Modifications of the Aerobic Respiratory Chain of Paracoccus Denitrificans in Response to Superoxide Oxidative Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Membrane Preparation and Protein Determination
2.3. Determination of Succinate
2.4. Enzyme Activities
2.5. Superoxide Production Assay
2.6. In Vitro Superoxide Exposure
2.7. Iron Sulfur Clusters Restoration
2.8. Intracellular Concentrations of NAD+ and NADH
2.9. qRT-PCR
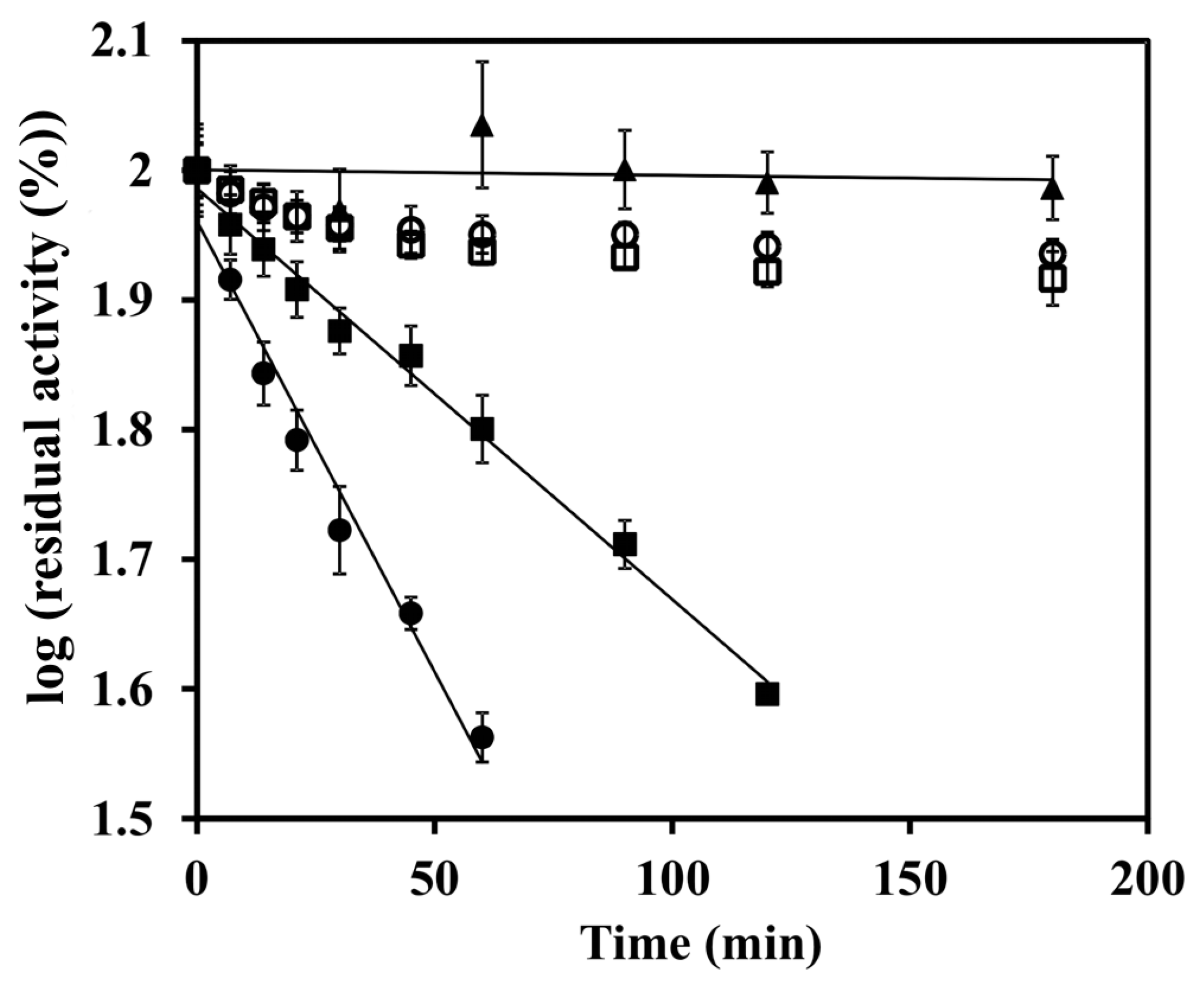
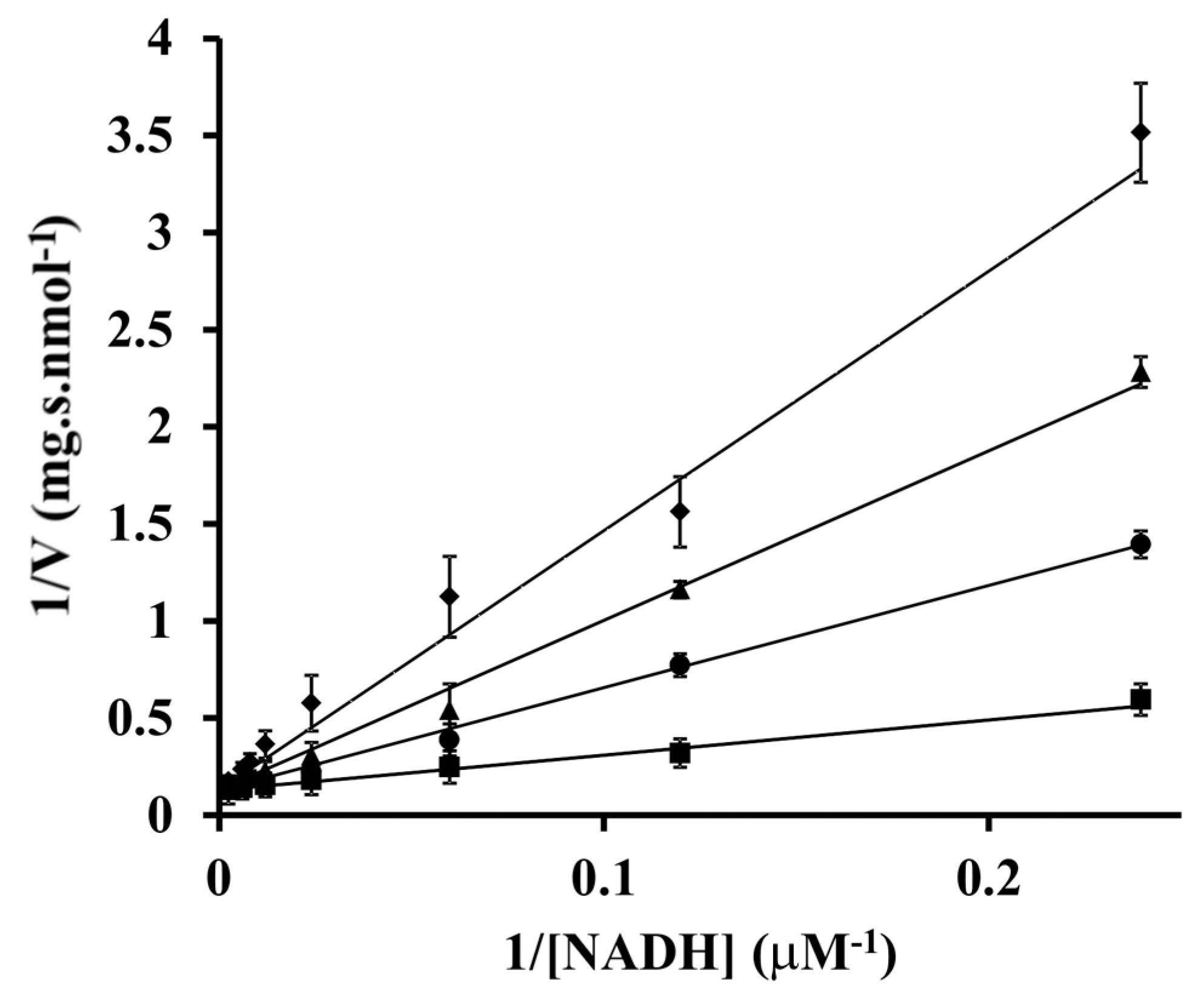
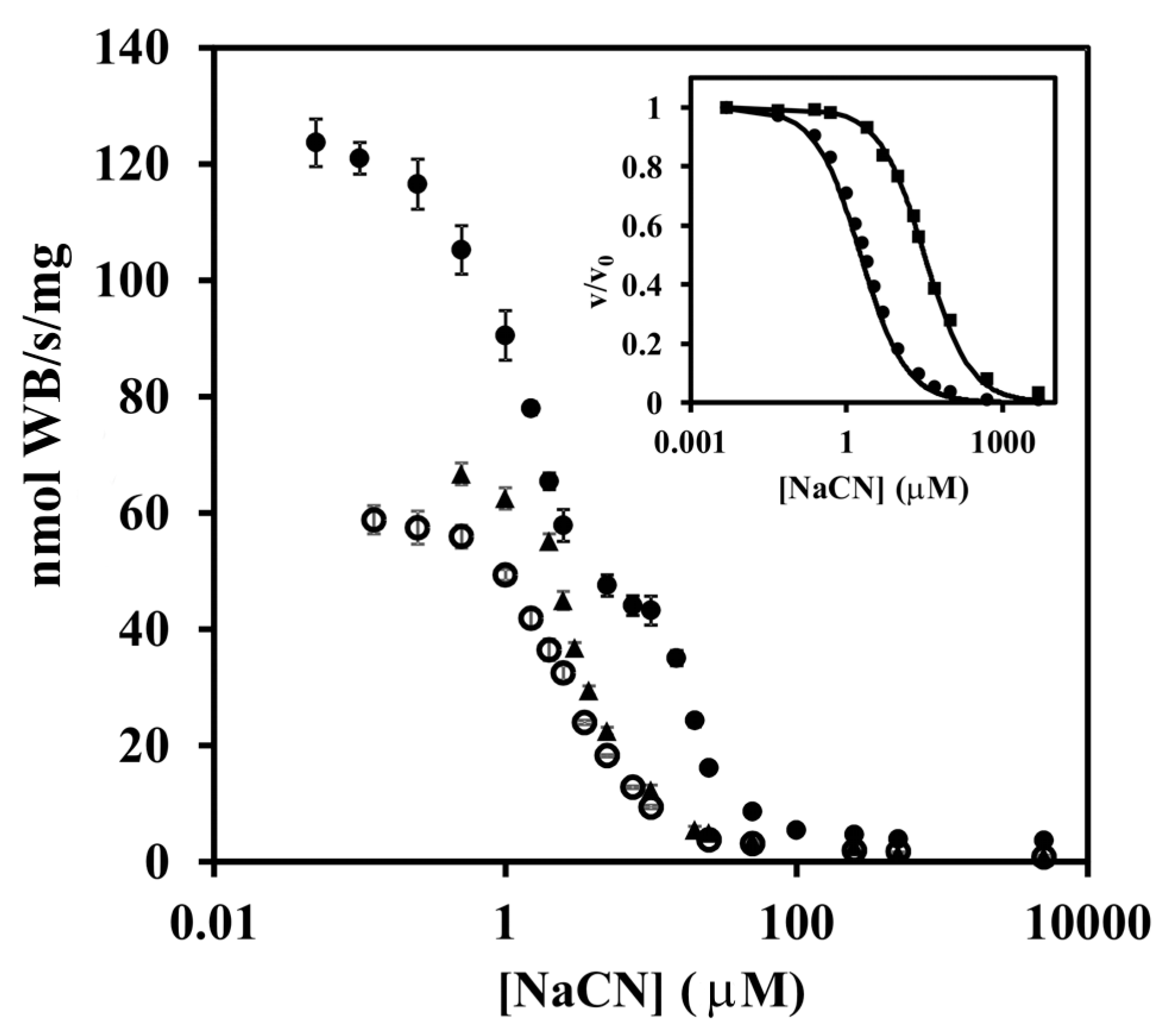
3. Results
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Campian, J.L.; Qian, M.Q.; Gao, X.H.; Eaton, J.W. Oxygen tolerance and coupling of mitochondrial electron transport. J. Biol. Chem. 2004, 279, 46580–46587. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A.; Fridovich, I. Assay of metabolic superoxide production in Escherichia coli. J. Biol. Chem. 1991, 266, 6957–6965. [Google Scholar]
- González-Flecha, B.; Demple, B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J. Biol. Chem. 1995, 270, 13681–13687. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Marcillat, O.; Giulivi, C.; Ernster, L.; Davies, K.J.A. The oxidative inactivation of mitochondrial electron-transport chain components and ATPase. J. Biol. Chem. 1990, 265, 16330–16336. [Google Scholar]
- Palmeira, C.M.; Moreno, A.J.; Madeira, V.M.C. Mitochondrial bioenergetics is affected by the herbicide paraquat. Bba-Bioenerg. 1995, 1229, 187–192. [Google Scholar] [CrossRef] [Green Version]
- Bosshard, F.; Bucheli, M.; Meur, Y.; Egli, T. The respiratory chain is the cell’s Achilles’ heel during UVA inactivation in Escherichia coli. Microbiol. Sgm 2010, 156, 2006–2015. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Vasquez, W.A.; Abarca-Lagunas, M.J.; Cornejo, F.A.; Pinto, C.A.; Arenas, F.A.; Vasquez, C.C. Tellurite-mediated damage to the Escherichia coli NDH-dehydrogenases and terminal oxidases in aerobic conditions. Arch. Biochem. Biophys. 2015, 566, 67–75. [Google Scholar] [CrossRef]
- John, P.; Whatley, F.R. Paracoccus denitrificans and evolutionary origin of mitochondrion. Nature 1975, 254, 495–498. [Google Scholar] [CrossRef]
- Xu, X.M.; Matsunoyagi, A.; Yagi, T. DNA sequencing of the seven remaining structural genes of the gene cluster encoding the energy-transducing NADH-quinone oxidoreductase of Paracoccus denitrificans. Biochemistry 1993, 32, 968–981. [Google Scholar] [CrossRef]
- Kurowski, B.; Ludwig, B. The genes of the Paracoccus denitrificans bc1 complex. Nucleotide sequence and homologies between bacterial and mitochondrial subunits. J. Biol. Chem. 1987, 262, 13805–13811. [Google Scholar]
- Raitio, M.; Jalli, T.; Saraste, M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. Embo J. 1987, 6, 2825–2833. [Google Scholar] [CrossRef]
- Rich, P.R.; Marechal, A. The mitochondrial respiratory chain. Essays Biochem. 2010, 47, 1–23. [Google Scholar]
- Otten, M.F.; Stork, D.R.; Reijnders, W.N.M.; Westerhoff, H.V.; van Spanning, R.J.M. Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur. J. Biochem. 2001, 268, 2486–2497. [Google Scholar] [CrossRef]
- De Gier, J.W.L.; Lubben, M.; Reijnders, W.N.M.; Tipker, C.A.; Slotboom, D.J.; van Spanning, R.J.M.; Stouthamer, A.H.; van der Oost, J. The terminal oxidases of Paracoccus denitrificans. Mol. Microbiol. 1994, 13, 183–196. [Google Scholar] [CrossRef]
- Zickermann, I.; Tautu, O.S.; Link, T.A.; Korn, M.; Ludwig, B.; Richter, O.M.H. Expression studies on the ba3 quinol oxidase from Paracoccus denitrificans. A bb3 variant is enzymatically inactive. Eur. J. Biochem. 1997, 246, 618–624. [Google Scholar] [CrossRef]
- Otten, M.F.; Reijnders, W.N.M.; Bedaux, J.J.M.; Westerhoff, H.V.; Krab, K.; van Spanning, R.J.M. The reduction state of the Q-pool regulates the electron flux through the branched respiratory network of Paracoccus denitrificans. Eur. J. Biochem. 1999, 261, 767–774. [Google Scholar] [CrossRef] [Green Version]
- Hutchings, M.I.; Crack, J.C.; Shearer, N.; Thompson, B.J.; Thomson, A.J.; Spiro, S. Transcription factor FnrP from Paracoccus denitrificans contains an iron-sulfur cluster and is activated by anoxia: Identification of essential cysteine residues. J. Bacteriol. 2002, 184, 503–508. [Google Scholar] [CrossRef] [Green Version]
- Crack, J.C.; Hutchings, M.I.; Thomson, A.J.; Le Brun, N.E. Biochemical properties of Paracoccus denitrificans FnrP: Reactions with molecular oxygen and nitric oxide. J. Biol. Inorg. Chem. 2016, 21, 71–82. [Google Scholar] [CrossRef] [Green Version]
- Van Spanning, R.J.M.; de Boer, A.P.N.; Reijnders, W.N.M.; Westerhoff, H.V.; Stouthamer, A.H.; van der Oost, J. FnrP and NNR of Paracoccus denitrificans are both members of the FNR family of transcriptional activators but have distinct roles in respiratory adaptation in response to oxygen limitation. Mol. Microbiol. 1997, 23, 893–907. [Google Scholar] [CrossRef]
- Ferguson, S.J. Paracoccus denitrificans oxidative phosphorylation: Retentions, gains, losses, and lessons en route to mitochondria. Iubmb Life 2018, 70, 1214–1221. [Google Scholar] [CrossRef] [Green Version]
- de Vries, G.E.; Harms, N.; Hoogendijk, J.; Stouthamer, A.H. Isolation and characterization of Paracoccus denitrificans mutants with increased conjugation frequencies and pleiotropic loss of a (nGATCn) DNA-modifying property. Arch. Microbiol. 1989, 152, 52–57. [Google Scholar] [CrossRef]
- Burnell, J.N.; John, P.; Whatley, F.R. Reversibility of active sulfate transport in membrane vesicles of Paracoccus denitrificans. Biochem. J. 1975, 150, 527–536. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.K.; Krohn, R.I.; Hermanson, G.T.; Mallia, A.K.; Gartner, F.H.; Provenzano, M.D.; Fujimoto, E.K.; Goeke, N.M.; Olson, B.J.; Klenk, D.C. Measurement of protein using bicinchoninic acid. Anal. Biochem. 1985, 150, 76–85. [Google Scholar] [CrossRef]
- Rothe, G.; Valet, G. Flow cytometric analysis of respiratory burst activity in phagocytes with hydroethidine and 2’,7’-dichlorofluorescin. J. Leukoc. Biol. 1990, 47, 440–448. [Google Scholar] [CrossRef]
- Kulzer, R.; Pils, T.; Kappl, R.; Huttermann, J.; Knappe, J. Reconstitution and characterization of the polynuclear iron-sulfur cluster in pyruvate formate-lyase-activating enzyme. Molecular properties of the holoenzyme form. J. Biol. Chem. 1998, 273, 4897–4903. [Google Scholar] [CrossRef] [Green Version]
- Bernofsky, C.; Swan, M. Improved cycling assay for nicotinamide adenine dinucleotide. Anal. Biochem. 1973, 53, 452–458. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, L.; Yang, F.; Lin, X.; Zhang, S.; Zhao, Z.K. Determining the extremes of the cellular NAD(H) level by using an Escherichia coli NAD(+)-auxotrophic mutant. Appl. Env. Microbiol. 2011, 77, 6133–6140. [Google Scholar] [CrossRef] [Green Version]
- Kucera, I.; Lampardova, L.; Dadak, V. Control of respiration rate in non-growing cells of Paracoccus denitrificans. Biochem. J. 1987, 246, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sedlacek, V.; Ptackova, N.; Rejmontova, P.; Kucera, I. The flavoprotein FerB of Paracoccus denitrificans binds to membranes, reduces ubiquinone and superoxide, and acts as an in vivo antioxidant. Febs J. 2015, 282, 283–296. [Google Scholar] [CrossRef]
- Bus, J.S.; Gibson, J.E. Paraquat: Model for oxidant-initiated toxicity. Env. Health Persp. 1984, 55, 37–46. [Google Scholar] [CrossRef]
- Flint, D.H.; Tuminello, J.F.; Emptage, M.H. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J. Biol. Chem. 1993, 268, 22369–22376. [Google Scholar]
- Flint, D.H.; Allen, R.M. Iron-sulfur proteins with nonredox functions. Chem. Rev. 1996, 96, 2315–2334. [Google Scholar] [CrossRef]
- Imlay, J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006, 59, 1073–1082. [Google Scholar] [CrossRef]
- Djaman, O.; Outten, F.W.; Imlay, J.A. Repair of oxidized iron-sulfur clusters in Escherichia coli. J. Biol. Chem. 2004, 279, 44590–44599. [Google Scholar] [CrossRef] [Green Version]
- Dunstan, R.H.; Whatley, F.R.; Greenaway, W. Growth of Paracoccus denitrificans on [2,3-13C]succinate and [1,4-13C]succinate. I. The flux of carbon in energy metabolism and the operation of the TCA cycle. Proc. R. Soc. Lond. BBiol. Sci. 1987, 231, 339–347. [Google Scholar]
- Mailloux, R.J.; Lemire, J.; Appanna, V.D. Metabolic networks to combat oxidative stress in Pseudomonas fluorescens. Antonie Van Leeuwenhoek 2011, 99, 433–442. [Google Scholar] [CrossRef]
- Massudi, H.; Grant, R.; Braidy, N.; Guest, J.; Farnsworth, B.; Guillemin, G.J. Age-associated changes in oxidative stress and NAD(+) metabolism in human tissue. PLos ONE 2012, 7, e42357. [Google Scholar] [CrossRef]
- Sedlacek, V.; Kucera, I. Functional and mechanistic characterization of an atypical flavin reductase encoded by the pden_5119 gene in Paracoccus denitrificans. Mol. Microbiol. 2019, 112, 166–183. [Google Scholar] [CrossRef]
- Crack, J.C.; Green, J.; Cheesman, M.R.; Le Brun, N.E.; Thomson, A.J. Superoxide-mediated amplification of the oxygen-induced switch from [4Fe-4S] to [2Fe-2S] clusters in the transcriptional regulator FNR. Proc. Natl. Acad. Sci. USA 2007, 104, 2092–2097. [Google Scholar] [CrossRef] [Green Version]
- Sutton, V.R.; Stubna, A.; Patschkowski, T.; Munck, E.; Beinert, H.; Kiley, P.J. Superoxide destroys the [2Fe-2S](2+) cluster of FNR from Escherichia coli. Biochemistry 2004, 43, 791–798. [Google Scholar] [CrossRef] [PubMed]
- Pernikarova, V.; Sedlacek, V.; Potesil, D.; Prochazkova, I.; Zdrahal, Z.; Bouchal, P.; Kucera, I. Proteomic responses to a methyl viologen-induced oxidative stress in the wild type and FerB mutant strains of Paracoccus denitrificans. J. Proteom. 2015, 125, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Preisig, O.; Zufferey, R.; ThonyMeyer, L.; Appleby, C.A.; Hennecke, H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 1996, 178, 1532–1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouchal, P.; Struharova, I.; Budinska, E.; Sedo, O.; Vyhlidalova, T.; Zdrahal, Z.; van Spanning, R.; Kucera, I. Unraveling an FNR based regulatory circuit in Paracoccus denitrificans using a proteomics-based approach. Bba-Proteins Proteom. 2010, 1804, 1350–1358. [Google Scholar] [CrossRef] [PubMed]
- Giannopoulos, G.; Sullivan, M.J.; Hartop, K.R.; Rowley, G.; Gates, A.J.; Watmough, N.J.; Richardson, D.J. Tuning the modular Paracoccus denitrificans respirome to adapt from aerobic respiration to anaerobic denitrification. Env. Microbiol. 2017, 19, 4953–4964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imlay, J.A. Transcription factors that defend bacteria against reactive oxygen species. Annu. Rev. Microbiol. 2015, 69, 93–108. [Google Scholar] [CrossRef] [Green Version]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sedláček, V.; Kučera, I. Modifications of the Aerobic Respiratory Chain of Paracoccus Denitrificans in Response to Superoxide Oxidative Stress. Microorganisms 2019, 7, 640. https://doi.org/10.3390/microorganisms7120640
Sedláček V, Kučera I. Modifications of the Aerobic Respiratory Chain of Paracoccus Denitrificans in Response to Superoxide Oxidative Stress. Microorganisms. 2019; 7(12):640. https://doi.org/10.3390/microorganisms7120640
Chicago/Turabian StyleSedláček, Vojtěch, and Igor Kučera. 2019. "Modifications of the Aerobic Respiratory Chain of Paracoccus Denitrificans in Response to Superoxide Oxidative Stress" Microorganisms 7, no. 12: 640. https://doi.org/10.3390/microorganisms7120640



