Comparison of Different Invasive and Non-Invasive Methods to Characterize Intestinal Microbiota throughout a Production Cycle of Broiler Chickens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Data Collection
2.3. Sampling Methods
2.4. DNA Extraction
2.5. 16S rRNA Gene Amplification Analysis
2.6. Statistical Data Analysis
3. Results
3.1. Technical Results of 16S rRNA Gene Sequencing
3.2. Alpha Diversity Analysis across Different Sample Types
3.3. Beta Diversity Analysis across Different Sample Types
3.4. The Effect of Age on the Bacterial Microbiota across Different Sample Types
3.5. The Effect of Farm on the Bacterial Microbiota across Different Sample Types
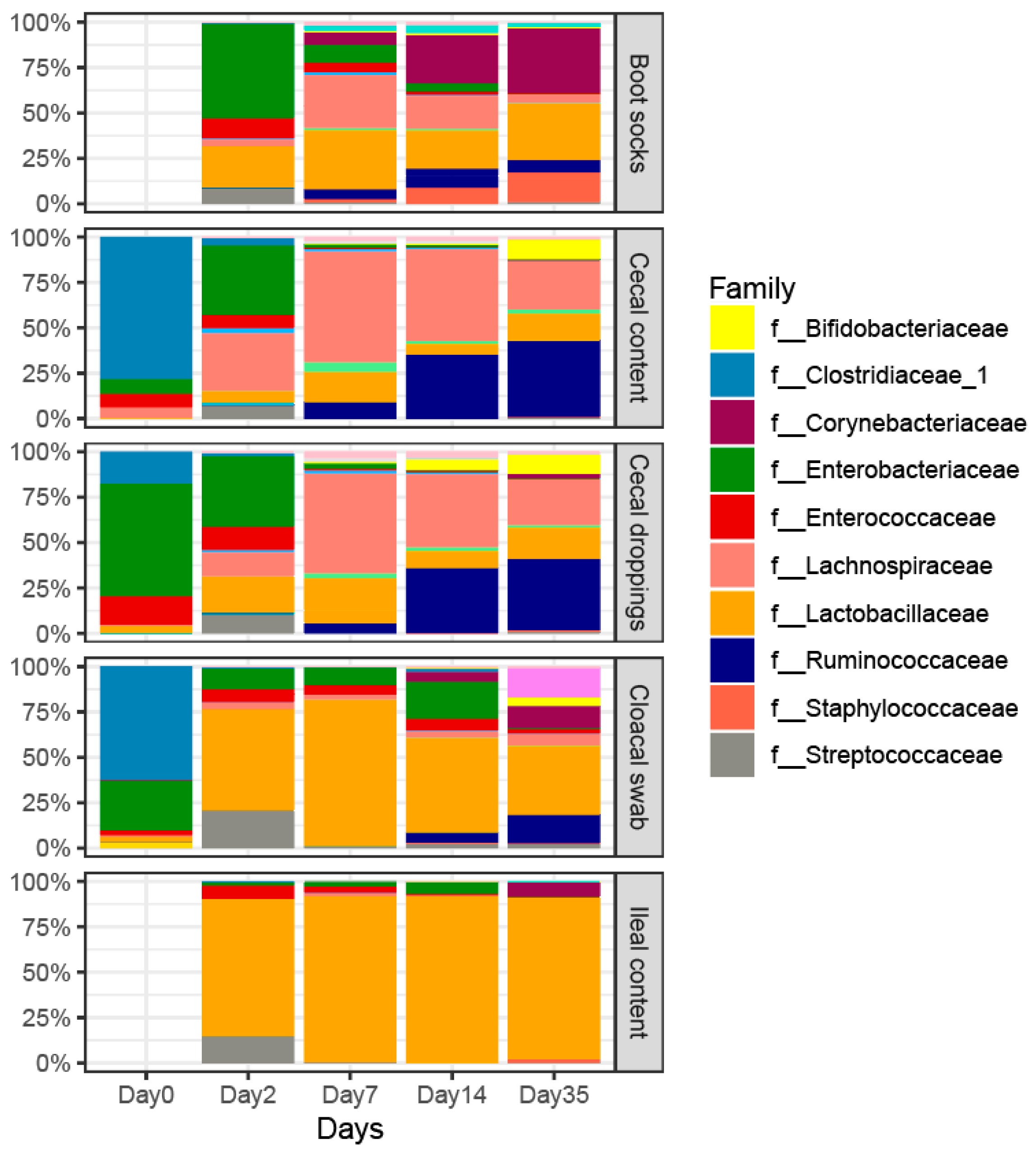
3.6. Microbial Taxa That Differ across the Invasive and Non-Invasive Sample Types
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Awad, W.A.; Mann, E.; Dzieciol, M.; Hess, C.; Schmitz-Esser, S.; Wagner, M.; Hess, M. Age-Related Differences in the Luminal and Mucosa-Associated Gut Microbiome of Broiler Chickens and Shifts Associated with Campylobacter jejuni Infection. Front. Cell. Infect. Microbiol. 2016, 6, 154. [Google Scholar] [CrossRef] [Green Version]
- Ijaz, U.Z.; Sivaloganathan, L.; McKenna, A.; Richmond, A.; Kelly, C.; Linton, M.; Stratakos, A.C.; Lavery, U.; Elmi, A.; Wren, B.W.; et al. Comprehensive Longitudinal Microbiome Analysis of the Chicken Cecum Reveals a Shift From Competitive to Environmental Drivers and a Window of Opportunity for Campylobacter. Front. Microbiol. 2018, 9, 2452. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Willer, T.; Pielsticker, C.; Gerzova, L.; Rychlik, I.; Rautenschlein, S. Differences in host breed and diet influence colonization by Campylobacter jejuni and induction of local immune responses in chicken. Gut Pathog. 2016, 8, 56. [Google Scholar] [CrossRef]
- Johnson, T.J.; Youmans, B.P.; Noll, S.; Cardona, C.; Evans, N.P.; Karnezos, T.P.; Ngunjiri, J.M.; Abundo, M.C.; Lee, C.W. A Consistent and Predictable Commercial Broiler Chicken Bacterial Microbiota in Antibiotic-Free Production Displays Strong Correlations with Performance. Appl. Environ. Microbiol. 2018, 84, e00362-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, D.; Denman, S.E.; Hughes, R.J.; Geier, M.S.; Crowley, T.M.; Chen, H.; Haring, V.R.; Moore, R.J. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl. Microbiol. Biotechnol. 2012, 96, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Yeoman, C.J.; Chia, N.; Jeraldo, P.; Sipos, M.; Goldenfeld, N.D.; White, B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health. Res. Rev. 2012, 13, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinttilä, T.; Apajalahti, J. Intestinal microbiota and metabolites-Implications for broiler chicken health and performance. J. Appl. Poult. Res. 2013, 22, 647–658. [Google Scholar] [CrossRef]
- Mohammed, A.A.; Jiang, S.; Jacobs, J.A.; Cheng, H.W. Effect of a synbiotic supplement on cecal microbial ecology, antioxidant status, and immune response of broiler chickens reared under heat stress. Poult. Sci. 2019, 98, 4408–4415. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Di Rienzi, S.C.; Poole, A.C.; Koren, O.; Walters, W.A.; Caporaso, J.G.; Knight, R.; Ley, R.E. Conducting a microbiome study. Cell 2014, 158, 250–262. [Google Scholar] [CrossRef]
- Corrigan, A.; de Leeuw, M.; Penaud-Frezet, S.; Dimova, D.; Murphy, R.A. Phylogenetic and functional alterations in bacterial community compositions in broiler ceca as a result of mannan oligosaccharide supplementation. Appl. Environ. Microbiol. 2015, 81, 3460–3470. [Google Scholar] [CrossRef]
- Ballou, A.L.; Ali, R.A.; Mendoza, M.A.; Ellis, J.C.; Hassan, H.M.; Croom, W.J.; Koci, M.D. Development of the Chick Microbiome: How Early Exposure Influences Future Microbial Diversity. Front. Vet. Sci. 2016, 3, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, A.J.; Vangay, P.; Al-Ghalith, G.A.; Hillmann, B.M.; Ward, T.L.; Shields-Cutler, R.R.; Kim, A.D.; Shmagel, A.K.; Syed, A.N.; Students, P.M.C. Daily Sampling Reveals Personalized Diet-Microbiome Associations in Humans. Cell Host Microbe 2019, 25, 789–802. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.C.; Berry, D. Microbial nutrient niches in the gut. Environ Microbiol. 2017, 19, 1366–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.H.; Kim, G.B.; Cha, C.J. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult. Sci. 2014, 93, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef] [PubMed]
- Svihus, B. Function of the digestive system. J. Appl. Poult. Res. 2014, 23, 306–314. [Google Scholar] [CrossRef]
- Ducatelle, R.; Goossens, E.; De Meyer, F.; Eeckhaut, V.; Antonissen, G.; Haesebrouck, F.; Van Immerseel, F. Biomarkers for monitoring intestinal health in poultry: Present status and future perspectives. Vet. Res. 2018, 49, 43. [Google Scholar] [CrossRef]
- Clench, M.H.; Mathias, J.R. The avian cecum: A review. Wilson Bull. 1995, 107, 93–121. [Google Scholar]
- Józefiak, D.; Rutkowski, A.; Martin, S. Carbohydrate fermentation in the avian ceca: A review. Anim. Feed. Sci. Technol. 2004, 113, 1–15. [Google Scholar] [CrossRef]
- Yegani, M.; Korver, D. Factors affecting intestinal health in poultry. Poult. Sci. 2008, 87, 2052–2063. [Google Scholar] [CrossRef]
- Stanley, D.; Geier, M.S.; Chen, H.; Hughes, R.J.; Moore, R.J. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. 2015, 15, 51. [Google Scholar] [CrossRef] [PubMed]
- Noy, Y.; Sklan, D. Digestion and absorption in the young chick. Poult. Sci. 1995, 74, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Zeevi, D.; Levy, M.; Segal, E.; Elinav, E. A day in the life of the meta-organism: Diurnal rhythms of the intestinal microbiome and its host. Gut Microbes 2015, 6, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Si, W.; Forster, R.J.; Huang, R.; Yu, H.; Yin, Y.; Yang, C.; Han, Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: From crops to ceca. FEMS Microbiol. Ecol. 2007, 59, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Owens, B.; Tucker, L.; Collins, M.A.; McCracken, K.J. Effects of different feed additives alone or in combination on broiler performance, gut microflora and ileal histology. Bri. Poult. Sci. 2008, 49, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, K.M.; Thompson, K.L.; Einstein, M.E.; Applegate, T.J.; Patterson, J.A. Influence of stressors on normal intestinal microbiota, intestinal morphology, and susceptibility to Salmonella Enteritidis colonization in broilers. Poult. Sci. 2008, 87, 1734–1741. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, J.; Taminiau, B.; Janssens, G.P.; De Beenhouwer, M.; Delhalle, L.; Daube, G.; Coopman, F. Cecal drop reflects the chickens’ cecal microbiome, fecal drop does not. J. Microbiol. Methods 2015, 117, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Videvall, E.; Strandh, M.; Engelbrecht, A.; Cloete, S.; Cornwallis, C.K. Measuring the gut microbiome in birds: Comparison of faecal and cloacal sampling. Mol. Ecol. Resour. 2018, 18, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Lennon, G.; O’Sullivan, O.; Docherty, N.; Balfe, A.; Maguire, A.; Mulcahy, H.E.; Doherty, G.; O’Donoghue, D.; Hyland, J.; et al. Spatial variation of the colonic microbiota in patients with ulcerative colitis and control volunteers. Gut 2015, 64, 1553–1561. [Google Scholar] [CrossRef] [Green Version]
- Vandeputte, D.; Falony, G.; Vieira-Silva, S.; Tito, R.Y.; Joossens, M.; Raes, J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016, 65, 57–62. [Google Scholar] [CrossRef]
- Wang, F.; Kaplan, J.L.; Gold, B.D.; Bhasin, M.K.; Ward, N.L.; Kellermayer, R.; Kirschner, B.S.; Heyman, M.B.; Dowd, S.E.; Cox, S.B.; et al. Detecting Microbial Dysbiosis Associated with Pediatric Crohn Disease Despite the High Variability of the Gut Microbiota. Cell Rep. 2016, 14, 945–955. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vazquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014, 15, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Geier, M.S.; Hughes, R.J.; Denman, S.E.; Moore, R.J. Highly variable microbiota development in the chicken gastrointestinal tract. PLoS ONE 2013, 8, e84290. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, A.; Fravalo, P.; Yergeau, E.; Arsenault, J.; Lahaye, L.; Letellier, A. Chicken Caecal Microbiome Modifications Induced by Campylobacter jejuni Colonization and by a Non-Antibiotic Feed Additive. PLoS ONE 2015, 10, e0131978. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Fron. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, E.E.; Stanley, D.; Hughes, R.J.; Moore, R.J. The time-course of broiler intestinal microbiota development after administration of cecal contents to incubating eggs. PeerJ 2017, 5, e3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedroso, A.A.; Menten, J.F.M.; Lambais, M.R. The structure of bacterial community in the intestines of newly hatched chicks. J. Appl. Poult. Res. 2005, 14, 232–237. [Google Scholar] [CrossRef]
- Williams, R. Epidemiological studies of coccidiosis in the domesticated fowl (Gallus gallus): II. Physical condition and survival of Eimeria acervulina oocysts in poultry-house litter. Appl. Parasitol. 1995, 36, 90–96. [Google Scholar]
- Skov, M.N.; Carstensen, B.; Tornoe, N.; Madsen, M. Evaluation of sampling methods for the detection of Salmonella in broiler flocks. J. Appl. Microbiol. 1999, 86, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Berghaus, R.D.; Thayer, S.G.; Law, B.F.; Mild, R.M.; Hofacre, C.L.; Singer, R.S. Enumeration of Salmonella and Campylobacter spp. in environmental farm samples and processing plant carcass rinses from commercial broiler chicken flocks. Appl. Environ. Microbiol. 2013, 79, 4106–4114. [Google Scholar] [CrossRef]
- Ramiro-Garcia, J.; Hermes, G.D.A.; Giatsis, C.; Sipkema, D.; Zoetendal, E.G.; Schaap, P.J.; Smidt, H. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Research 2016, 5, 1791. [Google Scholar] [CrossRef]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Lamot, D.M.; Stegeman, J.A.; Smidt, H. Take care of the enironment: Housing conditions affect the interplay of nutritional interventions and intestinal microbiota in broiler chickens. Anim. Microbiome 2019, 1, 1–14. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Faith, D.P. The Role of the Phylogenetic Diversity Measure, PD, in Bio-informatics: Getting the Definition Right. Evolut. Bioinform. 2006, 2, 277–283. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monographs 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Jaccard, P. The distribution of the flora in the alpine zone. New Phytol. 1912, 11, 37–50. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and qualitative β diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S.; Blake, T.; Salojarvi, J. Tools for Microbiome Analysis in R. Version 1.5.28. 2017. Available online: http://microbiome.github.com/microbiome (accessed on 19 February 2019).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; Vegan: Community Ecology Package. R Package Version 1.17-4. Available online: http://cran.r-project.org (accessed on 19 February 2019).
- van der Wielen, P.W.; Keuzenkamp, D.A.; Lipman, L.J.; van Knapen, F.; Biesterveld, S. Spatial and temporal variation of the intestinal bacterial community in commercially raised broiler chickens during growth. Microb. Ecol. 2002, 44, 286–293. [Google Scholar] [CrossRef]
- Gong, J.; Yu, H.; Liu, T.; Gill, J.J.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 2008, 104, 1372–1382. [Google Scholar] [CrossRef]
- Torok, V.A.; Hughes, R.J.; Ophel-Keller, K.; Ali, M.; MacAlpine, R. Influence of different litter materials on cecal microbiota colonization in broiler chickens. Poult. Sci. 2009, 88, 2474–2481. [Google Scholar] [CrossRef]
- Jones, N.R.; Millman, C.; van der Es, M.; Hukelova, M.; Forbes, K.J.; Glover, C.; Haldenby, S.; Hunter, P.R.; Jackson, K.; O’Brien, S.J. Novel sampling method for assessing human-pathogen interactions in the natural environment using boot socks and citizen scientists, with application to campylobacter seasonality. Appl. Environ. Microbiol. 2017, 83, e00162-00117. [Google Scholar] [CrossRef]
- Wang, L.; Lilburn, M.; Yu, Z. Intestinal Microbiota of Broiler Chickens As Affected by Litter Management Regimens. Front. Microbiol. 2016, 7, 593. [Google Scholar] [CrossRef] [Green Version]
- De Cesare, A.; Caselli, E.; Lucchi, A.; Sala, C.; Parisi, A.; Manfreda, G.; Mazzacane, S. Impact of a probiotic-based cleaning product on the microbiological profile of broiler litters and chicken caeca microbiota. Poult. Sci. 2019, 98, 3602–3610. [Google Scholar] [CrossRef]
- Knarreborg, A.; Simon, M.A.; Engberg, R.M.; Jensen, B.B.; Tannock, G.W. Effects of dietary fat source and subtherapeutic levels of antibiotic on the bacterial community in the ileum of broiler chickens at various ages. Appl. Environ. Microbiol. 2002, 68, 5918–5924. [Google Scholar] [CrossRef]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef]
- Videnska, P.; Faldynova, M.; Juricova, H.; Babak, V.; Sisak, F.; Havlickova, H.; Rychlik, I. Chicken faecal microbiota and disturbances induced by single or repeated therapy with tetracycline and streptomycin. BMC Vet. Res. 2013, 9, 30. [Google Scholar] [CrossRef]
- Broom, L.J. Host(-)Microbe Interactions and Gut Health in Poultry-Focus on Innate Responses. Microorganisms 2019, 7, 139. [Google Scholar] [CrossRef]


| Sample Type | Phylogenetic Diversity | Chao 1 | Shannon | ||||
|---|---|---|---|---|---|---|---|
| χ2 | p-Value | χ2 | p-Value | χ2 | p-Value | ||
| Cecal content versus cloacal swab (n = 100) | 10.633 | 0.001 | 15.076 | <0.0001 | 26.947 | 2.1 × 10−7 | |
| Ileal content versus cloacal swab (n = 80) | 18.501 | 1.7 × 10−5 | 0.245 | 0.620 | 3.379 | 0.066 | |
| Cecal content versus cecal droppings (n = 97) | 3.604 | 0.058 | 6.623 | 0.010 | 6.116 | 0.013 | |
| Cecal content versus boot socks (n = 80) | 2.582 | 0.108 | 0.241 | 0.624 | 1.447 | 0.229 | |
| Cecal content n = 50 | Age | 43.848 | 6.9 × 10−9 | 38.147 | 1.7 × 10−7 | 37.252 | 1.6 × 10−7 |
| Farm | 0.350 | 0.554 | 0.001 | 0.938 | 0.115 | 0.734 | |
| Ileal content n = 40 | Age | 3.902 | 0.272 | 3.547 | 0.315 | 11.545 | 0.009 |
| Farm | 0.026 | 0.871 | 0.751 | 0.386 | 1.230 | 0.267 | |
| Cloacal swab n = 50 | Age | 14.888 | 0.005 | 13.255 | 0.010 | 13.917 | 0.007 |
| Farm | 2.688 | 0.101 | 2.765 | 0.096 | 0.886 | 0.347 | |
| Cecal dropping n = 47 | Age | 38.679 | 8.1 × 10−8 | 33.46 | 9.4 × 10−7 | 32.865 | 1.3 × 10−6 |
| Farm | 0.007 | 0.932 | 0.009 | 0.924 | 0.016 | 0.898 | |
| Boot socks n = 40 | Age | 17.729 | 5.7 × 10−4 | 24.527 | 1.9 × 10−5 | 22.945 | 4.1 × 10−5 |
| Farm | 4.801 | 0.029 | 4.120 | 0.042 | 3.484 | 0.062 | |
| Samples | n | Bray–Curtis | Jaccard | Unweighted UniFrac | Weighted UniFrac | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R2 | p | FDis | R2 | p | FDis | R2 | p | FDis | R2 | p | FDis | ||
| Cecal content vs. cloacal swab | 100 | 6.6 | 1 × 10−4 | 0.042 | 4.7 | 1 × 10−4 | 0.034 | 10.3 | 1 × 10−4 | 0.318 | 14.6 | 1 × 10−4 | 0.627 |
| Ileal content vs. cloacal swab | 80 | 3.2 | 0.003 | 0.685 | 2.4 | 0.014 | 0.672 | 6.5 | 2 × 10−4 | 0.569 | 10.7 | 1 × 10−4 | 0.002 |
| Cecal content vs. cecal droppings | 97 | 3.4 | 3 × 10−4 | 0.001 | 2.6 | 2 × 10−4 | 0.001 | 1.6 | 0.155 | 0.169 | 2.9 | 0.028 | 0.370 |
| Cecal content vs. boot socks | 80 | 8.5 | 1 × 10−4 | 0.001 | 6.2 | 1 × 10−4 | 0.001 | 10.7 | 1 × 10−4 | 0.626 | 22.7 | 1 × 10−4 | 0.682 |
| Cecal content Farm | 50 | 3.5 | 0.039 | 0.135 | 3.1 | 0.029 | 0.188 | 2.9 | 0.165 | 0.825 | 1.6 | 0.519 | 0.990 |
| Cecal content Age | 50 | 37.8 | 1 × 10−4 | 0.041 | 26.9 | 1 × 10−4 | 0.058 | 55.7 | 1 × 10−4 | 0.001 | 62.0 | 1 × 10−4 | 0.564 |
| Ileal content Farm | 40 | 6.5 | 0.006 | 0.884 | 5.5 | 0.005 | 0.887 | 3.4 | 0.174 | 0.012 | 5.1 | 0.019 | 0.970 |
| Ileal content Age | 40 | 23.0 | 1 × 10−4 | 0.005 | 18.8 | 1 × 10−4 | 0.007 | 29.6 | 1 × 10−4 | 0.959 | 18.3 | 1 × 10−4 | 0.245 |
| Cloacal swab Farm | 50 | 4.1 | 0.019 | 0.866 | 3.6 | 0.014 | 0.871 | 3.4 | 0.065 | 0.678 | 3.4 | 0.131 | 0.497 |
| Cloacal swab Age | 50 | 30.3 | 1 × 10−4 | 0.009 | 23.4 | 1 × 10−4 | 0.010 | 34.4 | 1 × 10−4 | 0.755 | 38.7 | 1 × 10−4 | 0.027 |
| Cecal dropping Farm | 47 | 3.8 | 0.061 | 0.827 | 3.5 | 0.033 | 0.841 | 3.9 | 0.110 | 0.782 | 2.0 | 0.364 | 0.674 |
| Cecal dropping Age | 47 | 42.7 | 1 × 10−4 | 0.937 | 30.5 | 1 × 10−4 | 0.882 | 64.5 | 1 × 10−4 | 0.006 | 68.9 | 1 × 10−4 | 0.206 |
| Boot sock Farm | 40 | 10.0 | 0.002 | 0.932 | 8.9 | 6 × 10−4 | 0.939 | 9.2 | 0.004 | 0.966 | 11.4 | 0.004 | 0.850 |
| Boot sock Age | 40 | 50.6 | 1 × 10−4 | 0.005 | 38.6 | 0.387 | 0.041 | 51.0 | 1 × 10−4 | 0.034 | 55.9 | 1 × 10−4 | 0.001 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kers, J.G.; Fischer, E.A.J.; Stegeman, J.A.; Smidt, H.; Velkers, F.C. Comparison of Different Invasive and Non-Invasive Methods to Characterize Intestinal Microbiota throughout a Production Cycle of Broiler Chickens. Microorganisms 2019, 7, 431. https://doi.org/10.3390/microorganisms7100431
Kers JG, Fischer EAJ, Stegeman JA, Smidt H, Velkers FC. Comparison of Different Invasive and Non-Invasive Methods to Characterize Intestinal Microbiota throughout a Production Cycle of Broiler Chickens. Microorganisms. 2019; 7(10):431. https://doi.org/10.3390/microorganisms7100431
Chicago/Turabian StyleKers, Jannigje G., Egil A.J. Fischer, J. Arjan Stegeman, Hauke Smidt, and Francisca C. Velkers. 2019. "Comparison of Different Invasive and Non-Invasive Methods to Characterize Intestinal Microbiota throughout a Production Cycle of Broiler Chickens" Microorganisms 7, no. 10: 431. https://doi.org/10.3390/microorganisms7100431