Genetic and Phenotypic Characterisation of a Saccharomyces cerevisiae Population of ‘Merwah’ White Wine
Abstract
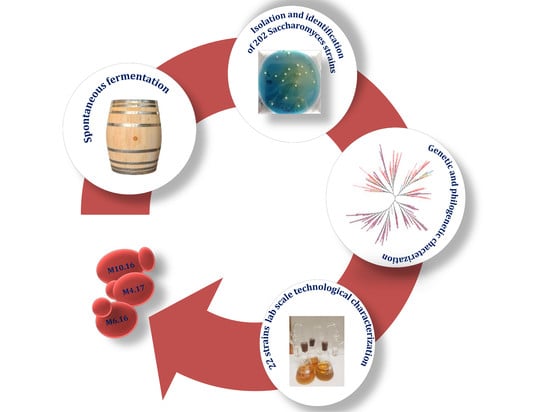
:1. Introduction
2. Materials and Methods
2.1. Winery and Winemaking Process
2.2. Sample Collection, Isolation and Morphological Identification
2.3. DNA Extraction
2.4. Interdelta PCR Typing
2.5. ITS-PCR
2.6. Microsatellite Amplification
2.7. Microfermentation and Phenotypic Analysis
- H2S production on BiGGY agar (Bismuth Sulphite Glucose Glycine Yeast, Difco, Sparks (MD), USA). The quantities of H2S produced by the yeast strains were evaluated qualitatively by colony colour formation, with scoring of the degree of browning (1–6) associated with the yeast growth, according to the following scale: white, 1; cream, 2; light brown, 3; brown, 4; dark-brown, 5; black, 6 [38].
- CO2 production (g/100mL) using the gravimetric method [36].
- Volatile acidity, using enzymatic reaction kits (Cat. No. 10148261035; Boehringer Mannheim, R-Biopharm, Darmstadt, Germany) with a double-beam UV/Vis spectrophotometer (UV S100; Shimadzu, Duisburg, Germany) ), and expressed as g/L acetic acid.
- Residual sugar by UV-visible spectrophotometry, with the dinitrosalicylic acid method, and expressed as g/L [39].
- Ethanol concentration by considering the theoretical yeast yield ~16.83 g/L to produce 1% alcohol: (initial sugar concentration—Residual sugar)/16.83 [40] and expressed as %.
- Total and free SO2 using the modified Ripper iodometric method and expressed as mg/L [41].
- Total acidity, with titration using 0.1 M NaOH to pH 7.00 ± 0.05, with the concentration determined here by acid-base titration and expressed as g/L sulphuric acid.
- The pH, with a pH meter (ST3000; Ohaus Co., Parsippany, NJ, USA).
2.8. Statistical Analysis of the Micro-Fermentations
3. Results
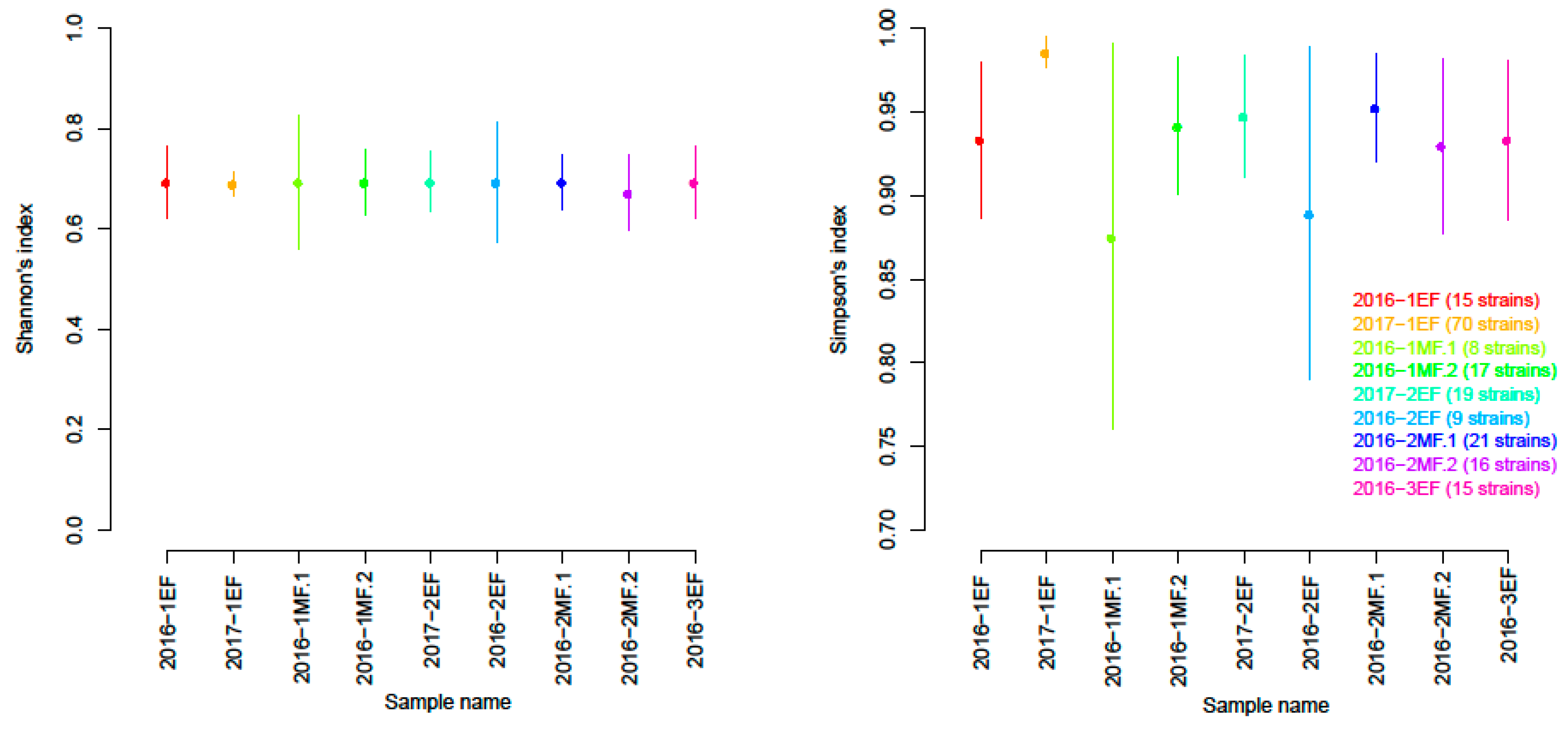
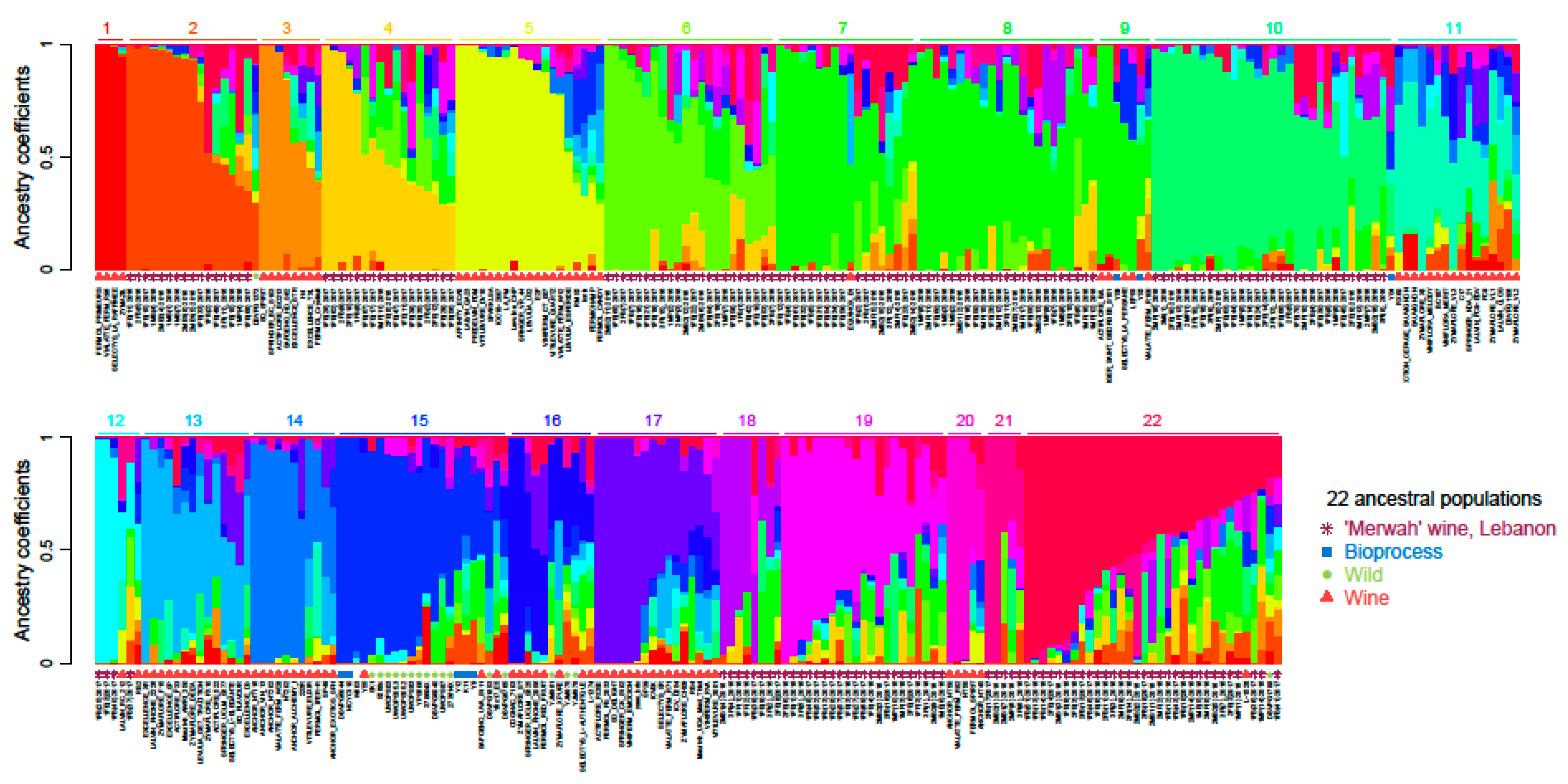
3.1. Genetic Characterisation
3.1.1. Genotyping by Interdelta PCR
3.1.2. Biodiversity of S. cerevisiae Strains According to Microsatellite Markers
3.2. Technological Characterisation
3.2.1. Kinetics of the 22 ‘Merwah’ Wine S. cerevisiae during Alcoholic Fermentation in Synthetic Grape Juice
3.2.2. Phenotypic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Capece, A.; Romaniello, R.; Siesto, G.; Romano, P. Diversity of Saccharomyces cerevisiae yeasts associated to spontaneously fermenting grapes from an Italian ‘heroic vine-growing area’. Food Microbiol. 2012, 31, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Tristezza, M.; Fantastico, L.; Vetrano, C.; Bleve, G.; Corallo, D.; Grieco, F.; Mita, G.; Grieco, F. Molecular and technological characterization of Saccharomyces cerevisiae strains isolated from natural fermentation of Susumaniello grape must in Apulia, southern Italy. Int. J. Microbiol. 2014, 11. [Google Scholar] [CrossRef]
- Capece, A.; Granchi, L.; Guerrini, S.; Mangani, S.; Romaniello, R.; Vincenzini, M.; Romano, P. Diversity of Saccharomyces cerevisiae strains isolated from two Italian wine-producing regions. Front. Microbiol. 2016, 7, 1018. [Google Scholar] [CrossRef] [PubMed]
- Setati, M.E.; Jacobson, D.; Andong, U.-C.; Bauer, F. The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 2012, 7, e52609. [Google Scholar] [CrossRef] [PubMed]
- Schuller, D.; Cardoso, F.; Sousa, S.; Gomes, P.; Gomes, A.C.; Santos, M.A. Genetic diversity and population structure of Saccharomyces cerevisiae strains isolated from different grape varieties and winemaking regions. PLoS ONE 2012, 7, e32507. [Google Scholar] [CrossRef] [PubMed]
- Milanović, Z.; Pantelić, S.; Trajković, N.; Sporiš, G.; Kostić, R.; James, N. Age-related decrease in physical activity and functional fitness among elderly men and women. Clin. Interv. Aging 2013, 8, 549–556. [Google Scholar] [CrossRef] [Green Version]
- Franco-Duarte, R.; Bigey, F.; Carreto, L.; Mendes, I.; Dequin, S.; AS Santos, M.; Pais, C.; Schuller, D. Intrastrain genomic and phenotypic variability of the commercial Saccharomyces cerevisiae strain Zymaflore VL1 reveals microevolutionary adaptation to vineyard environments. FEMS Yeast Res. 2015, 15, fov063. [Google Scholar] [CrossRef]
- Ciani, M.; Capece, A.; Comitini, F.; Canonico, L.; Siesto, G.; Romano, P. Yeast interactions in inoculated wine fermentation. Front. Microbiol. 2016, 7, 555. [Google Scholar] [CrossRef]
- Francesca, N.; Gaglioa, R.; Alfonzoa, A.; Settannia, L.; Coronaa, O.; Mazzei, P.; Romano, R.; Piccolo, A.; Moschettia, G. The wine: Typicality or mere diversity? The effect of spontaneous fermentations and biotic factors on the characteristics of wine. Agric. Agric. Sci. Procedia 2016, 8, 769–773. [Google Scholar] [CrossRef]
- Varela, C.; Bomeman, R.A. Yeast found in vineyards and wineries. Yeast 2017, 34, 111–128. [Google Scholar] [CrossRef]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Ayoub, M.J.; Legras, J.-L.; Saliba, R.; Gaillardin, C. Application of multi-locus sequence typing to the analysis of the biodiversity of indigenous Saccharomyces cerevisiae wine yeasts from Lebanon. J. Appl. Microbiol. 2006, 100, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Chalak, L.; Touma, S.; Rahme, S.; Azzi, R.; Guiberteau, F.; Touma, J.-A. Assessment of the Lebanese grapevine germplasm reveals a substantial diversity and a high potential for selection. 39th World Congress of Vine and Wine. BIO Web Conf. 2016, 7, 01020. [Google Scholar] [CrossRef]
- Sun, Y.; Qin, Y.; Pei, Y.; Wang, G.; Joseph, C.M.; Bisson, L.F.; Liu, Y. Evaluation of Chinese Saccharomyces cerevisiae wine strains from different geographical origins. Am. J. Enol. Vitic. 2017, 68, 1. [Google Scholar] [CrossRef]
- Pallmann, C.L.; Brown, J.A.; Olineka, T.L.; Cocolin, L.; Mills, D.A.; Bisson, L.F. Use of WL medium to profile native flora fermentations. Am. J. Enol. Vitic. 2001, 52, 198–203. [Google Scholar]
- De Celis, M.; Ruiz, J.; Martin-Santamaria, M.; Alonso, A.; Marquina, D.; Navascués, E.; Gómez-Flechoso, M.A.; Belda, I.; Santos, A. Diversity of Saccharomyces cerevisiae yeasts associated to spontaneous and inoculated fermenting grapes from Spanish vineyards. Appl. Microbiol. 2019, 68, 580–588. [Google Scholar] [CrossRef]
- Kumar, M.; Shukla, P.K. Use of PCR targeting of internal transcribed spacer regions and-stranded conformation polymorphism analysis of sequence variation in different regions of rRNA genes in fungi for rapid diagnosis of mycotic keratitis. J. Clin. Microbiol. 2005, 43, 662–668. [Google Scholar] [CrossRef]
- Harju, S.; Fedosyuk, H.; Peterson, K.R. Rapid isolation of yeast genomic DNA: Bust n’ Grab. BMC Biotechnol. 2004, 4, 8. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D. Purification of nucleic acids by extraction with phenol:chloroform. Cold Spring Harb. Protoc. 2006, 2006. [Google Scholar] [CrossRef]
- Xufre, A.; Albergaria, H.; Girio, F.; Spencer-Martins, I. Use of interdelta polymorphisms of Saccharomyces cerevisiae strains to monitor population evolution during wine fermentation. J. Ind. Microbiol. Biotechnol. 2011, 38, 127–132. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.-L.; Ruh, O.; Merdinoglu, D.; Karst, F. Selection of hypervariable microsatellite loci for the characterization of Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 2005, 102, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Wills, C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. USA 1998, 95, 1647–1652. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez Techera, A.; Jubany, S.; Carrau, F.; Gaggero, C. Differentiation of industrial wine yeast strains using microsatellite markers. Lett. Appl. Microbiol. 2001, 33, 71–75. [Google Scholar] [CrossRef]
- Hennequin, C.; Thierry, A.; Richard, G.F.; Lecointre, G.; Nguyen, H.V.; Gaillardin, C.; Dujon, B. Microsatellite typing as a new tool for identification of Saccharomyces cerevisiae strains. J. Clin. Microbiol. 2001, 39, 551–559. [Google Scholar] [CrossRef]
- R Development Core Team (2010); Version 2.6.2 (2008-02-08); R: A language and environment for statistical computing; R Foundation for Statistical Computing: Vienna, Austria, 2010; Available online: http://softlibre.unizar.es/manuales/aplicaciones/r/fullrefman.pdf (accessed on 26 October 2019).
- Bruvo, R.; Michiels, N.K.; D’Souza, T.G.; Schulenburg, H. A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molec. Ecol. 2004, 13, 2101–2106. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef]
- Kamvar, Z.N.; Brooks, J.C.; Grünwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E.; Claude, J.; Strimmer, K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef]
- Frichot, E.; Francois, O. LEA: An R package for landscape and ecological association studies. Methods Ecol. Evol. 2015, 6, 925–929. [Google Scholar] [CrossRef]
- Frichot, E.; Mathieu, F.; Trouillon, T.; Bouchard, G.; François, O. Fast and efficient estimation of individual ancestry coefficients. Genetics 2014, 196, 973–983. [Google Scholar] [CrossRef]
- Clark, D.L.V.; Jasieniuk, V. POLYSAT: An R package for polyploidy microsatellite analysis. Mol. Ecol. Resour. 2011, 11, 562–566. [Google Scholar] [CrossRef] [PubMed]
- Bely, M.; Sablayrolles, J.-M.; Barre, P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 1990, 70, 246–252. [Google Scholar] [CrossRef]
- Martini, A. Ecofisiologia dei lieviti vinari. In Microbiologia del vino, 1st ed.; Vincenzini, M., Romano, P., Farris, G.A., Eds.; AMV Ediciones: Madrid, Spain, 2005; pp. 63–81. [Google Scholar]
- Albertin, W.; Miot-Sertier, C.; Bely, M.; Marullo, P.; Coulon, J.; Moine, V.; Colonna-Ceccaldi, B.; Masneuf-Pomarede, I. Oenological prefermentation practices strongly impact yeast population dynamics and alcoholic fermentation kinetics in Chardonnay grape must. Int. J. Food Microbiol. 2014, 178, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Sipiczki, M. Diversity, variability and fast adaptive evolution of the wine yeast (Saccharomyces cerevisiae) genome—A review. Ann. Microbiol. 2011, 61, 85–93. [Google Scholar] [CrossRef]
- Garriga, M.; Almaraz, M.; Marchiaro, A. Determination of reducing sugars in extracts of Undaria pinnatifida (harvey) algae by UV-visible spectrophotometry (DNS method). Actas. de. Ingeniería. 2015, 3, 173–179. [Google Scholar]
- Cowey, G. Predicting Alcohol Levels. Available online: https://www.awri.com.au/information_services/publications/ask-the-awri/ (accessed on 26 October 2019).
- Rizk, Z.; El Rayess, Y.; Ghanem, C.; Mathieu, F.; Taillendier, P.; Nehme, N. Impact of inhibitory peptides released by Saccharomyces cerevisiae BDX on the malolactic fermentation performed by Oenococcus oeni Vitilactic F. Int. J. Food Microbiol. 2016, 233, 90–96. [Google Scholar] [CrossRef]
- Magurran, A.E. Introduction: Measurement of (biological) diversity. Measuring Biol. Divers. 2004, 1–17. [Google Scholar]
- Börlin, M.; Venet, P.; Claisse, O.; Salin, F.; Legras, J.-L.; Masneuf-Pomarede, I. Cellar-associated Saccharomyces cereviciae population structure revealed high-level diversity and perennial persistence at Sauterners Wine Estates. Appl. Environ. Microbiol. 2016, 82, 2909–2917. [Google Scholar] [CrossRef]
- Hartl, D.; Clark, A. Principles of Population Genetics, 4th ed.; Sinauer Associates, Inc. Publishers: Sunderland, MA, USA, 1997; pp. 3–38. [Google Scholar]
- De Mendiburu, F.; Simon, R. Agricolae-Ten years of an open source statistical tool for experiments in breeding, agriculture and biology. PeerJ PrePrints 2015, 3, e1404v1. [Google Scholar] [CrossRef]
- Caridi, A. Il metabolismo dei lieviti vinari, I edition. In Microbiologia del vino, 1st ed.; Vicenzini, M., Romano, P., Farris, G.A., Eds.; AMV Ediciones: Madrid, Spain, 2005; pp. 83–98. [Google Scholar]
- Wine Production and Quality. International Code of Oenological Practices; Organisation Internationale de la Vigne et du Vin (OIV): Paris, France, 2015; ISBN 979-10-91799-73-7. Available online: http://www.oiv.int/public/medias/5119/code-2017-en.pdf (accessed on 26 October 2019).
- Garofalo, C.; Tristezza, M.; Grieco, F.; Spano, G.; Capozzi, V. From grape berries to wine: Population dynamics of cultivable yeasts associated to “Nero di Troia” autochthonous grape cultivar. World J. Microbiol. Biotechnol. 2016, 32, 59. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Liu, Y.L. Dynamic study of yeast species and Saccharomyces cerevisiae strains during the spontaneous fermentations of Muscat blanc in Jingyang, China. Food Microbiol. 2013, 33, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Santamaría, P.; Garijo, P.; López, R.; Tenorio, C.; Gutiérrez, A.R. Analysis of yeast population during spontaneous alcoholic fermentation: Effect of the age of the cellar and the practice of inoculation. Int. J. Food Microbiol. 2005, 103, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Viel, A.; Legras, J.-L.; Nadai, C.; Carlot, M.; Lombardi, A.; Crespan, M.; Migliaro, D.; Giacomini, A.; Corich, V. The geographic distribution of Saccharomyces cerevisiae isolates within three Italian neighboring winemaking regions reveals strong differences in yeast abundance, genetic diversity and industrial strain dissemination. Front. Microbiol. 2017, 8, 1595. [Google Scholar] [CrossRef]
- Sicard, D.; Legras, J.-L. Bread, beer and wine: Yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011, 334, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Grangeteau, C.; Roullier-Gall, C.; Rousseaux, S.; Gougeon, R.; Schmitt-Kopplin, P.; Alexandre, H.; Guilloux-Benatier, M. Wine microbiology is driven by vineyard and winery anthropogenic factors. Micriobial. Biotechnol. 2017, 10, 354–370. [Google Scholar] [CrossRef]
- Zara, G.; Angelozzi, D.; Belviso, S.; Bardi, L.; Goffrini, P.; Lodi, T.; Budroni, M.; Mannazzu, I. Oxygen is required to restore flor strain viability and lipid biosynthesis under fermentative conditions. FEMS Yeast Res. 2009, 9, 217–225. [Google Scholar] [CrossRef] [Green Version]
- Zara, G.; Budroni, M.; Mannazzu, I.; Zara, S. Air-liquid biofilm formation is dependent on ammonium depletion in a Saccharomyces cerevisiae flor strain. Yeast 2011, 28, 809–81414. [Google Scholar] [CrossRef]
- Gilbert, J.; van der Leliec, D.; Zarraonaindia, I. Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. USA 2014, 11, 5–6. [Google Scholar] [CrossRef]
- Capozzi, V.; Garofalo, C.; Chiriatti, M.A.; Grieco, F.; Spano, G. Microbial terroir and food innovation: The case of yeast biodiversity in wine. Microbiol. Res. 2015, 181, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Osborne, J.; Edwards, C. Inhibition of malolactic fermentation by Saccharomyces during alcoholic fermentation under low- and high-nitrogen conditions: A study in synthetic media. Austral. J. Grape Wine Res. 2008, 12, 69–78. [Google Scholar] [CrossRef]
- Nikolaou, E.; Soufleros, E.H.; Bouloumpasi, E.; Tzanetakis, N. Selection of indigenous Saccharomyces cerevisiae strains according to their oenological characteristics and vinification results. Food Microbiol. 2006, 23, 205–211. [Google Scholar] [CrossRef] [PubMed]





| Mix | Site Name | Multiplexed Primers | Motif and Type | Fluorescent Dye | Reference |
|---|---|---|---|---|---|
| #1 | C5-F | TGACACAATAGCAATGGCCTTCA | GT | 5′-YAKYE | [22] |
| C5-R | GCAAGCGACTAGAACAACAATCACA | ||||
| SCYOR267C-F | TACTAACGTCAACACTGCTGCCAA | TGT | 5′-YAKYE | [23,24] | |
| SCYOR267C-R | GGATCTACTTGCAGTATACGGG | ||||
| C8-F | CAGGTCGTTCTAACGTTGGTAAAATG | TAA | 5′-FAM | [22] | |
| C8-R | GCTGTTGCTGTTGGTAGCATTACTGT | ||||
| C11-F | TTCCATCATAACCGTCTGGGATT | GT | 5′-FAM | [25] | |
| C11-R | TGCCTTTTTCTTAGATGGGCTTTC | ||||
| SCAAT2-F | CAGTCTTATTGCCTTGAACGA | TAA | 5′-AT565 | [24] | |
| SCAAT2-R | GTCTCCATCCTCCAAACAGCC | ||||
| #2 | C9-F | AAGGGTTCGTAAACATATAACTGGCA | TAA | 5′-AT550 | [22] |
| C9-R | TATAAGGGAAAAGAGCACGATGGC | ||||
| C4-F | AGGAGAAAAATGCTGTTTATTCTGACC | TAA + TAG | 5′-AT550 | [22] | |
| C4-R | TTTTCCTCCGGGACGTGAAATA | ||||
| SCAAT5-F | AGCATAATTGGAGGCAGTAAAGCA | TAA | 5′-AT550 | [22] | |
| SCAAT5-R | TCTCCGTCTTTTTTGTACTGCGTG | ||||
| SCAAT1-F | AAAGCGTAAGCAATGGTGTAGATACTT | TTA | 5′-YAKYE | [22,23,24] | |
| SCAAT1-R | CAAGCCTCTTCAAGCATGACCTTT | ||||
| C6-F | GTGGCATCATATCTGTCAATTTTATCAC | CA | 5′-YAKYE | [22] | |
| C6-R | CAATCAAGCAAAAGATCGGCCT | ||||
| YKL172W-F | CAGGACGCTACCGAAGCTCAAAAG | GAA | 5′-FAM | [25] | |
| YKL172W-R | ACTTTTGGCCAATTTCTCAAGAT | ||||
| YPL009c-F | AACCCATTGACCTCGTTACTATCGT | CTT | 5′-FAM | [23,24] | |
| YPL009c-R | TTCGATGGCTCTGATAACTCCATTC |
| Sample Code | Sample Name | Isolate Number | Harvesting Year |
|---|---|---|---|
| M.1.16 | 2016-1EF | 3 | 2016 |
| M.2.16 | 2016-1EF | 16 | 2016 |
| M.3.16 | 2016-1EF | 6 | 2016 |
| M.4.16 | 2016-2EF | 1 | 2016 |
| M.5.16 | 2016-1EF | 2 | 2016 |
| M.6.16 | 2016-2EF | 3 | 2016 |
| M.7.16 | 2016-2EF | 5 | 2016 |
| M.8.16 | 2016-2EF | 8 | 2016 |
| M.9.16 | 2016-3EF | 9 | 2016 |
| M.10.16 | 2016-3EF | 6 | 2016 |
| M.1.17 | 2017-1EF | 3 | 2017 |
| M.2.17 | 2017-1EF | 20 | 2017 |
| M.3.17 | 2017-1EF | 21 | 2017 |
| M.4.17 | 2017-1EF | 39 | 2017 |
| M.5.17 | 2017-1EF | 49 | 2017 |
| M.6.17 | 2017-1EF | 54 | 2017 |
| M.7.17 | 2017-1EF | 66 | 2017 |
| M.8.17 | 2017-2EF | 4 | 2017 |
| M.9.17 | 2017-2EF | 5 | 2017 |
| M.10.17 | 2017-2EF | 9 | 2017 |
| M.11.17 | 2017-2EF | 16 | 2017 |
| M.12.17 | 2017-2EF | 17 | 2017 |
| Sample Name * | Vineyard Location | Number of Isolates | Mean Fermentation Temperature (°C) | Density | Sugar Concentration (g/L) | pH | Total Acidity (g/L H2SO4) |
|---|---|---|---|---|---|---|---|
| 2016-1MF | Wata el Jozz | 25 | 17 | 1.038 | 95 | 3.20 | 4.12 |
| 2016-1EF | Wata el Jozz | 19 | 17 | 0.998 | 8 | 3.28 | 4.31 |
| 2016-2MF | Bekaatet Achout | 40 | 17 | 1.038 | 103 | 3.28 | 3.62 |
| 2016-2EF | Bekaatet Achout | 12 | 17 | 0.998 | 8 | 3.29 | 3.32 |
| 2016-3MF | Wata el Jozz Bekaatet Achout | 1 | 17 | 1.040 | 96 | 3.32 | 3.68 |
| 2016-3EF | Wata el Jozz Bekaatet Achout | 15 | 17 | 0.994 | 4 | 3.13 | 3.23 |
| 2017-1EF | Wata el Jozz Bekaatet Achout | 71 | 18 | 0.996 | 8 | 3.15 | 4.21 |
| 2017-2EF | Wata el Jozz Bekaatet Achout | 19 | 16 | 0.994 | 4 | 3.29 | 4.26 |
| Population | Fixation Index (Fst) According to Yeast Strain Population | |||
|---|---|---|---|---|
| Bioprocess | NA | 0.056 | 0.026 | 0.017 |
| Wild | 0.056 | NA | 0.041 | 0.028 |
| Wine (industrial) | 0.026 | 0.041 | NA | 0.100 |
| Lebanon ‘Merwah’ wine | 0.017 | 0.028 | 0.100 | NA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feghali, N.; Albertin, W.; Tabet, E.; Rizk, Z.; Bianco, A.; Zara, G.; Masneuf-Pomarede, I.; Budroni, M. Genetic and Phenotypic Characterisation of a Saccharomyces cerevisiae Population of ‘Merwah’ White Wine. Microorganisms 2019, 7, 492. https://doi.org/10.3390/microorganisms7110492
Feghali N, Albertin W, Tabet E, Rizk Z, Bianco A, Zara G, Masneuf-Pomarede I, Budroni M. Genetic and Phenotypic Characterisation of a Saccharomyces cerevisiae Population of ‘Merwah’ White Wine. Microorganisms. 2019; 7(11):492. https://doi.org/10.3390/microorganisms7110492
Chicago/Turabian StyleFeghali, Nadine, Warren Albertin, Edouard Tabet, Ziad Rizk, Angela Bianco, Giacomo Zara, Isabelle Masneuf-Pomarede, and Marilena Budroni. 2019. "Genetic and Phenotypic Characterisation of a Saccharomyces cerevisiae Population of ‘Merwah’ White Wine" Microorganisms 7, no. 11: 492. https://doi.org/10.3390/microorganisms7110492
APA StyleFeghali, N., Albertin, W., Tabet, E., Rizk, Z., Bianco, A., Zara, G., Masneuf-Pomarede, I., & Budroni, M. (2019). Genetic and Phenotypic Characterisation of a Saccharomyces cerevisiae Population of ‘Merwah’ White Wine. Microorganisms, 7(11), 492. https://doi.org/10.3390/microorganisms7110492