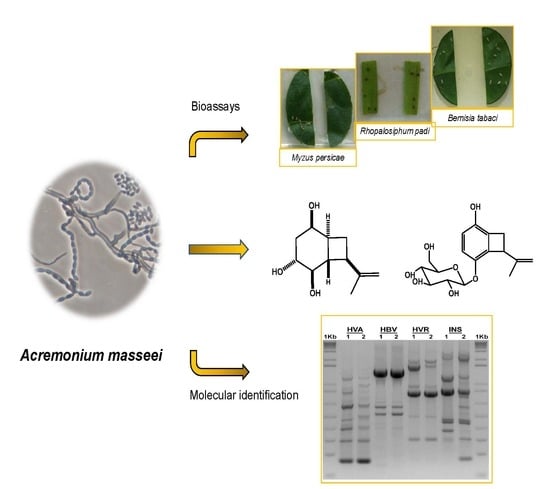
Identification of Insect-Deterrent Metabolites from Acremonium masseei strain CICY026, a Saprophytic Fungus from a Sinkhole in Yucatán
Abstract
:1. Introduction
2. Materials and Methods
2.1. General
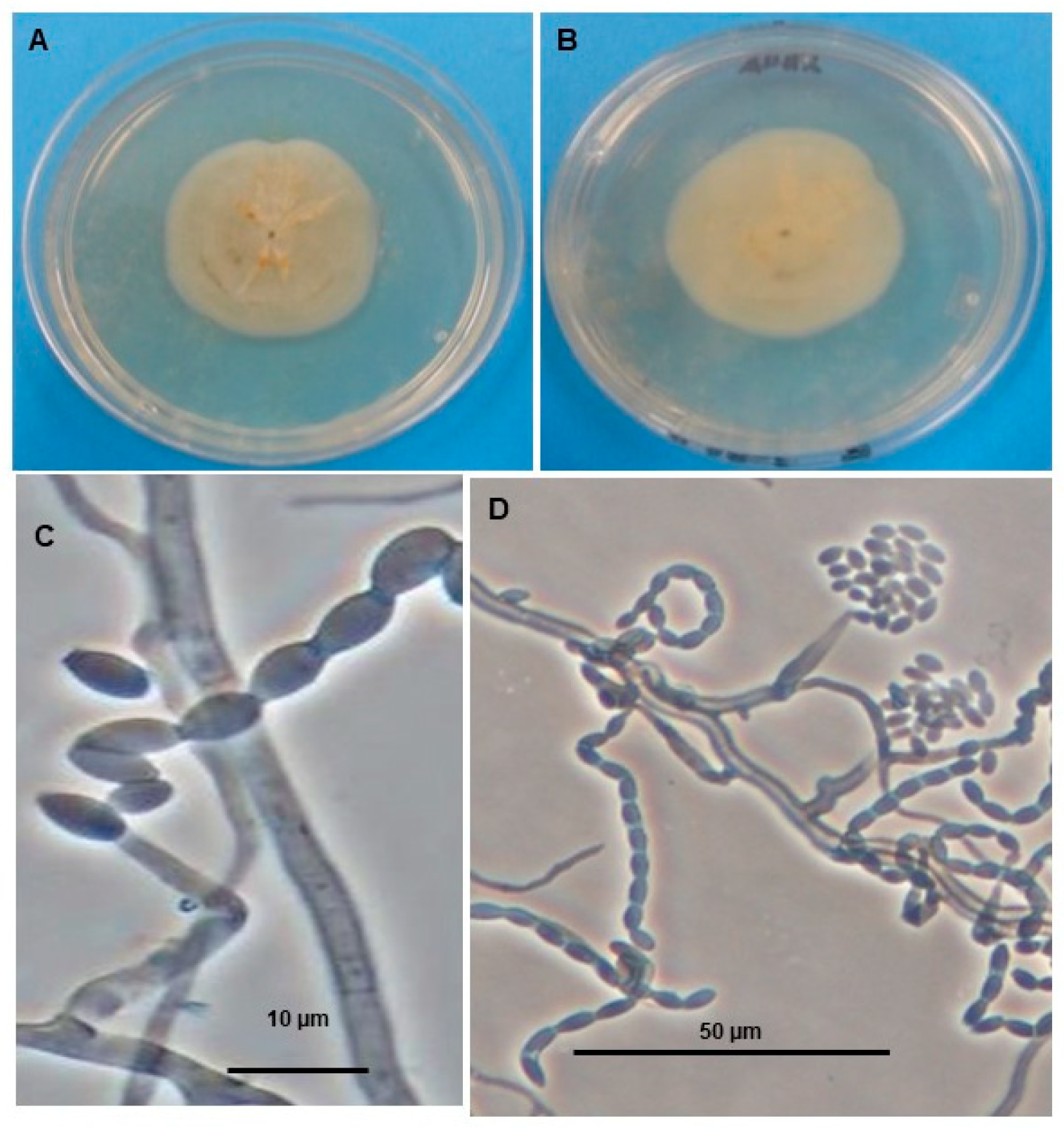
2.2. Fungal Material
2.3. Molecular Identification of Fungal Strains
2.3.1. Isolation of DNA
2.3.2. PCR Amplification and Sequencing of 5.8S-ITS of rDNA
2.4. PCR Fingerprint Analyses
2.4.1. Directed Amplification of Minisatellite DNA (DAMD) Reactions
2.4.2. Arbitrarily Primed PCR (AP-PCR)
2.5. Fungal Extraction and Purification of Compounds
2.6. Computational Details
2.7. Insecticidal Activity
2.7.1. Bioassay on Oviposition Inhibition on Bemisia tabaci
2.7.2. Bioassay of Settling Inhibition of Myzus persicae and Rhopalosiphum padi
3. Results and Discussion
3.1. Molecular Identification of Acremonium masseei Strain CICY026
3.2. Isolation and Structural Identification of Compounds 1 and 2
3.3. Insecticidal Activity of Acremonium masseei CICY026
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Blackman, R.L.; Eastop, V.F. Aphids on the World’s Crops: An Identification and Information Guide, 2nd ed.; Wiley and Sons: Chichester, UK, 2000; 466p. [Google Scholar]
- Oliveira, M.R.V.; Henneberry, T.J.; Anderson, P. History, current status, and collaborative research projects for Bemisia tabaci. Crop. Prot. 2001, 20, 709–723. [Google Scholar] [CrossRef] [Green Version]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Murilo Zerbini, F.; Adkins, S.; et al. World Management of Geminiviruses. Ann. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Jiménez, A.L.; González-Coloma, A.; Andrés-Yeves, M.F.; Ruiz-Sánchez, E.; Heredia, G.; Peraza-Sánchez, S.R.; Medina-Baizabal, I.L.; Reyes-Estebanez, M.; Canto-Canché, B.; Gamboa-Angulo, M. Insecticidal and nematotoxic effects of tropical fungal extracts and deterrent activity of Gliomastix masseei on aphids. Rev. Arg. Microbiol. 2017, 49, 83–92. [Google Scholar]
- Gamboa-Angulo, M.; de la Rosa-García, S.C.; Heredia-Abarca, G.; Medina-Baizabal, I.L. Antimicrobial screening of tropical microfungi isolated from sinkholes located in the Yucatán peninsula, México. Afr. J. Microbial. Res. 2012, 6, 2305–2312. [Google Scholar] [CrossRef]
- Summerbell, R.C.; Gueidan, C.; Schroers, H.-J.; de Hoog, G.S.; Starink, M.; Arocha Rosete, Y.; Guarro, J.; Scott, J.A. Acremonium phylogenetic overview and revision of Gliomastix, Sarocladium, and Trichothecium. Stud. Mycol. 2011, 68, 139–162. [Google Scholar] [CrossRef]
- Index Fungorum. Available online: http://www.indexfungorum.org/Names/NamesRecord.asp?RecordID=314510 (accessed on 31 August 2019).
- Tian, J.; Lai, D.; Zhou, L. Secondary metabolites from Acremonium fungi: Diverse structures and bioactivities. Mini. Rev. Med. Chem. 2017, 17, 603–632. [Google Scholar] [CrossRef]
- Rowan, D.D.; Hunt, M.B.; Gaynor, D.L. Peramine, a novel insect feeding deterrent from ryegrass infected with the endophyte Acremonium loliae. J. Chem. Soc. Chem. Commun. 1986, 935–936. [Google Scholar] [CrossRef]
- Schiell, M.; Hofmann, J.; Kurz, M.; Schmidt, F.R.; Vertesy, L.; Vogel, M.; Wink, J.; Seibert, G. Cephaibols, new peptaibol antibiotics with anthelmintic properties from Acremonium tubakii DSM 12774. J. Antibiot. 2001, 54, 220–233. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Zhang, P.; Liu, T.; Wang, X.-F.; Li, Z.-X.; Li, W.; Wang, F.L. Derivatives isolated from a marine alga-derived endophytic fungus, Acremonium vitellinum, against the cotton bollworm, Helicoverpa armigera (Hϋbner) (Lepidoptera: Noctuidae). Molecules 2018, 23, 2995. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Estebanez, M.; Herrera-Parra, E.; Cristóbal-Alejo, J.; Heredia-Abarca, G.; Canto-Canché, B.; Medina-Baizabal, I.; Gamboa-Angulo, M. Antimicrobial and nematicidal screening of anamorphic fungi isolated from plant debris of tropical areas in Mexico. Afr. J. Microbiol. Res. 2011, 5, 1083–1089. [Google Scholar] [CrossRef]
- Moo-Koh, F.A.; Cristóbal-Alejo, J.; Reyes-Ramírez, A.; Tun-Suárez, J.M.; Sandoval-Luna, R.; Ramírez-Pool, J.A. In vitro activity of an aqueous extract of Bonellia flammea against phytopathogenic fungi. Agrociencia 2014, 48, 833–845. [Google Scholar]
- White, T.; Bruns, T.; Lee, S.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White , T.J., Eds.; Academic Press: New York, NY, USA, 1990; Volume 31, pp. 315–322. [Google Scholar]
- BioEdit version 7.0.5. Available online: http://www.mbio.ncsu.edu/BioEdit/ (accessed on 16 December 2019).
- Green, M.R.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor: New York, NY, USA, 2012. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Talhinhas, P.; Sreenivasaprasad, S.; Neves-Martin, J.; Oliveira, H. Genetic and morphological characterization of Colletotrichum acutatum causing anthracnose of lupins. Phytopathology 2002, 92, 986–996. [Google Scholar] [CrossRef] [Green Version]
- Bridge, P.D.; Pearce, D.A.; Rivera, A.; Rutherford, M.A. VNTR derived oligonucleotides as PCR primers for population studies in filamentous fungi. Lett. Appl. Microbiol. 1997, 24, 426–430. [Google Scholar] [CrossRef]
- Zhou, Z.; Bebeli, P.J.; Somers, D.J.; Gustafson, J.P. Direct amplification of minisatellite-region DNA with VNTR core sequences in the genus Oryza. Theor. Appl. Genet. 1997, 95, 942–949. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, C.1; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Hehre, W.; Ohlinger, S. Spartan’16 Tutorial and User’s Guide; Wavefunction, Inc.: Irvine, CA, USA, 2017. [Google Scholar]
- Cruz-Estrada, A.; Gamboa-Angulo, M.; Borges-Argáez, R.; Ruiz-Sánchez, E. Insecticidal effects of plant extracts on immature whitefly Bemisia tabaci Genn. (Hemiptera: Aleyroideae). Electron. J. Biotechnol. 2013, 16. [Google Scholar] [CrossRef]
- Cruz-Estrada, A.; Ruiz-Sánchez, E.; Medina-Baizabal, I.L.; Balam-Uc, E.; Gamboa-Angulo, M. Effect of Eugenia winzerlingii extracts on Bemisia tabaci and evaluation of its nursery propagation. Phyton J. Exp. Bot. 2019, 88, 161–170. [Google Scholar]
- González-Coloma, A.; Gutiérrez, C.; del Corral, J.M.; Gordaliza, M.; de la Puente, M.L.; San Feliciano, A. Structure and species-dependent insecticidal effects of neoclerodane diterpenes. J. Agric. Food Chem. 2000, 48, 3677–3681. [Google Scholar] [CrossRef]
- Abdel-Lateff, A.; König, G.M.; Fisch, K.M.; Höller, U.; Jones, P.G.; Wright, A.D. New antioxidant hydroquinone derivatives from the algicolous marine fungus Acremonium sp. J. Nat. Prod. 2002, 65, 1605–1611. [Google Scholar] [CrossRef]
- Kara, Y.; Balci, M. A new and stereospecific synthesis of an inositol analogue: bis-homoinositol. Tetrahedron 2003, 59, 2063–2066. [Google Scholar] [CrossRef]
- Deyrup, S.T.; Swenson, D.C.; Gloer, J.B.; Wicklow, D.T. Caryophyllene sesquiterpenoids from a fungicolous isolate of Pestalotiopsis disseminata. J. Nat. Prod. 2006, 69, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.H.; Wiclkow, D.T.; Gloer, J.B. New punctaporins from two fungicolous isolates of Pestalotiopsis sp. Phytochem Lett. 2016, 16, 257–262. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, Q.; Liu, X.; Che, Y. Punctaporonins N–S, New Caryophyllene Sesquiterpenoids from Cytospora sp. BioMed Res. Int. 2017, 2017, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudlicky, T.; Entwistle, D.A.; Pitzer, K.K.; Thorpe, A.J. Modern methods of monosaccharide synthesis from non-carbohydrate sources. Chem. Rev. 1996, 96, 1195–1220. [Google Scholar] [CrossRef]
- Santana, O.; Reina, M.; Fraga, B.M.; Sanz, J.; González-Coloma, A. Antifeedant activity of fatty acid esters and phytosterols from Echium wildpretii. Chem. Biodivers. 2012, 9, 567–576. [Google Scholar] [CrossRef]
- Castillo, L.; Díaz, M.; González-Coloma, A.; González, A.; Alonso-Paz, E.; Bassagoda, M.J.; Rossini, C. Clytostoma callistegioides (Bignoniaceae) wax extract with activity on aphid settling. Phytochemistry 2010, 71, 2052–2057. [Google Scholar] [CrossRef]
- Schuster, D.J.; Thompson, S.; Ortega, L.D.; Polston, J.E. Laboratory evaluation of products to reduce settling of sweetpotato whitefly adults. J. Econ. Entomol. 2009, 102, 1482–1489. [Google Scholar] [CrossRef]
- Nerio, L.S.; Olivero-Verbel, J.; Stashenko, E. Repellent activity of essential oils: A review. Bioresour. Technol. 2010, 101, 372–378. [Google Scholar] [CrossRef]
- Strasser, H.; Vey, A.; Butt, T.M. Are there any risks in using entomopathogenic fungi for pest control, with particular reference to the bioactive metabolites of Metarhizium, Tolypocladium and Beauveria species? Biocontrol Sci. Tech. 2010, 10, 717–735. [Google Scholar] [CrossRef]





| Position | 13C ppm | δ 1H, m, J = Hz | COSY | HMBC |
|---|---|---|---|---|
| 1 | 43.5 (d) | 2.04, ddd, 10.2, 10.2, 11.8 | 2, 6, 8 | C-2, C-5 |
| 2 | 78.9 (d) | 3.49, dd, 8.2, 10.5 | 1, 3 | C-3 |
| 3 | 72.7 (d) | 3.73, ddd, 5.1, 8.2, 11.4 | 2, 4a, 4b | |
| 4 | 41.7 (t) | a. 2.14, ddd, 2.6, 5.0, 13.9 | 3, 4b, 5 | C-3 |
| b. 1.59, ddd, 2.8, 11.4, 14.0 | 3, 4a, 5 | C-3 | ||
| 5 | 66.6 (d) | 4.04, dd, 2.5, 5.0 | 4a, 4b | C-1, C-3, C-4 |
| 6 | 41.3 (d) | 1.75, m | 1, 5, 7a, 7b | C-8, C-5 |
| 7 | 28.6 (t) | a. 1.62, ddd, 9.2, 9.2, 10.9 | 6, 7b, 8 | |
| b. 1.99, ddd, 6.4, 9.2, 12.9 | 6, 7a, 8 | |||
| 8 | 46.4 (d) | 2.54, bdd, 9.2, 15.8 | 1, 7a | |
| 9 | 147.3 (s) | |||
| 10 | 109.3 (t) | a. 4.80, bs | 8, 10b, 11 | C-8, C-9, C-11 |
| b. 4.74, dd, 1.4, 2.9 | 10a, 11 | C-8, C-9, C-11 | ||
| 11 | 21.0 (q) | 1.72, s | 10a, 10b | C-8, C-9, C-10 |
| Carbon No. | δ 13C | |
|---|---|---|
| Experimental | Theoretical | |
| C1 | 66.6 | 65.7 |
| C2 | 41.7 | 41.0 |
| C3 | 72.7 | 72.8 |
| C4 | 78.9 | 78.9 |
| C5 | 43.5 | 44.9 |
| C6 | 41.3 | 40.5 |
| C7 | 28.6 | 24.9 |
| C8 | 46.4 | 44.9 |
| C9 | 147.3 | 144.6 |
| C10 | 109.3 | 110.3 |
| C11 | 21.0 | 21.1 |
| rms | 1.60 | |
| Sample | % OI B. tabaci | % SI M. persicae | % SI R. padi |
|---|---|---|---|
| EtOAc extract | 35.7 ± 11.4 | 67.5 ± 7.4 | 75.3 ± 5.0 |
| Fraction 1A (AcN fraction) | 31.6 ± 10.7 | 76.1 ± 5.8 | 48.6 ± 6.9 |
| Fraction 1B (hexane fraction) | 43.8 ± 14.8 | 67.8 ± 7.0 | 41.5 ± 7.5 |
| 1 | 24.4 ± 8.6 | 55.6 ± 5.4 | 59.0 ± 4.5 |
| 2 | 19.6 ± 6.2 | 67.2 ± 4.8 | 48.1 ± 4.9 |
| Blank (fermented rice) | 34.1 ± 12.8 | 34 ± 8.2 | 48.4 ± 7.3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Jiménez, A.L.; Ruiz-Sánchez, E.; Heredia, G.; Tapia-Tussell, R.; González-Coloma, A.; Peraza-Jiménez, K.; Moo-Koh, F.A.; Medina-Baizabal, I.L.; Hernández-Romero, Y.; Mena-Rejón, G.J.; et al. Identification of Insect-Deterrent Metabolites from Acremonium masseei strain CICY026, a Saprophytic Fungus from a Sinkhole in Yucatán. Microorganisms 2019, 7, 712. https://doi.org/10.3390/microorganisms7120712
Ruiz-Jiménez AL, Ruiz-Sánchez E, Heredia G, Tapia-Tussell R, González-Coloma A, Peraza-Jiménez K, Moo-Koh FA, Medina-Baizabal IL, Hernández-Romero Y, Mena-Rejón GJ, et al. Identification of Insect-Deterrent Metabolites from Acremonium masseei strain CICY026, a Saprophytic Fungus from a Sinkhole in Yucatán. Microorganisms. 2019; 7(12):712. https://doi.org/10.3390/microorganisms7120712
Chicago/Turabian StyleRuiz-Jiménez, Ana L., Esaú Ruiz-Sánchez, Gabriela Heredia, Raúl Tapia-Tussell, Azucena González-Coloma, Karla Peraza-Jiménez, Felicia A. Moo-Koh, Irma L. Medina-Baizabal, Yanet Hernández-Romero, Gonzalo J. Mena-Rejón, and et al. 2019. "Identification of Insect-Deterrent Metabolites from Acremonium masseei strain CICY026, a Saprophytic Fungus from a Sinkhole in Yucatán" Microorganisms 7, no. 12: 712. https://doi.org/10.3390/microorganisms7120712
APA StyleRuiz-Jiménez, A. L., Ruiz-Sánchez, E., Heredia, G., Tapia-Tussell, R., González-Coloma, A., Peraza-Jiménez, K., Moo-Koh, F. A., Medina-Baizabal, I. L., Hernández-Romero, Y., Mena-Rejón, G. J., Quijano-Quiñones, R. F., & Gamboa-Angulo, M. (2019). Identification of Insect-Deterrent Metabolites from Acremonium masseei strain CICY026, a Saprophytic Fungus from a Sinkhole in Yucatán. Microorganisms, 7(12), 712. https://doi.org/10.3390/microorganisms7120712