HP1717 Contributes to Streptococcus suis Virulence by Inducing an Excessive Inflammatory Response and Influencing the Biosynthesis of the Capsule
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Growth Conditions
2.2. Cell Culture
2.3. Purification of the HP1717 Protein
2.4. Flow Cytometry Analysis
2.5. RNA Extraction and qRT-PCR
2.6. Examination of Cytokines by ELISA
2.7. Determination of HP1717 Recognition Receptor
2.8. Analysis of the HP1717-Induced Cell Signal Transduction Pathway
2.9. Mutant Construction
2.10. Gram Staining and Transmission Electron Microscope (TEM) Observation
2.11. Whole-Blood Bactericidal Assay
2.12. In Vivo Experiments and Immunohistochemistry
2.13. Zymogram Analysis
2.14. Statistical Analysis
3. Results
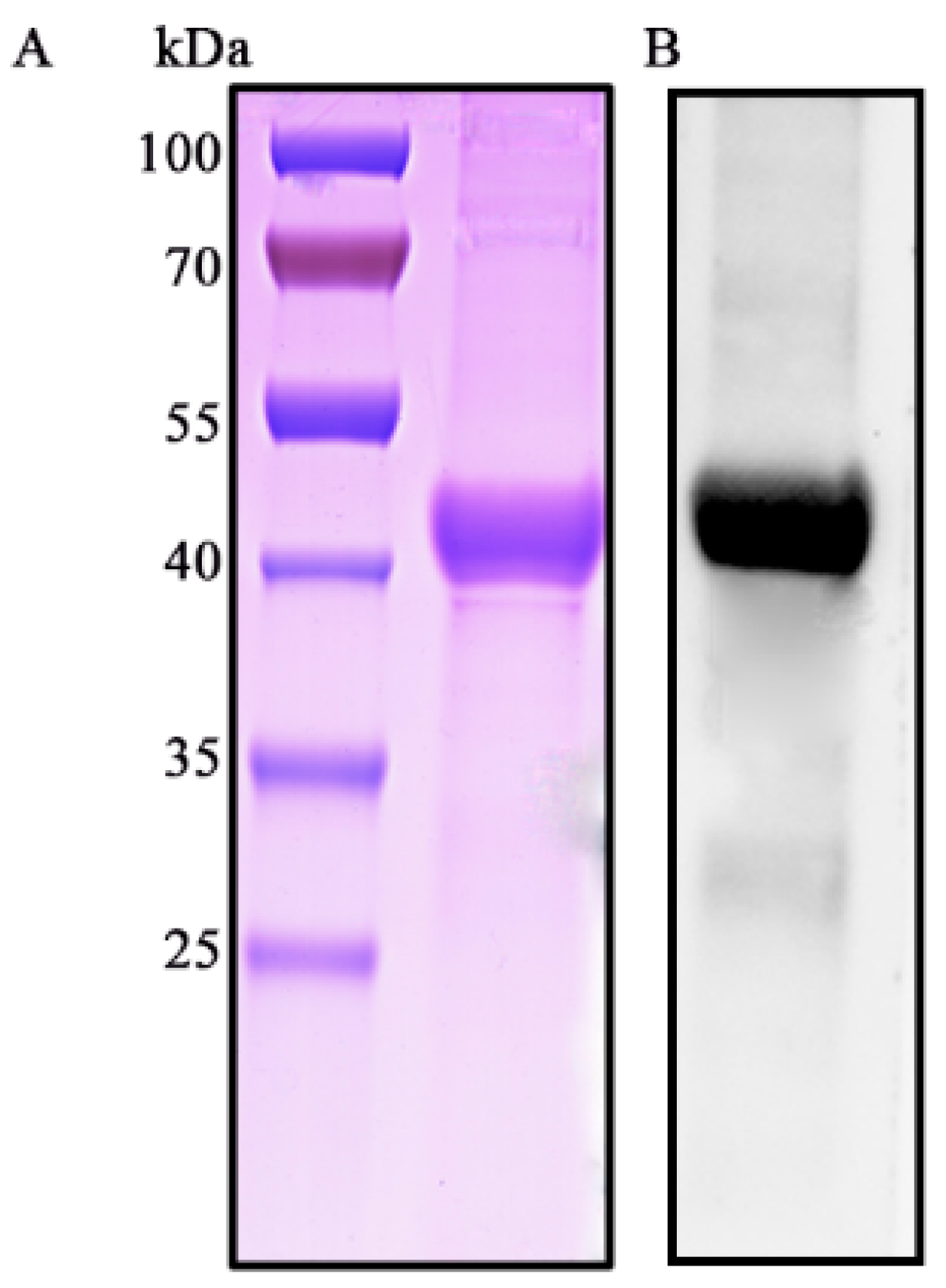
3.1. Characterization of S. suis 2 HP1717
3.2. HP1717 is Expressed on the Surface of S. suis 2
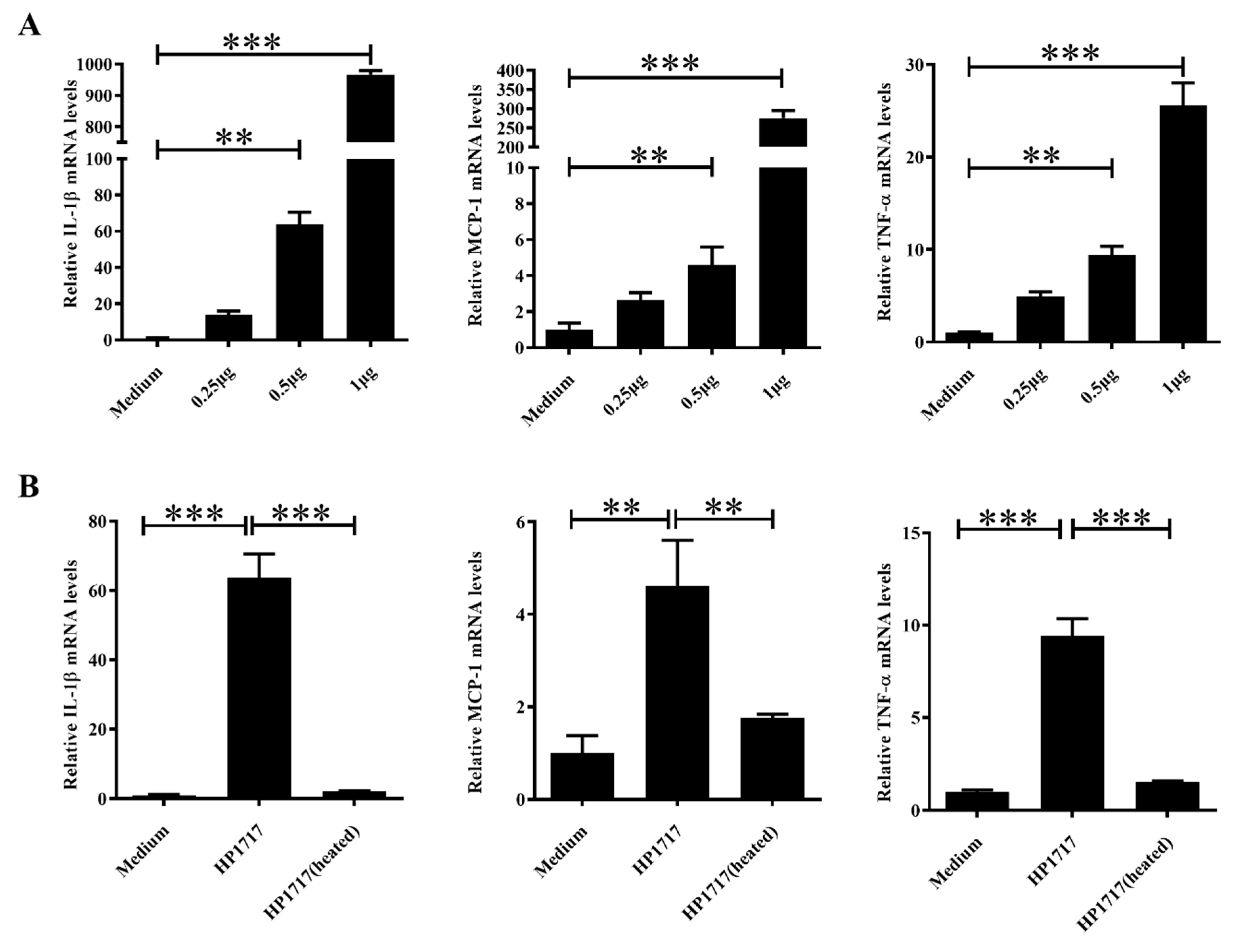
3.3. HP1717 Induces Potent Expression of Pro-Inflammatory Cytokines in RAW264.7 Cells
3.4. The Pro-Inflammatory Activity of HP1717 is Dose-Dependent and Heat-Sensitive
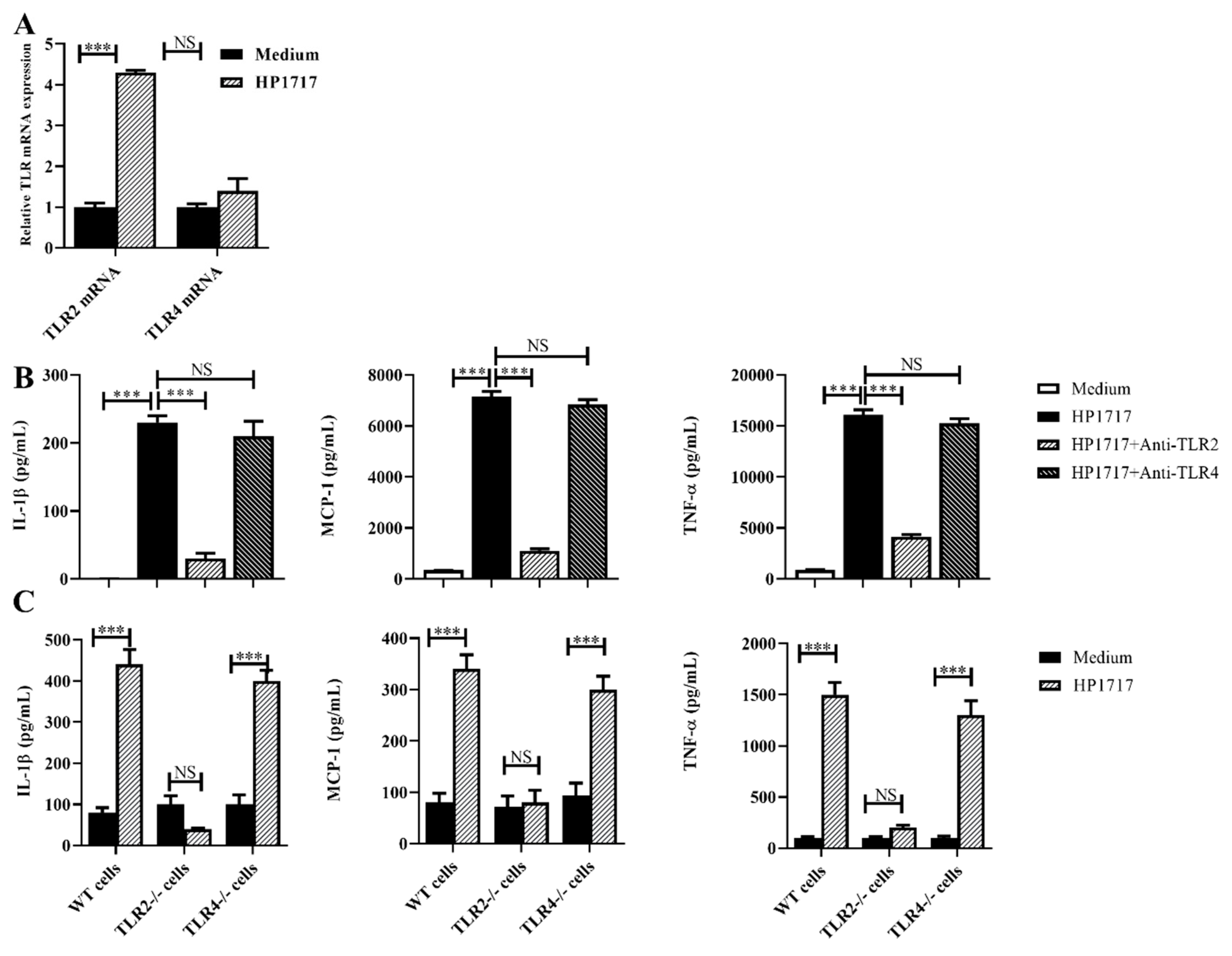
3.5. The Pro-Inflammatory Role of HP1717 Depends on the Recognition of TLR2
3.6. The Pro-Inflammatory Role of HP1717 Depends on the Phosphorylation of NF-κB and ERK1/2
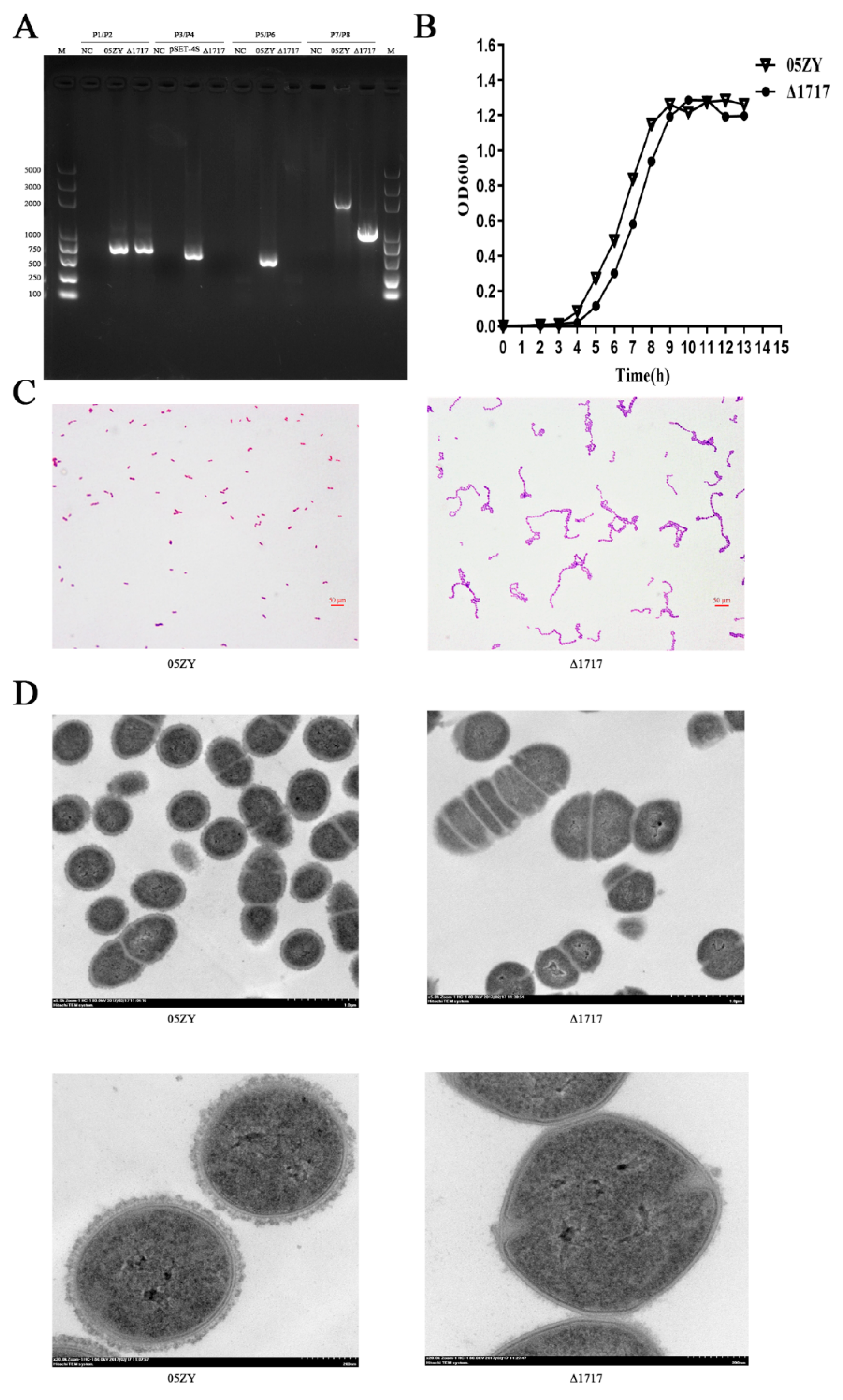
3.7. Construction and Characterization of Mutant Strain Δ1717
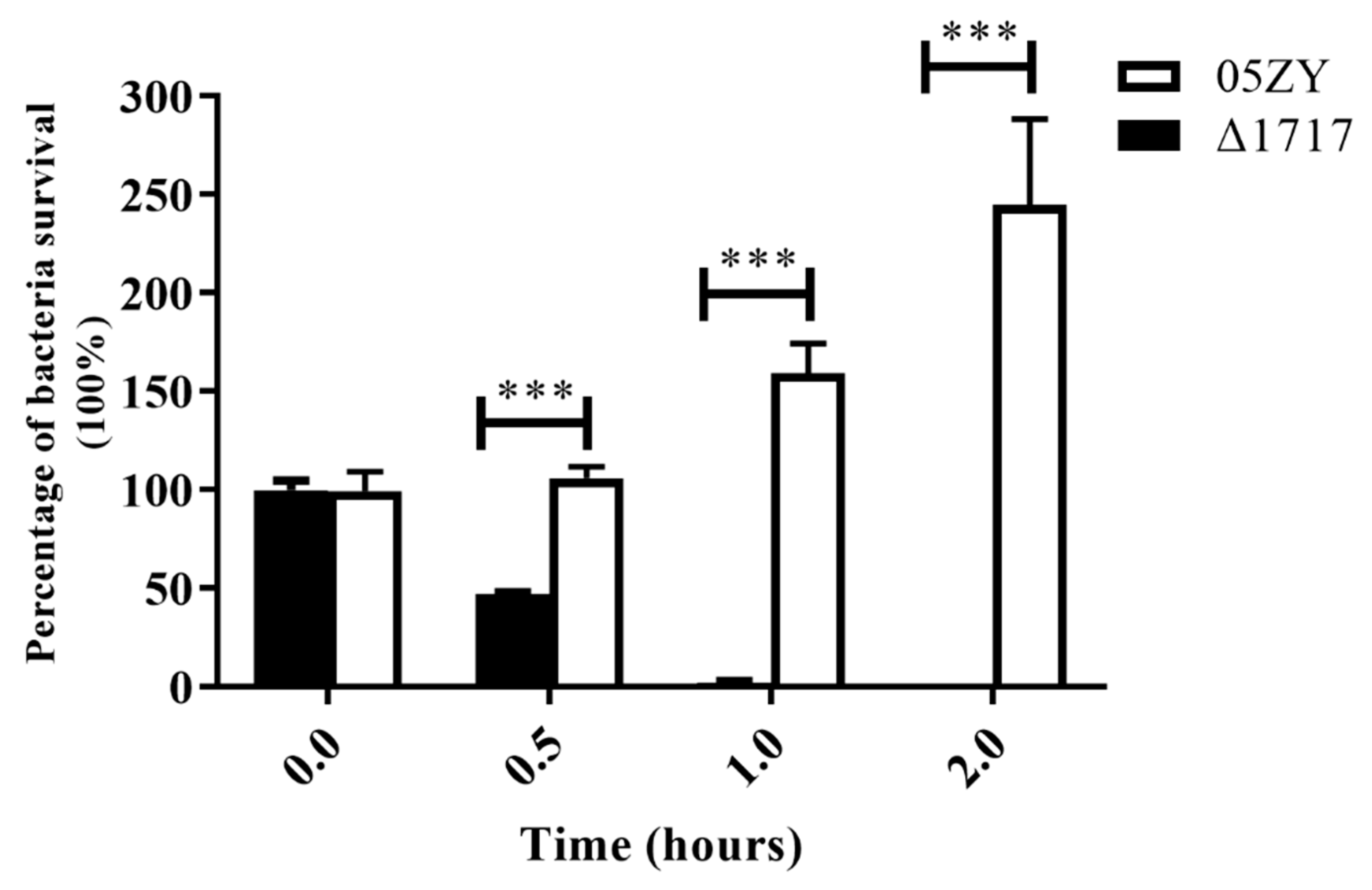
3.8. Δ1717 Displays Reduced Resistance to Whole-Blood Killing
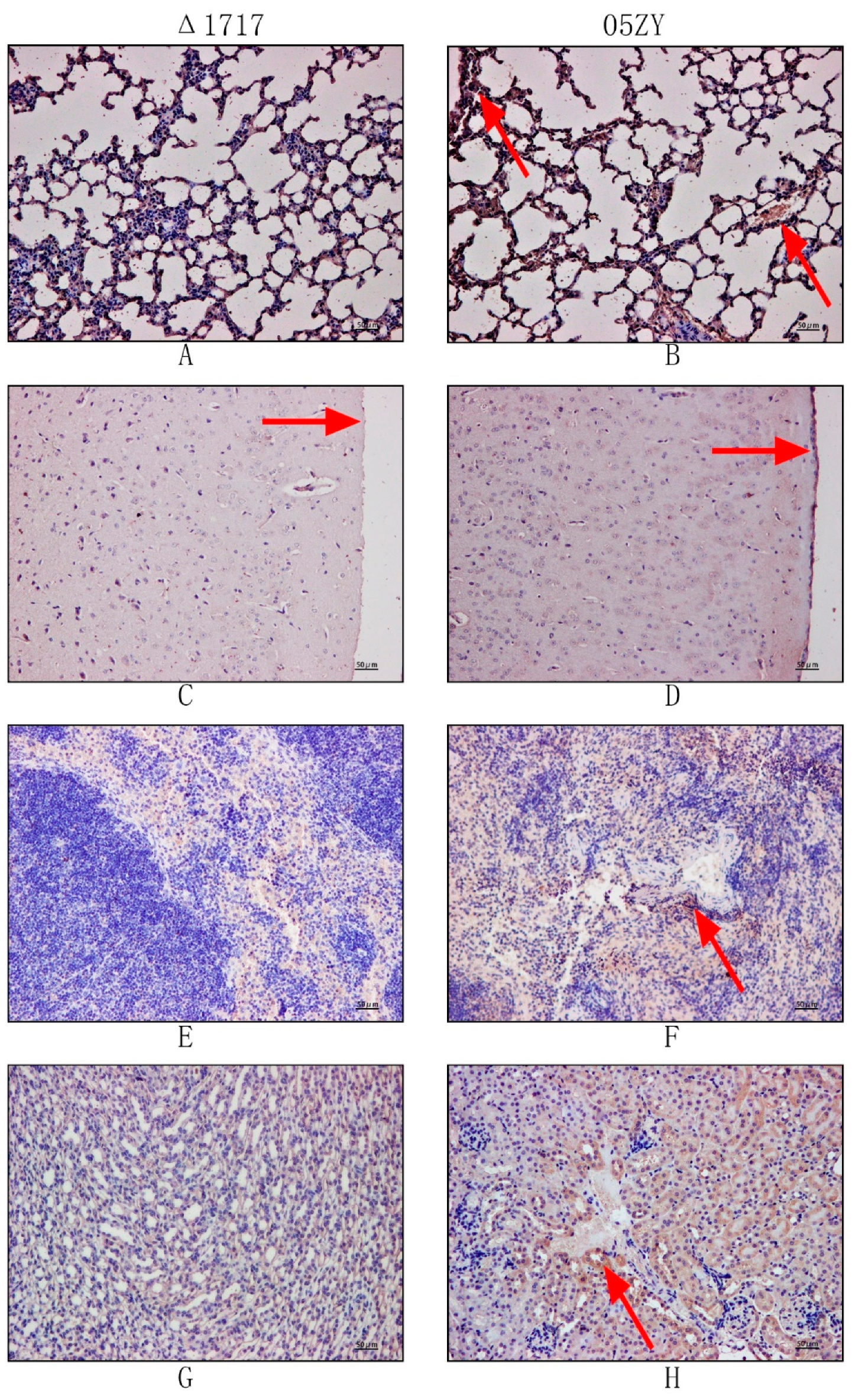
3.9. HP1717 Deficiency Leads to Attenuated Virulence, Decreased Pro-Inflammatory Ability, and Reduced Bacterial Loads in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Votsch, D.; Willenborg, M.; Weldearegay, Y.B.; Valentin-Weigand, P. Streptococcus suis—The “Two Faces” of a Pathobiont in the Porcine Respiratory Tract. Front. Microbiol. 2018, 9, 480. [Google Scholar] [CrossRef]
- Tien Le, H.T.; Nishibori, T.; Nishitani, Y.; Nomoto, R.; Osawa, R. Reappraisal of the taxonomy of Streptococcus suis serotypes 20, 22, 26, and 33 based on DNA—DNA homology and sodA and recN phylogenies. Vet. Microbiol. 2013, 162, 842–849. [Google Scholar] [CrossRef]
- Goyette-Desjardins, G.; Auger, J.P.; Xu, J.; Segura, M.; Gottschalk, M. Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 2014, 3, e45. [Google Scholar] [CrossRef]
- Chen, C.; Tang, J.; Dong, W.; Wang, C.; Feng, Y.; Wang, J.; Zheng, F.; Pan, X.; Liu, D.; Li, M.; et al. A glimpse of streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE 2007, 2, e315. [Google Scholar] [CrossRef]
- Tang, J.; Wang, C.; Feng, Y.; Yang, W.; Song, H.; Chen, Z.; Yu, H.; Pan, X.; Zhou, X.; Wang, H.; et al. Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 2006, 3, e151. [Google Scholar] [CrossRef]
- Lun, Z.R.; Wang, Q.P.; Chen, X.G.; Li, A.X.; Zhu, X.Q. Streptococcus suis: An emerging zoonotic pathogen. Lancet Infect. Dis. 2007, 7, 201–209. [Google Scholar] [CrossRef]
- Kerdsin, A.; Dejsirilert, S.; Puangpatra, P.; Sripakdee, S.; Chumla, K.; Boonkerd, N.; Polwichai, P.; Tanimura, S.; Takeuchi, D.; Nakayama, T.; et al. Genotypic profile of Streptococcus suis serotype 2 and clinical features of infection in humans, Thailand. Emerg. Infect. Dis. 2011, 17, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Punaro, M.C.; Segura, M.; Plante, M.M.; Lacouture, S.; Rivest, S.; Gottschalk, M. Streptococcus suis serotype 2, an important swine and human pathogen, induces strong systemic and cerebral inflammatory responses in a mouse model of infection. J. Immunol. 2007, 179, 1842–1854. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, B.; Zhang, Q.; Liu, L.; Zhang, A.; Yang, Y.; Huang, K.; Yan, S.; Yu, J.; Sun, X.; et al. Streptococcus suis 2 Transcriptional Regulator TstS Stimulates Cytokine Production and Bacteremia to Promote Streptococcal Toxic Shock-Like Syndrome. Front. Microbiol. 2018, 9, 1309. [Google Scholar] [CrossRef]
- Zhao, J.; Pan, S.; Lin, L.; Fu, L.; Yang, C.; Xu, Z.; Wei, Y.; Jin, M.; Zhang, A. Streptococcus suis serotype 2 strains can induce the formation of neutrophil extracellular traps and evade trapping. FEMS Microbiol. Lett. 2015, 362, fnv022. [Google Scholar] [CrossRef]
- Segura, M.; Fittipaldi, N.; Calzas, C.; Gottschalk, M. Critical Streptococcus suis Virulence Factors: Are They All Really Critical? Trends Microbiol. 2017, 25, 586–599. [Google Scholar] [CrossRef] [PubMed]
- Dutkiewicz, J.; Zajac, V.; Sroka, J.; Wasinski, B.; Cisak, E.; Sawczyn, A.; Kloc, A.; Wojcik-Fatla, A. Streptococcus suis: A re-emerging pathogen associated with occupational exposure to pigs or pork products. Part II—Pathogenesis. Ann. Agric. Environ. Med. 2018, 25, 186–203. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Huang, J.; Yu, J.; Xu, Z.; Liu, L.; Song, Y.; Sun, X.; Zhang, A.; Jin, M. HP1330 Contributes to Streptococcus suis Virulence by Inducing Toll-Like Receptor 2-and ERK1/2-Dependent Pro-Inflammatory Responses and Influencing in Vivo S. suis Loads. Front. Immunol. 2017, 8, 869. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Matsumura, T.; Ikebe, T.; Ichibayashi, R.; Fukui, Y.; Satoh, T.; Tsubota, T.; Honda, M.; Ishii, Y.; Tateda, K.; et al. Streptococcal toxic shock syndrome caused by beta-hemolytic streptococci: Clinical features and cytokine and chemokine analyses of 15 cases. J. Infect. Chemother. 2019, 25, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Gottschalk, M.; Xu, J.; Calzas, C.; Segura, M. Streptococcus suis: A new emerging or an old neglected zoonotic pathogen? Future Microbiol. 2010, 5, 371–391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yang, Y.; Yan, S.; Liu, J.; Xu, Z.; Yu, J.; Song, Y.; Zhang, A.; Jin, M. A novel pro-inflammatory protein of Streptococcus suis 2 induces the Toll-like receptor 2-dependent expression of pro-inflammatory cytokines in RAW 264.7 macrophages via activation of ERK1/2 pathway. Front. Microbiol. 2015, 6, 178. [Google Scholar] [CrossRef]
- Takamatsu, D.; Osaki, M.; Sekizaki, T. Thermosensitive suicide vectors for gene replacement in Streptococcus suis. Plasmid 2001, 46, 140–148. [Google Scholar] [CrossRef]
- Zheng, F.; Shao, Z.Q.; Hao, X.; Wu, Q.; Li, C.; Hou, H.; Hu, D.; Wang, C.; Pan, X. Identification of oligopeptide-binding protein (OppA) and its role in the virulence of Streptococcus suis serotype 2. Microb. Pathog. 2018, 118, 322–329. [Google Scholar] [CrossRef]
- Hayashi, T.; Tsukagoshi, H.; Sekizuka, T.; Ishikawa, D.; Imai, M.; Fujita, M.; Kuroda, M.; Saruki, N. Next-generation DNA sequencing analysis of two Streptococcus suis ST28 isolates associated with human infective endocarditis and meningitis in Gunma, Japan: A case report. Infect. Dis. 2019, 51, 62–66. [Google Scholar] [CrossRef]
- Tan, M.F.; Liu, W.Q.; Zhang, C.Y.; Gao, T.; Zheng, L.L.; Qiu, D.X.; Li, L.; Zhou, R. The involvement of MsmK in pathogenesis of the Streptococcus suis serotype 2. MicrobiologyOpen 2017, 6, e00433. [Google Scholar] [CrossRef]
- Pian, Y.; Gan, S.; Wang, S.; Guo, J.; Wang, P.; Zheng, Y.; Cai, X.; Jiang, Y.; Yuan, Y. Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 2012, 80, 2402–2413. [Google Scholar] [CrossRef] [PubMed]
- Bartual, S.G.; Straume, D.; Stamsas, G.A.; Munoz, I.G.; Alfonso, C.; Martinez-Ripoll, M.; Havarstein, L.S.; Hermoso, J.A. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae. Nat. Commun. 2014, 5, 3842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Cheng, G.; Wang, C.; Pan, X.; Cong, Y.; Pan, Q.; Wang, J.; Zheng, F.; Hu, F.; Tang, J. Identification and experimental verification of protective antigens against Streptococcus suis serotype 2 based on genome sequence analysis. Curr. Microbiol. 2009, 58, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.S.; Yuan, Z.Z.; Li, J.T.; Zhang, Q.; Xu, Z.M.; Yan, S.X.; Zhang, A.D.; Jin, M.L. Identification and characterisation a surface-associated arginine peptidase in Streptococcus suis serotype 2. Microbiol. Res. 2015, 170, 168–176. [Google Scholar] [CrossRef]
- Normile, D. Infectious diseases. WHO probes deadliness of China’s pig-borne disease. Science 2005, 309, 1308–1309. [Google Scholar] [CrossRef]
- Ye, C.; Zheng, H.; Zhang, J.; Jing, H.; Wang, L.; Xiong, Y.; Wang, W.; Zhou, Z.; Sun, Q.; Luo, X.; et al. Clinical, experimental, and genomic differences between intermediately pathogenic, highly pathogenic, and epidemic Streptococcus suis. J. Infect. Dis. 2009, 199, 97–107. [Google Scholar] [CrossRef]
- Bi, Y.H.; Li, J.; Yang, L.M.; Zhang, S.; Li, Y.; Jia, X.J.; Sun, L.; Yin, Y.B.; Qin, C.; Wang, B.N.; et al. Assessment of the pathogenesis of Streptococcus suis type 2 infection in piglets for understanding streptococcal toxic shock-like syndrome, meningitis, and sequelae. Vet. Microbiol. 2014, 173, 299–309. [Google Scholar] [CrossRef]
- Martinon, F.; Tschopp, J. NLRs join TLRs as innate sensors of pathogens. Trends Immunol. 2005, 26, 447–454. [Google Scholar] [CrossRef]
- Kahn, F.; Morgelin, M.; Shannon, O.; Norrby-Teglund, A.; Herwald, H.; Olin, A.I.; Bjorck, L. Antibodies against a surface protein of Streptococcus pyogenes promote a pathological inflammatory response. PLoS Pathog. 2008, 4, e1000149. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, J.; Lin, L.; Pan, S.; Fu, L.; Han, L.; Jin, M.; Zhou, R.; Zhang, A. Targeting TREM-1 Signaling in the Presence of Antibiotics is Effective Against Streptococcal Toxic-Shock-Like Syndrome (STSLS) Caused by Streptococcus suis. Front. Cell. Infect. Microbiol. 2015, 5, 79. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.E.; Damman, M.; van der Velde, J.; Wagenaar, F.; Wisselink, H.J.; Stockhofe-Zurwieden, N.; Smits, M.A. Identification and characterization of the cps locus of Streptococcus suis serotype 2: The capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 1999, 67, 1750–1756. [Google Scholar] [PubMed]
- Segura, M.; Gottschalk, M.; Olivier, M. Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect. Immun. 2004, 72, 5322–5330. [Google Scholar] [CrossRef] [PubMed]
- Graveline, R.; Segura, M.; Radzioch, D.; Gottschalk, M. TLR2-dependent recognition of Streptococcus suis is modulated by the presence of capsular polysaccharide which modifies macrophage responsiveness. Int. Immunol. 2007, 19, 375–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanabe, S.; Bonifait, L.; Fittipaldi, N.; Grignon, L.; Gottschalk, M.; Grenier, D. Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can. J. Vet. Res. 2010, 74, 65–70. [Google Scholar]
- Yunck, R.; Cho, H.; Bernhardt, T.G. Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Mol. Microbiol. 2016, 99, 700–718. [Google Scholar] [CrossRef]
- Tsui, H.C.; Zheng, J.J.; Magallon, A.N.; Ryan, J.D.; Yunck, R.; Rued, B.E.; Bernhardt, T.G.; Winkler, M.E. Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol. Microbiol. 2016, 100, 1039–1065. [Google Scholar] [CrossRef]
- Gao, T.; Tan, M.; Liu, W.; Zhang, C.; Zhang, T.; Zheng, L.; Zhu, J.; Li, L.; Zhou, R. GidA, a tRNA Modification Enzyme, Contributes to the Growth, and Virulence of Streptococcus suis Serotype 2. Front. Cell. Infect. Microbiol. 2016, 6, 44. [Google Scholar] [CrossRef]



| Group | Name | Characteristics and Functions | Sources or References |
|---|---|---|---|
| Strains | |||
| 05ZY | Streptococcus suis serotype 2 wild type | Laboratory collection | |
| Δ1717 | Δ1717-deletion mutant strain | This study | |
| Escherichia coli DH5α | Cloning host for recombinant vector | TIANGEN | |
| E. coli BL21 | Expression host for recombinant protein | TIANGEN | |
| Plasmids | |||
| pSET4s | E. coli–S. suis shuttle vector; Spcr | Laboratory collection [17] | |
| pET28a | Expression vector; Kan | TIANGEN [13] | |
| Primers | Primers Sequence (5′-3′) | Functions or PCR Product |
|---|---|---|
| hp1717-1 | CCCGAATTCATGTCGATTGTTGTAGTGGCA(EcoRI) | For amplification of the hp1717 ORF gene |
| hp1717-2 | CCCAAGCTTTTACTCATTATTAAGATGTGCATTTAC(HindIII) | |
| hp1717L1 | CGCGGATCCCTACTGGGTTGTCGGTGGT(BamHI) | Upstream border of hp1717 |
| hp1717L2 | AACACATTGTCTCTGTTATCATTCTTAAAACAAAATTATGTGGTT | |
| hp1717R1 | GATAACAGAGACAATGTGTTTAGCGGCATTATCTTGTTTT | Downstream border of hp1717 |
| hp1717R2 | CCCAAGCTTTAAGGGACAGGGAGTGGG(HindIII) | |
| MCP-1-1 | AGAAGGAATGGGTCCAGACATA | For qRT-PCR assay |
| MCP-1-2 | GTGCTTGAGGTGGTTGTGGA | |
| TNF-α-1 | GAGTGACAAGCCTGTAGCCC | For qRT-PCR assay |
| TNF-α-2 | GACAAGGTACAACCCATCGG | |
| IL-1β-1 | TCATTGTGGCTGTGGAGAAGC | For qRT-PCR assay |
| IL-1β-2 | TCATCTCGGAGCCTGTAGTGC | |
| GAPDH-1 | TGGCCTTCCGTGTTCCTAC | For qRT-PCR assay |
| GAPDH-2 | TGAAGTCGCAGGAGACAACC | |
| P1 | TGGAAATGTTCAAGTCAACC | For PCR to detect the gdh |
| P2 | CGTTTTTCTTTGATGTCCAC | |
| P3 | CTACGAACTGCTAACA | For PCR to detect the pSET4s |
| P4 | GAATACATACGAACAAAT | |
| P5 | CCAAAGATGCCAAGGT | For PCR to detect the hp1717 |
| P6 | ATCGCCAAAGCACTTC | |
| P7 | ATTCGTGGATTACCTG | External primers of hp1717 ORF |
| P8 | TACCATCAAGCTCGTC | |
| 16s-1 | CAGAAAGGGACGGCTAA | For amplification of the 16s gene |
| 16s-2 | CGGCTGGCTCCTAAAA | |
| 16s-rt-1 | AGATGGACCTGCGTTGTATT | For qRT-PCR assay |
| 16s-rt-2 | TCCGAAAACCTTCTTCACTC | |
| 1717rt-1 | TGGCATCTTTGGTTGA | For qRT-PCR assay |
| 1717rt-2 | CGAAAGGCTTGGACTA |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.; Zhang, Q.; Xu, Z.; Huang, J.; Zhu, W.; Zhang, A.; Sun, X.; Jin, M. HP1717 Contributes to Streptococcus suis Virulence by Inducing an Excessive Inflammatory Response and Influencing the Biosynthesis of the Capsule. Microorganisms 2019, 7, 522. https://doi.org/10.3390/microorganisms7110522
Liu L, Zhang Q, Xu Z, Huang J, Zhu W, Zhang A, Sun X, Jin M. HP1717 Contributes to Streptococcus suis Virulence by Inducing an Excessive Inflammatory Response and Influencing the Biosynthesis of the Capsule. Microorganisms. 2019; 7(11):522. https://doi.org/10.3390/microorganisms7110522
Chicago/Turabian StyleLiu, Liang, Qiang Zhang, Zhongmin Xu, Jingjing Huang, Weifeng Zhu, Anding Zhang, Xiaomei Sun, and Meilin Jin. 2019. "HP1717 Contributes to Streptococcus suis Virulence by Inducing an Excessive Inflammatory Response and Influencing the Biosynthesis of the Capsule" Microorganisms 7, no. 11: 522. https://doi.org/10.3390/microorganisms7110522




