimination by probiotic Bacillus via signalling interference. Nature 2018, 562, 532. [Google Scholar]
- Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- Servin, A.L. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 2004, 28, 405–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aw, W.; Fukuda, S. Protective effects of bifidobacteria against enteropathogens. Microb. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Iaconelli, C.; Lemetais, G.; Kechaou, N.; Chain, F.; Bermúdez-Humarán, L.G.; Langella, P.; Gervais, P.; Beney, L. Drying process strongly affects probiotics viability and functionalities. J. Biotechnol. 2015, 214, 17–26. [Google Scholar] [CrossRef]
- Marcial-Coba, M.S.; Knochel, S.; Nielsen, D.S. Low-moisture food matrices as probiotic carriers. FEMS Microbiol. Lett. 2019, 366. [Google Scholar] [CrossRef]
- Sanchez, B.; Champomier-Vergès, M.C.; Collado Mdel, C.; Anglade, P.; Baraige, F.; Sanz, Y.; de los Reyes-Gavilán, C.G.; Margolles, A.; Zagorec, M. Low-pH adaptation and the acid tolerance response of Bifidobacterium longum biotype longum. Appl. Environ. Microbiol. 2007, 73, 6450–6459. [Google Scholar] [CrossRef] [PubMed]
- Bron, P.A.; Marco, M.; Hoffer, S.M.; Van Mullekom, E.; De Vos, W.M.; Kleerebezem, M. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 2004, 186, 7829–7835. [Google Scholar] [CrossRef] [PubMed]
- Justice, S.S.; Harrison, A.; Becknell, B.; Mason, K.M. Bacterial differentiation, development, and disease: Mechanisms for survival. FEMS Microbiol. Lett. 2014, 360, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wingender, J.; Neu, T.R.; Flemming, H.C. What are Bacterial Extracellular Polymeric Substances? In Microbial Extracellular Polymeric Substances; Springer: Berlin/Heidelberg, Germany, 1999; pp. 1–19. [Google Scholar]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Webb, J.S.; Givskov, M.; Kjelleberg, S. Bacterial biofilms: Prokaryotic adventures in multicellularity. Curr. Opin. Microbiol. 2003, 6, 578–585. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.B.; Firestone, M.K. Relationship between Desiccation and Exopolysaccharide Production in a Soil Pseudomonas sp. Appl. Environ. Microbiol. 1992, 58, 1284–1291. [Google Scholar] [PubMed]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial Extracellular Polysaccharides in Biofilm Formation and Function. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilinskaya, O.N.; Ulyanova, V.V.; Yarullina, D.R.; Gataullin, I.G. Secretome of Intestinal Bacilli: A Natural Guard against Pathologies. Front. Microbiol. 2017, 8, 1666. [Google Scholar] [CrossRef] [PubMed]
- Batista, M.T.; Souza, R.D.; Paccez, J.D.; Luiz, W.B.; Ferreira, E.L.; Cavalcante, R.C.; Ferreira, R.C.; Ferreira, L.C. Gut Adhesive Bacillus subtilis Spores as a Platform for Mucosal Delivery of Antigens. Infect. Immun. 2014, 82, 1414–1423. [Google Scholar] [CrossRef] [Green Version]
- Shemesh, M.; Chai, Y. A combination of glycerol and manganese promotes biofilm formation in Bacillus subtilis via histidine kinase KinD signaling. J. Bacteriol. 2013, 195, 2747–2754. [Google Scholar] [CrossRef]
- Pasvolsky, R.; Zakin, V.; Ostrova, I.; Shemesh, M. Butyric acid released during milk lipolysis triggers biofilm formation of Bacillus species. Int. J. Food Microbiol. 2014, 181, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Vlamakis, H.; Chai, Y.; Beauregard, P.; Losick, R.; Kolter, R. Sticking together: building a biofilm the Bacillus subtilis way. Nat. Rev. Microbiol. 2013, 11, 157–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefevre, M.; Racedo, S.M.; Ripert, G.; Housez, B.; Cazaubiel, M.; Maudet, C.; Jüsten, P.; Marteau, P.; Urdaci, M.C. Probiotic strain Bacillus subtilis CU1 stimulates immune system of elderly during common infectious disease period: a randomized, double-blind placebo-controlled study. Immun. Ageing. 2015, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Lefevre, M.; Racedo, S.M.; Denayrolles, M.; Ripert, G.; Desfougères, T.; Lobach, A.R.; Simon, R.; Pélerin, F.; Jüsten, P.; Urdaci, M.C. Safety assessment of Bacillus subtilis CU1 for use as a probiotic in humans. Regul. Toxicol. Pharmacol. 2017, 83, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosoi, T.; Ametani, A.; Kiuchi, K.; Kaminogawa, S. Improved growth and viability of lactobacilli in the presence of Bacillus subtilis (natto), catalase, or subtilisin. Can. J. Microbiol. 2000, 46, 892–897. [Google Scholar] [CrossRef]
- Urdaci, M.C.; Pinchuk, I. Antimicrobial activity of Bacillus probiotics. In Bacterial Spore Formers–Probiotics and Emerging Applications; Horizon Bioscience: Norfolk, UK, 2004; pp. 171–182. [Google Scholar]
- Khochamit, N.; Siripornadulsil, S.; Sukon, P.; Siripornadulsil, W. Antibacterial activity and genotypic-phenotypic characteristics of bacteriocin-producing Bacillus subtilis KKU213: Potential as a probiotic strain. Microbiol. Res. 2015, 170, 36–50. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, L.; Xu, X.; Jiang, C.; Shi, J.; Zhang, Y.; Liu, L.; Lei, S.; Shao, D.; Huang, Q. Potential of Bacillus subtilis lipopeptides in anti-cancer I: induction of apoptosis and paraptosis and inhibition of autophagy in K562 cells. AMB Express 2018, 8, 78. [Google Scholar] [CrossRef]
- Zhao, H.; Shao, D.; Jiang, C.; Shi, J.; Li, Q.; Huang, Q.; Rajoka, M.S.R.; Yang, H.; Jin, M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017, 101, 5951–5960. [Google Scholar] [CrossRef]
- Zeriouh, H.; de Vicente, A.; Pérez-García, A.; Romero, D. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 2014, 16, 2196–2211. [Google Scholar] [CrossRef]
- López, D.; Fischbach, M.A.; Chu, F.; Losick, R.; Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2009, 106, 280–285. [Google Scholar] [CrossRef]
- Yahav, S.; Berkovich, Z.; Ostrov, I.; Reifen, R.; Shemesh, M. Encapsulation of beneficial probiotic bacteria in extracellular matrix from biofilm-forming Bacillus subtilis. Artif. Cells Nanomed. Biotechnol. 2018, 46, 974–982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, X.C.; Stanton, C.; Fitzgerald, G.F.; Daly, C.; Ross, R.P. Anhydrobiotics: The challenges of drying probiotic cultures. Food Chem. 2008, 106, 1406–1416. [Google Scholar] [CrossRef]
- Chai, Y.; Chu, F.; Kolter, R.; Losick, R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 67, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.A.; Herman, N.; White, P.A.; Vesey, G. Preservation of micro-organisms by drying; a review. J. Microbiol. Methods 2006, 66, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, W.; Zhu, X.; Zhao, H.; Lu, Y.; Zhang, C.; Lu, Z. Surfactin effectively inhibits Staphylococcus aureus adhesion and biofilm formation on surfaces. Appl. Microbiol. Biotechnol. 2019, 103, 4565–4574. [Google Scholar] [CrossRef] [PubMed]
- Flores-Belmonta, I.A.; Palou, E.; López-Malo, A.; Jiménez-Munguía, M.T. Simple and double microencapsulation of Lactobacillus acidophilus with chitosan using spray drying. Int. J. Food Stud. 2015. [Google Scholar] [CrossRef]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol 2000, 182, 2675–2679. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The Microbial “Protective Clothing” in Extreme Environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Pasvolsky, R.; Zakin, V. External pH is a cue for the behavioral switch that determines surface motility and biofilm formation of Alicyclobacillus acidoterrestris. J. Food Prot 2014, 77, 1418–1423. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Kolter, R.; Losick, R. The Biocide Chlorine Dioxide Stimulates Biofilm Formation in Bacillus subtilis by Activation of the Histidine Kinase KinC. J. Bacteriol. 2010, 192, 6352–6356. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Razafindralambo, H.; Blecker, C.; N’Yapo, C.; Thonart, P.; Delvigne, F. Stochastic exposure to sub-lethal high temperature enhances exopolysaccharides (EPS) excretion and improves Bifidobacterium bifidum cell survival to freeze-drying. Biochem. Eng. J. 2014, 88, 85–94. [Google Scholar] [CrossRef]
- Carvalho, A.S.; Silva, J.; Ho, P.; Teixeira, P.; Malcata, F.X.; Gibbs, P. Relevant factors for the preparation of freeze-dried lactic acid bacteria. Int. Dairy J. 2004, 14, 835–847. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Truong, D.H.; Kouhoundé, S.; Ly, S.; Razafindralambo, H.; Delvigne, F. Biochemical Engineering Approaches for Increasing Viability and Functionality of Probiotic Bacteria. Int. J. Mol. Sci. 2016, 17, 867. [Google Scholar] [CrossRef] [PubMed]
- Suva, M.A.; Sureja, V.P.; Kheni, D.B. Novel insight on probiotic Bacillus subtilis: Mechanism of action and clinical applications. J. Curr. Res. Sci. Med. 2016, 2, 65. [Google Scholar] [CrossRef]
- Gonzalez, D.J.; Haste, N.M.; Hollands, A.; Fleming, T.C.; Hamby, M.; Pogliano, K.; Dorrestein, P.C. Microbial competition between Bacillus subtilis and Staphylococcus aureus monitored by imaging mass spectrometry. Microbiol. Sgm. 2011, 157, 2485–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimelman, H.; Shemesh, M. Probiotic Bifunctionality of Bacillus subtilis—Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus. Microorganisms 2019, 7, 407. https://doi.org/10.3390/microorganisms7100407
Kimelman H, Shemesh M. Probiotic Bifunctionality of Bacillus subtilis—Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus. Microorganisms. 2019; 7(10):407. https://doi.org/10.3390/microorganisms7100407
Chicago/Turabian StyleKimelman, Hadar, and Moshe Shemesh. 2019. "Probiotic Bifunctionality of Bacillus subtilis—Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus" Microorganisms 7, no. 10: 407. https://doi.org/10.3390/microorganisms7100407
APA StyleKimelman, H., & Shemesh, M. (2019). Probiotic Bifunctionality of Bacillus subtilis—Rescuing Lactic Acid Bacteria from Desiccation and Antagonizing Pathogenic Staphylococcus aureus. Microorganisms, 7(10), 407. https://doi.org/10.3390/microorganisms7100407




