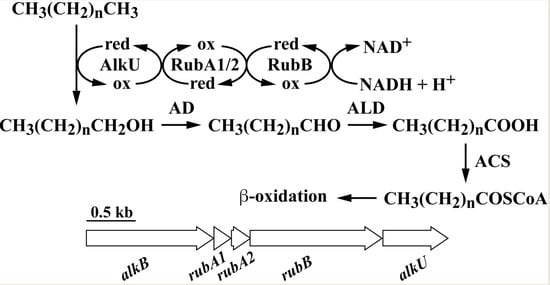
Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1
Abstract
Share and Cite
Gibu, N.; Kasai, D.; Ikawa, T.; Akiyama, E.; Fukuda, M. Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms 2019, 7, 479. https://doi.org/10.3390/microorganisms7110479
Gibu N, Kasai D, Ikawa T, Akiyama E, Fukuda M. Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms. 2019; 7(11):479. https://doi.org/10.3390/microorganisms7110479
Chicago/Turabian StyleGibu, Namiko, Daisuke Kasai, Takumi Ikawa, Emiko Akiyama, and Masao Fukuda. 2019. "Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1" Microorganisms 7, no. 11: 479. https://doi.org/10.3390/microorganisms7110479
APA StyleGibu, N., Kasai, D., Ikawa, T., Akiyama, E., & Fukuda, M. (2019). Characterization and Transcriptional Regulation of n-Alkane Hydroxylase Gene Cluster of Rhodococcus jostii RHA1. Microorganisms, 7(11), 479. https://doi.org/10.3390/microorganisms7110479