Internal and External Microbial Community of the Thitarodes Moth, the Host of Ophiocordyceps sinensis
Abstract
:1. Introduction
2. Materials and Methods
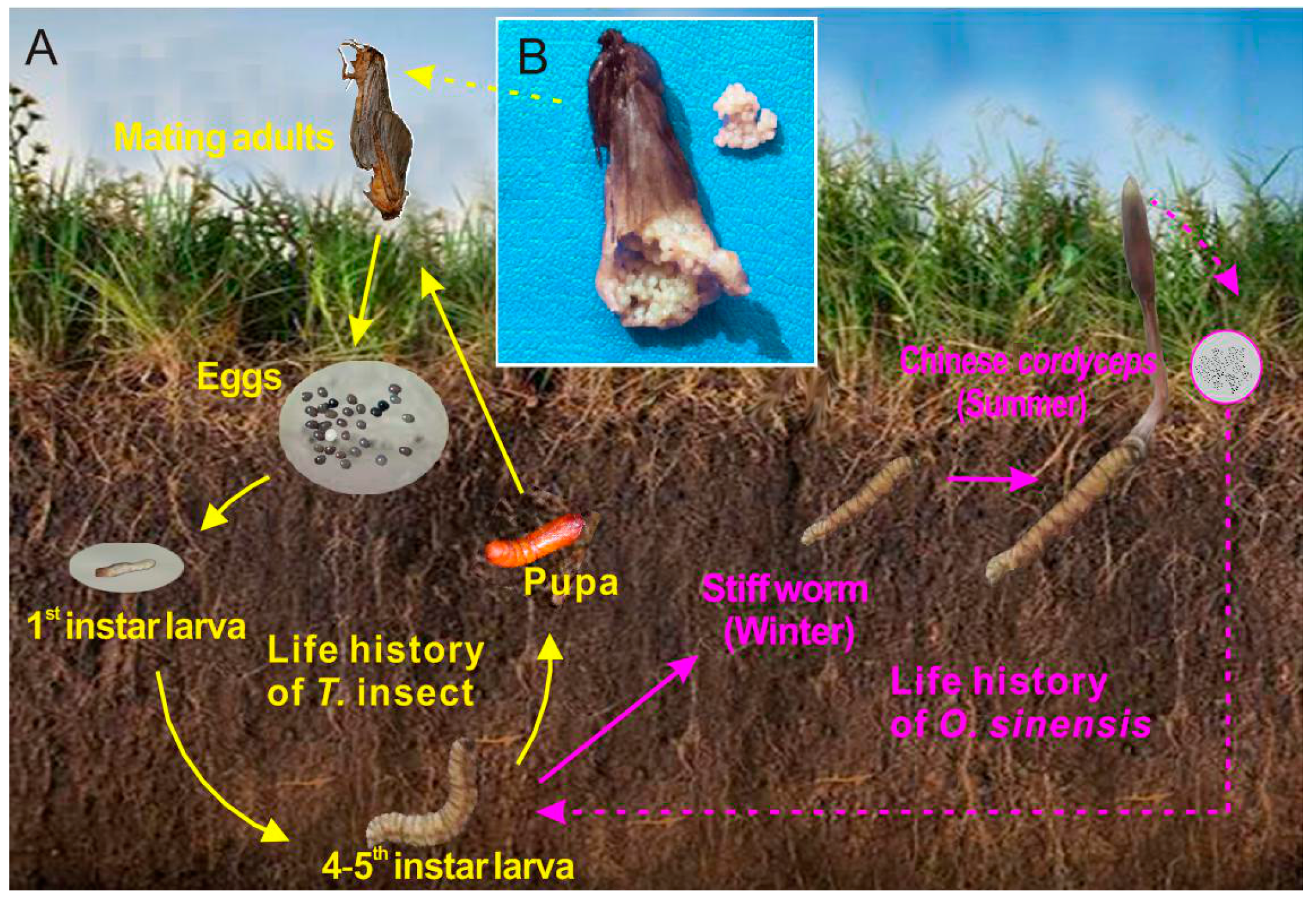
2.1. Field Site Description and Sample Collection
2.2. DNA Extraction, PCR, Library Preparation, Sequencing, and Data Analysis
2.3. Data Normalization and Statistical Analysis
3. Results
3.1. Microbial Diversities
3.2. Bacterial and Fungal Structure
3.3. Differential OTUs Related with the Occurrence of Chinese cordyceps
4. Discussion
4.1. The Internal Microbial Community is Significantly Different from that in the External Soil Environment
4.2. Internal Microbial Composition in the Unfertilized Eggs of Thitarodes
4.3. Discovery of Cordyceps-Related Fungi in the Unfertilized Eggs of Thitarodes
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Sample ID | Number of Sequences (ITS) | Number of OTUs (ITS) | Number of Sequences (16S) | Number of OTUs (16S) |
|---|---|---|---|---|
| E-A1 | 114,096 | 199 | 25,063 | 78 |
| E-A2 | 63,095 | 149 | 32,326 | 60 |
| E-A3 | 49,102 | 190 | 28,918 | 139 |
| E-A4 | 34,788 | 133 | 20,079 | 123 |
| E-A5 | 12,341 | 169 | 30,337 | 68 |
| E-B1 | 24,247 | 235 | 30,383 | 90 |
| E-B2 | 3664 | 87 | 22,309 | 43 |
| E-B3 | 6392 | 152 | 22,996 | 81 |
| E-B4 | 63,019 | 184 | 19,477 | 42 |
| E-B5 | 74,894 | 153 | 31,458 | 49 |
| E-C1 | 15,188 | 215 | 36,614 | 1167 |
| E-C2 | 17,619 | 203 | 22,164 | 106 |
| E-C3 | 34,708 | 157 | 29,450 | 80 |
| E-C4 | 88,818 | 226 | 21,937 | 87 |
| S-A1 | 18,835 | 916 | 53,307 | 2891 |
| S-A2 | 32,131 | 1014 | 54,504 | 3045 |
| S-A3 | 24,957 | 926 | 59,174 | 3008 |
| S-B1 | 40,748 | 1224 | 66,007 | 3135 |
| S-B2 | 93,904 | 1489 | 54,588 | 2928 |
| S-B3 | 36,089 | 1107 | 35,083 | 2529 |
| S-C1 | 29,793 | 908 | 50,667 | 3110 |
| S-C2 | 22,007 | 839 | 52,630 | 3072 |
| S-C3 | 19,069 | 791 | 51,873 | 3117 |
| Ranked List | Annotations in Figure 4e | Detailed Taxonomic Information |
|---|---|---|
| 1st | Anaplasmataceae | Proteobacteria; Alphaproteobacteria; Rickettsiales; Anaplasmataceae |
| 2nd | Unclassfied_38 | Firmicutes; Unclassfied; Unclassfied; Unclassfied |
| 3rd | Unclassfied_52 | Proteobacteria; Gammaproteobacteria; Unclassfied; Unclassfied |
| 4th | Spiroplasmataceae | Tenericutes; Mollicutes; Entomoplasmatales; Spiroplasmataceae |
| 5th | Carnobacteriaceae | Firmicutes; Bacilli; Lactobacillales; Carnobacteriaceae |
| 6th | Sphingomonadaceae | Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae |
| 7th | Moraxellaceae | Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae |
| 8th | Burkholderiales_incertae_sedis | Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiales_incertae_sedis |
| 9th | Methylobacteriaceae | Proteobacteria; Alphaproteobacteria; Rhizobiales; Methylobacteriaceae |
| 10th | Pseudomonadaceae | Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae |
| 11th | Actinomycetales | Actinobacteria; Actinobacteria; Actinobacteridae; Actinomycetales |
| 12th | Burkholderiaceae | Proteobacteria; Betaproteobacteria; Burkholderiales; Burkholderiaceae |
| 13th | Bacillaceae 1 | Bacilli; Bacillales; Bacillaceae 1 |
| 14th | Comamonadaceae | Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae |
| 15th | Enterobacteriaceae | Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae |
| 16th | Rhodocyclaceae | Proteobacteria; Betaproteobacteria; Rhodocyclales; Rhodocyclaceae |
| 17th | Xanthomonadaceae | Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae |
| 18th | Unclassfied_41 | Unclassfied; Unclassfied; Unclassfied; Unclassfied |
| 19th | Planctomycetaceae | Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae |
| 20th | Unclassfied_14 | Acidobacteria; Acidobacteria_Gp4; Gp4; Unclassfied |
| Others | Others |
| Ranked List | Annotations in Figure 4f | Detailed Taxonomic Information |
|---|---|---|
| 1st | [Chthoniobacteraceae] | Verrucomicrobia; [Spartobacteria]; [Chthoniobacterales]; [Chthoniobacteraceae] |
| 2nd | Unclassfied_25 | Acidobacteria; Acidobacteria-6; iii1-15; Unclassfied |
| 3rd | Unclassfied_214 | Planctomycetes; Phycisphaerae; WD2101; Unclassfied |
| 4th | Pirellulaceae | Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae |
| 5th | Unclassfied_106 | Chloroflexi; Ellin6529; Unclassfied; Unclassfied |
| 6th | Gemmataceae | Planctomycetes; Planctomycetia; Gemmatales; Gemmataceae |
| 7th | Ellin6075 | Acidobacteria; [Chloracidobacteria]; RB41; Ellin6075 |
| 8th | Chitinophagaceae | Bacteroidetes; [Saprospirae]; [Saprospirales]; Chitinophagaceae |
| 9th | Thermogemmatisporaceae | Chloroflexi; Ktedonobacteria; Thermogemmatisporales; Thermogemmatisporaceae |
| 10th | [Kouleothrixaceae] | Chloroflexi; Chloroflexi; [Roseiflexales]; [Kouleothrixaceae] |
| 11th | Hyphomicrobiaceae | Proteobacteria; Alphaproteobacteria; Rhizobiales; Hyphomicrobiaceae |
| 12th | Rhodobacteraceae | Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae |
| 13th | Anaeroplasmataceae | Tenericutes; Mollicutes; Anaeroplasmatales; Anaeroplasmataceae |
| 14th | Flavobacteriaceae | Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae |
| 15th | Moraxellaceae | Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae |
| 16th | Comamonadaceae | Proteobacteria; Betaproteobacteria; Burkholderiales; Comamonadaceae |
| 17th | Verrucomicrobiaceae | Verrucomicrobia; Verrucomicrobiae; Verrucomicrobiales; Verrucomicrobiaceae |
| 18th | FFCH4570 | Chloroflexi; TK10; B07_WMSP1; FFCH4570 |
| 19th | RB40 | Acidobacteria; Acidobacteria-6; iii1-15; RB40 |
| 20th | Planctomycetaceae | Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae |
| Others | Others |
| Ranked List | Annotations in Figure 4f | Detailed Taxonomic Information |
|---|---|---|
| 1st | Unclassfied_23 | Basidiomycota; Unclassfied; Unclassfied; Unclassfied |
| 2nd | Unclassfied_27 | Unclassfied; Unclassfied; Unclassfied; Unclassfied |
| 3rd | Dothioraceae | Ascomycota; Dothideomycetes; Dothideales; Dothioraceae |
| 4th | Unclassfied_5 | Ascomycota; Eurotiomycetes; Chaetothyriales; Unclassfied |
| 5th | Unclassfied_14 | Ascomycota; Unclassfied; Unclassfied; Unclassfied |
| 6th | Unclassfied_1 | Ascomycota; Archaeorhizomycetes; Unclassfied; Unclassfied |
| 7th | Gloniaceae | Ascomycota; Dothideomycetes; Hysteriales; Gloniaceae |
| 8th | Unclassfied_2 | Ascomycota; Dothideomycetes; Capnodiales; Unclassfied |
| 9th | Unclassfied_21 | Basidiomycota; Agaricomycetes; Unclassfied; Unclassfied |
| 10th | Unclassfied_12 | Ascomycota; Leotiomycetes; Helotiales; Unclassfied |
| 11th | Davidiellaceae | Ascomycota; Dothideomycetes; Capnodiales; Davidiellaceae |
| 12th | Unclassfied_6 | Ascomycota; Eurotiomycetes; Eurotiales; Unclassfied |
| 13th | Unclassfied_18 | Ascomycota; Sordariomycetes; Unclassfied; Unclassfied |
| 14th | Incertae_sedis_13 | Ascomycota; Dothideomycetes; Pleosporales; Incertae_sedis_13 |
| 15th | Helotiaceae | Ascomycota; Leotiomycetes; Helotiales; Helotiaceae |
| 16th | Incertae_sedis_2 | Ascomycota; Leotiomycetes; Helotiales; Incertae_sedis_2 |
| 17th | Herpotrichiellaceae | Ascomycota; Eurotiomycetes; Chaetothyriales; Herpotrichiellaceae |
| 18th | Incertae_sedis_5 | Basidiomycota; Incertae_sedis_4; Malasseziales; Incertae_sedis_5 |
| 19th | Incertae_sedis_1 | Ascomycota; Leotiomycetes; Incertae_sedis; Incertae_sedis_1 |
| 20th | Incertae_sedis_12 | Basidiomycota; Tremellomycetes; Tremellales; Incertae_sedis_12 |
| Others | Others |
| Ranked List | Annotations in Figure 4h | Detailed Taxonomic Information |
|---|---|---|
| 1st | Unclassfied_22 | Basidiomycota; Unclassfied; Unclassfied; Unclassfied |
| 2nd | Unclassfied_1 | Unclassfied; Unclassfied; Unclassfied; Unclassfied |
| 3rd | Helotiales_family_Incertae_sedis | Ascomycota; Leotiomycetes; Helotiales; Helotiales_family_Incertae_sedis |
| 4th | Pyronemataceae | Ascomycota; Pezizomycetes; Pezizales; Pyronemataceae |
| 5th | Hygrophoraceae | Basidiomycota; Agaricomycetes; Agaricales; Hygrophoraceae |
| 6th | Leptosphaeriaceae | Ascomycota; Dothideomycetes; Pleosporales; Leptosphaeriaceae |
| 7th | Unclassfied_2 | Ascomycota; Unclassfied; Unclassfied; Unclassfied |
| 8th | Pleosporales_family_Incertae_sedis | Ascomycota; Dothideomycetes; Pleosporales; Pleosporales_family_Incertae_sedis |
| 9th | Ascomycota_family_Incertae_sedis | Ascomycota; Ascomycota_class_Incertae_sedis; Ascomycota_order_Incertae_sedis;family_Incertae_sedis |
| 10th | Nectriaceae | Ascomycota; Sordariomycetes; Hypocreales; Nectriaceae |
| 11th | Unclassfied_12 | Ascomycota; Leotiomycetes; Helotiales; |
| 12th | Mortierellaceae | Zygomycota; Zygomycota_class_Incertae_sedis; Mortierellales; Mortierellaceae |
| 13th | Unclassfied_11 | Ascomycota; Leotiomycetes; Unclassfied; Unclassfied |
| 14th | Chaetomiaceae | Ascomycota; Sordariomycetes; Sordariales; Chaetomiaceae |
| 15th | Davidiellaceae | Ascomycota; Dothideomycetes; Capnodiales; Davidiellaceae |
| 16th | Venturiaceae | Ascomycota; Dothideomycetes; Venturiales; Venturiaceae |
| 17th | Unclassfied_14 | Ascomycota; Leotiomycetes; Thelebolales; Unclassfied |
| 18th | Unclassfied_23 | Basidiomycota; Agaricomycetes; Unclassfied; Unclassfied |
| 19th | Myxotrichaceae | Ascomycota; Leotiomycetes; Leotiomycetes_order_Incertae_sedis; Myxotrichaceae |
| 20th | Strophariaceae | Basidiomycota; Agaricomycetes; Agaricales; Strophariaceae |
| Others | Others |
References
- Zhang, Y.J.; Li, E.W.; Wang, C.S.; Li, Y.L.; Liu, X.Z. Ophiocordyceps sinensis, the flagship fungus of China: Terminology, life strategy and ecology. Mycology 2012, 3, 2–10. [Google Scholar]
- Xia, E.H.; Yang, D.R.; Jiang, J.J.; Zhang, Q.J.; Liu, Y.; Liu, Y.L.; Zhang, Y.; Zhang, H.B.; Shi, C.; Tong, Y.; et al. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 2017, 7, 1806. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.L.; Yu, Y.X.; Liu, L.X.; Zhang, C.X.; Fang, C.X. A draft genome of the ghost moth, Thitarodes (Hepialus) sp., a medicinal caterpillar fungus. Insect Sci. 2016, 2323, 326–329. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.C.; Hsieh, C.; Lin, F.Y.; Hsu, T.H. A systematic review of the mysterious caterpillar fungus Ophiocordyceps sinensis in Dong-Chong-Xia-Cao (Dōng Chóng Xià Căo) and related bioactive ingredients. J. Tradit. Complement. Med. 2013, 3, 16–32. [Google Scholar] [CrossRef]
- Xu, J.; Huang, Y.; Chen, X.X.; Zheng, S.C.; Chen, P.; Mo, M.H. The mechanisms of pharmacological activities of Ophiocordyceps sinensis fungi. Phytother. Res. 2016, 30, 1572–1583. [Google Scholar] [CrossRef]
- Baral, B.; Shrestha, B.; da Silva, J.A.T. A review of Chinese Cordyceps with special reference to Nepal, focusing on conservation. Environ. Exp. Biol. 2015, 13, 6173. [Google Scholar]
- Guo, L.X.; Zhang, G.W.; Wang, J.T.; Zhong, Y.P.; Huang, Z.G. Determination of arsenic species in Ophiocordyceps sinensis from major habitats in China by HPLC-ICP-MS and the edible hazard assessment. Molecules 2018, 23, 1012. [Google Scholar] [CrossRef]
- Quan, Q.M.; Chen, L.L.; Wang, X.; Li, S.; Yang, X.L.; Zhu, Y.G.; Wang, M.; Cheng, Z. Genetic diversity and distribution patterns of host insects of caterpillar fungus Ophiocordyceps sinensis in the Qinghai-Tibet Plateau. PLoS ONE 2014, 9, e92293. [Google Scholar] [CrossRef]
- Shrestha, U.B.; Bawa, K.S. Impact of climate change on potential distribution of Chinese caterpillar fungus (Ophiocordyceps sinensis) in Nepal Himalaya. PLoS ONE 2014, 9, e106405. [Google Scholar] [CrossRef]
- He, J. Harvest and trade of caterpillar mushroom (Ophiocordyceps sinensis) and the implications for sustainable use in the Tibet region of southwest China. J. Ethnopharmacol. 2018, 221, 86–90. [Google Scholar] [CrossRef]
- Li, X.; Liu, Q.; Li, W.J.; Li, Q.P.; Qian, Z.G.; Liu, X.Z.; Dong, C.Y. A breakthrough in the artificial cultivation of Chinese Cordyceps on a large-scale and its impact on science, the economy, and industry. Crit. Rev. Biotechnol. 2019, 39, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Baral, B. Entomopathogenicity and biological attributes of Himalayan treasured fungus Ophiocordyceps sinensis (Yarsagumba). J. Fungi 2017, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Shao, J.L.; Lai, B.; Jiang, W.; Wang, J.T.; Hong, Y.H.; Chen, F.B.; Tan, S.Q.; Guo, L.X. Diversity and co-occurrence patterns of soil bacterial and fungal communities of Chinese Cordyceps habitats at Shergyla Mountain, Tibet: Implications for the occurrence. Microorganisms 2019, 7, 284. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.X.; Xu, X.M.; Liang, F.R.; Yuan, J.P.; Peng, J.; Wu, C.F.; Wang, J.H. Morphological observations and fatty acid composition of indoor-cultivated Cordyceps sinensis at a high-altitude laboratory on Sejila Mountain, Tibet. PLoS ONE 2015, 10, e0126095. [Google Scholar] [CrossRef]
- Yu, H.W.; Wang, Z.K.; Liu, L.; Xia, Y.X.; Cao, Y.Q.; Yin, Y.P. Analysis of the intestinal microflora in Hepialus gonggaensis larvae using 16S rRNA sequences. Curr. Microbiol. 2008, 56, 391–396. [Google Scholar] [CrossRef]
- Yu, H.W.; Wang, Z.K.; Liu, L.; Xia, Y.X.; Yin, Y.P. Analysis of fungal diversity in intestines of Hepialus gonggaensis larva. Acta Microbiol. Sin. 2008, 48, 439–445. [Google Scholar]
- Wang, J.G.; Wei, H.Y. Anatomy of internal structure of the larva of Hapialus hunanensis Chu et Wang (Lepidoptera, Hepialidae). Acta Agric. Univ. Jiangxi 1997, 19, 5–8. [Google Scholar]
- Chen, J.; Gao, Z.X.; Yu, H. Observation on egg’s feature and chorionic ultrastructure of swiftmoth (Hepialus oblifurcus Chu et Wang). Acta Agric. Univ. Zhejiang 1991, 39, 379–383. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tindall, B.J.; Rossello-Mora, R.; Busse, H.J.; Ludwig, W.; Kampfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. Microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Knight, R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Joop, G.; Vilcinskas, A. Coevolution of parasitic fungi and insect hosts. Zoology 2016, 119, 350–358. [Google Scholar] [CrossRef]
- Douglas, A. Lessons from studying insect symbioses. Cell Host Microbe 2011, 10, 359–367. [Google Scholar] [CrossRef]
- Chen, D.; Yuan, J.P.; Xu, S.P.; Zhou, X.G.; Zhang, Y.; Xu, X.M.; Zou, Z.W.; Zhang, G.R.; Wang, J.H. Stable carbon isotope evidence for tracing the diet of the host Hepialus, larva of Cordyceps sinensis, in the Tibetan Plateau. Sci. China Ser. D 2009, 52, 655–659. [Google Scholar] [CrossRef]
- Li, J.G.; Zou, Z.W.; Liu, X.; Zhang, G.R. Biology of Thitarodes pui (Lepidoptera, Hepialidae), a host species of Ophiocordyceps sinensis. J. Environ. Entomol. 2011, 33, 195–202. [Google Scholar]
- Li, Y.; Guo, L.X.; Zhou, Q.Z.; Chen, D.; Liu, J.Z.; Xu, X.M.; Wang, J.H. Characterization of humic substances in the soils of Ophiocordyceps sinensis habitats in the Sejila Mountain, Tibet: Implication for the food source of Thitarodes larvae. Molecules 2019, 24, 246. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, Z.K.; Yu, H.W.; Mu, D.D.; Yuan, Q.; Yin, Y.P. Effects of feeding on tetracycline and streptomycin on the growth and gut digestive enzymes of Hepialus gonggaensis larvae. Chin. J. Appl. Entomol. 2008, 45, 272–275. [Google Scholar]
- Wang, Z.L.; Lu, J.D.; Feng, M.G. Primary roles of two dehydrogenases in the mannitol metabolism and multi-stress tolerance of entomopathogenic fungus Beauveria bassiana. Environ. Microbiol. 2012, 14, 2139–2150. [Google Scholar] [CrossRef]
- Shokal, U.; Yadav, S.; Atri, J.; Accetta, J.; Kenney, E.; Banks, K.; Katakam, A.; Jaenike, J.; Eleftherianos, I. Effects of co-occurring Wolbachia and Spiroplasma endosymbionts on the Drosophila immune response against insect pathogenic and non-pathogenic bacteria. BMC Microbiol. 2016, 16, 16. [Google Scholar] [CrossRef] [PubMed]
- Goto, S.; Anbutsu, H.; Fukatsu, T. Asymmetrical interactions between Wolbachia and Spiroplasma endosymbionts coexisting in the same insect host. Appl. Environ. Microbiol. 2006, 72, 4805–4810. [Google Scholar] [CrossRef]
- Mu, D.D.; Wang, Z.K.; Yin, Y.P. Effects of feeding Carnobacterium hg4-03 on bacterial diversity in the larval intestine of Hepialus gonggaensis. Acta Microbiol. Sin. 2010, 50, 251–255. [Google Scholar]
- Prasongsuk, S.; Lotrakul, P.; Ali, I.; Bankeeree, W.; Punnapayak, H. The current status of Aureobasidium pullulans in biotechnology. Folia Microbiol. 2017, 63, 129–140. [Google Scholar] [CrossRef]
- Deshmukh, P.; Rai, M.K.; Kövics, G.J.; Irinyi, L.; Sándor, E. Phomas: Can These Fungi be Used as Biocontrol Agents and Sources of Secondary Metabolites. In Proceedings of the 4th International Plant Protection Symposium at Debrecen University & 11th Trans-tisza Plant Protection Forum, Debrecen, Hungary, 18–19 October 2006; pp. 224–234. [Google Scholar]
- Dantur, K.I.; Enrique, R.; Welin, B.; Castagnaro, A.P. Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express 2015, 5, 1–11. [Google Scholar] [CrossRef]
- Rodrigues, A.; Cable, R.N.; Mueller, U.G.; Bacci, M.; Pagnocca, F.C. Antagonistic interactions between garden yeasts and microfungal garden pathogens of leaf-cutting ants. Antonie van Leeuwenhoek 2009, 96, 331342. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.X.; Hong, Y.H.; Zhou, Q.Z.; Zhu, Q.; Xu, X.M.; Wang, J.H. Fungus-larva relation in the formation of Cordyceps sinensis as revealed by stable carbon isotope analysis. Sci. Rep. 2017, 7, 7789. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, Y.; Xiao, G.; Zheng, P.; Xia, Y.; Zhang, X.; St Leger, R.J.; Liu, X.; Wang, C. Genome survey uncovers the secrets of sex and lifestyle in caterpillar fungus. Chin. Sci. Bull. 2013, 58, 2846–2854. [Google Scholar] [CrossRef] [Green Version]
- Suh, S.O.; Noda, H.; Blackwell, M. Insect symbiosis: Derivation of yeast-like endosymbionts within an entomopathogenic filamentous lineage. Mol. Biol. Evol. 2001, 18, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the Clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef] [PubMed]
- Méndez, C.; Salas, J.A. ABC transporters in antibiotic-producing actinomycetes. FEMS Microbiol. Lett. 2010, 158, 1–8. [Google Scholar] [CrossRef]
- Gupta, V.; Haider, S.; Sood, U.; Gilbert, J.A.; Ramjee, M.; Forbes, K.; Singh, Y.; Lopes, B.S.; Lal, R. Comparative genomic analysis of novel acinetobacter symbionts: A combined systems biology and genomics approach. Sci. Rep. 2016, 6, 29043. [Google Scholar] [CrossRef]
- Su, S.; Zeng, X.; Bai, L.; Williams, P.N.; Wang, Y.; Zhang, L.; Wu, C. Inoculating chlamydospores of Trichoderma asperellum SM-12F1 changes arsenic availability and enzyme activity in soils and improves water spinach growth. Chemosphere 2017, 175, 497–504. [Google Scholar] [CrossRef]
- Zhang, X.P.; Deng, W.; Yang, X.M. The background concentrations of 13 soil trace elements and their relationships to parent materials and vegetation in Xizang (Tibet), China. J. Asian Earth Sci. 2002, 21, 167–174. [Google Scholar] [CrossRef]
- Amend, A. From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathog. 2014, 10, e1004277. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Guo, D.F. Soil fungal diversity and its relationship with soil physical and chemical properties in saline alkali soil of Yellow River Delta. Acta Agric. Zhejiangensis 2016, 28, 1901–1907. [Google Scholar]
- Gasparich, G.E. Spiroplasmas: Evolution, adaptation and diversity. Front. Biosci. 2002, 7, d619–d640. [Google Scholar]
- Sanadamorimura, S.; Matsumura, M.; Noda, H. Male killing caused by a Spiroplasma symbiont in the small brown planthopper, Laodelphax striatellus. J. Hered. 2013, 104, 821–829. [Google Scholar] [CrossRef] [PubMed]
- Tabata, J.; Hattori, Y.; Sakamoto, H.; Yukuhiro, F.; Fujii, T.; Kugimiya, S.; Mochizuki, A.; Ishikawa, Y.; Kageyama, D. Male killing and incomplete inheritance of a novel Spiroplasma in the moth Ostrinia zaguliaevi. Microb. Ecol. 2011, 61, 254–263. [Google Scholar] [CrossRef] [PubMed]





| Classified | Sample Sites | Number of Sequences | Number of OTUs | Shannon | Simpson | Chao1 |
|---|---|---|---|---|---|---|
| Bacteria | Egg A | 27,345 ± 4854 | 94 ± 35 | 1.47 ± 1.34 | 0.37 ± 0.32 | 112.25 ± 39.36 |
| Egg B | 25,325 ± 5290 | 61 ± 23 | 0.78 ± 0.66 | 0.23 ± 0.23 | 86.54 ± 14.91 | |
| Egg C | 27,541 ± 6983 | 360 ± 538 | 3.35 ± 2.19 # | 0.64 ± 0.27 # | 382.7 ± 561.93 | |
| Total | 373,511 | 2213 | / | / | / | |
| Soil A | 55,662 ± 3100 | 2981 ± 80 | 8.83 ± 0.13 | 0.99 ± 0.00 | 3437.59 ± 64.92 | |
| Soil B | 51,893 ± 15,637 | 2864 ± 308 | 8.52 ± 0.01 * | 0.99 ± 0.00 * | 3413.66 ± 60.17 | |
| Soil C | 51,723 ± 990 | 3100 ± 24 | 9.25 ± 0.07 *,# | 1 ± 0.00 *,# | 3586.21 ± 14.8 *,# | |
| Total | 477,833 | 26,835 | / | / | / | |
| Fungi | Egg A | 54,684 ± 38,146 | 168 ± 28 | 0.91 ± 1.41 | 0.21 ± 0.35 | 39.2 ± 33.75 |
| Egg B | 34,443 ± 32,753 | 162 ± 54 | 2.22 ± 1.95 | 0.48 ± 0.42 | 94.6 ± 72.78 | |
| Egg C | 39,083 ± 34,275 | 200 ± 30 | 1.93 ± 1.6 | 0.43 ± 0.37 | 82.5 ± 25.75 | |
| Total | 601,971 | 2452 | / | / | / | |
| Soil A | 25,308 ± 6655 | 952 ± 54 | 6.1 ± 0.47 | 0.92 ± 0.02 | 1229.76 ± 41.22 | |
| Soil B | 56,914 ± 32,119 | 1298 ± 270 | 6.97 ± 0.58 | 0.96 ± 0.03 | 1312.59 ± 46.79 | |
| Soil C | 23,623 ± 5542 | 846 ± 59 # | 6.55 ± 0.14 | 0.96 ± 0.00 * | 1133.13 ± 18.88 *,# | |
| Total | 317,533 | 9219 | / | / | / |
| ANOSIM * | Adonis | MRPP | |||||||
|---|---|---|---|---|---|---|---|---|---|
| R | P | F | R2 | P | Observed Delta (δ) | Expected Delta (δ) | Effect Size (A) | P | |
| Bacteria for unfertilized eggs | 0.5112 | 0.003 | 3.1266 | 0.3624 | 0.005 | 0.6818 | 0.8126 | 0.1609 | 0.013 |
| Bacteria for soils | 1.0000 | 0.003 | 23.117 | 0.8851 | 0.009 | 0.1940 | 0.4793 | 0.5952 | 0.004 |
| Fungi for unfertilized eggs | −0.1053 | 0.807 | 0.78622 | 0.1251 | 0.639 | 0.8316 | 0.8074 | −0.0299 | 0.690 |
| Fungi for soils | 1.0000 | 0.009 | 11.057 | 0.7866 | 0.003 | 0.3285 | 0.6432 | 0.4893 | 0.005 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, Y.; Hong, Y.; Mai, Z.; Zhu, Q.; Guo, L. Internal and External Microbial Community of the Thitarodes Moth, the Host of Ophiocordyceps sinensis. Microorganisms 2019, 7, 517. https://doi.org/10.3390/microorganisms7110517
Liang Y, Hong Y, Mai Z, Zhu Q, Guo L. Internal and External Microbial Community of the Thitarodes Moth, the Host of Ophiocordyceps sinensis. Microorganisms. 2019; 7(11):517. https://doi.org/10.3390/microorganisms7110517
Chicago/Turabian StyleLiang, Yi, Yuehui Hong, Zhanhua Mai, Qijiong Zhu, and Lianxian Guo. 2019. "Internal and External Microbial Community of the Thitarodes Moth, the Host of Ophiocordyceps sinensis" Microorganisms 7, no. 11: 517. https://doi.org/10.3390/microorganisms7110517
APA StyleLiang, Y., Hong, Y., Mai, Z., Zhu, Q., & Guo, L. (2019). Internal and External Microbial Community of the Thitarodes Moth, the Host of Ophiocordyceps sinensis. Microorganisms, 7(11), 517. https://doi.org/10.3390/microorganisms7110517





