The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Microfungal Isolation
2.2. Molecular Experiment and Phylogenetic Analysis for Identification
2.3. Effect of Fungal Metabolites on PM Growth
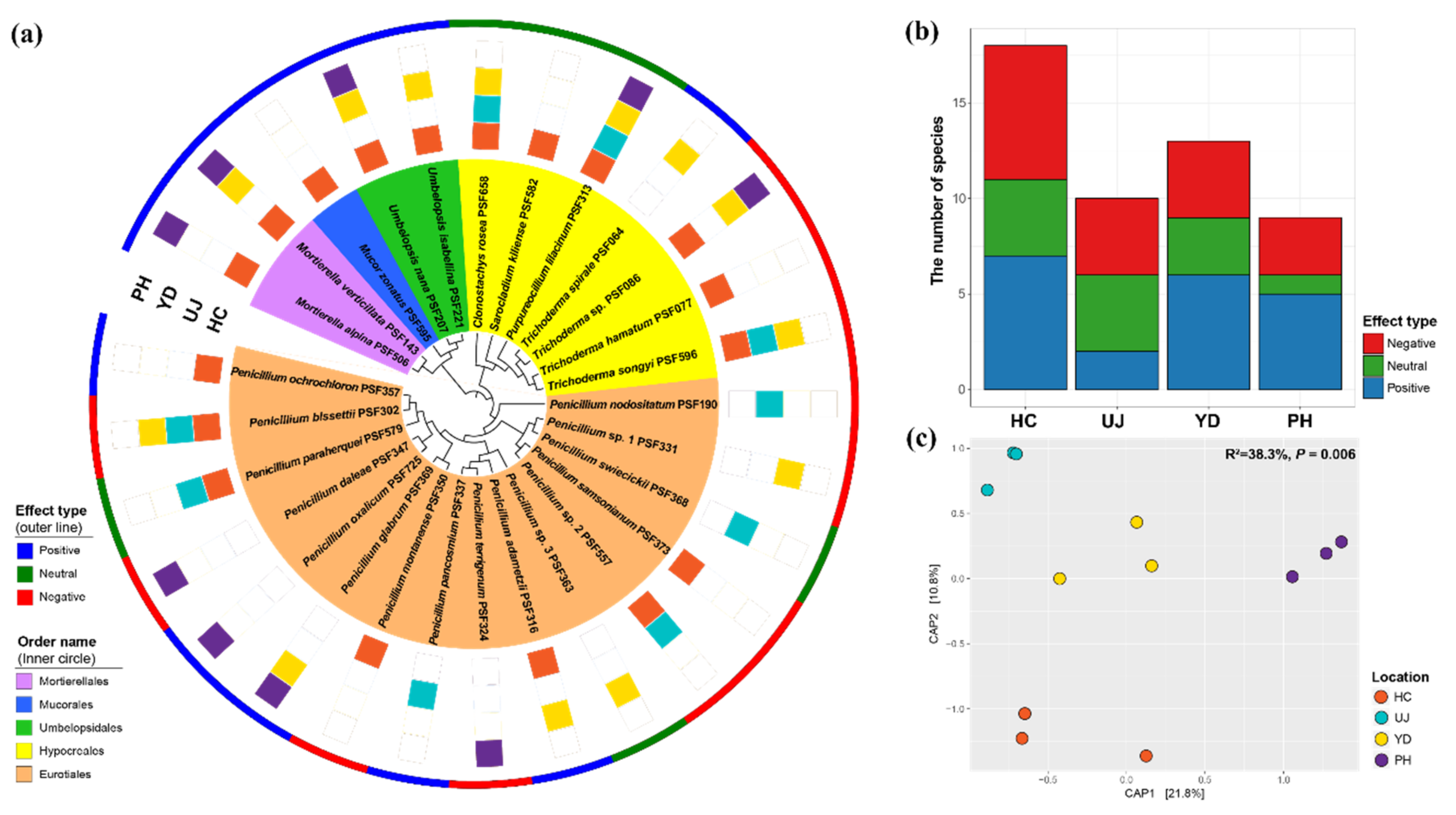
3. Results
3.1. Species Identification and Composition
3.2. Effect of Fungal Metabolite on Mycelial Growth of PM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Landeweert, R.; Hoffland, E.; Finlay, R.D.; Kuyper, T.W.; van Breemen, N. Linking plants to rocks: Ectomycorrhizal fungi mobilize nutrients from minerals. Trends Ecol. Evol. 2001, 16, 248–254. [Google Scholar] [CrossRef]
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P. At the interface between mycorrhizal fungi and plants: The structural organization of cell wall, plasma membrane and cytoskeleton. In The Mycota IX: Fungal Associations; Hock, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2001; pp. 45–61. [Google Scholar]
- Garbaye, J. Helper bacteria: A new dimension to the mycorrhizal symbiosis. New Phytol. 1994, 128, 197–210. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Garbaye, J.; Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 2007, 176, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A.; et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, G.A.; Kiers, E.T.; Gardner, A.; West, S.A. A biological market analysis of the plant-mycorrhizal symbiosis. Evolution 2014, 68, 2603–2618. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Pärtel, K.; Jairus, T.; Gates, G.; Põldmaa, K.; Tamm, H. Ascomycetes associated with ectomycorrhizas: Molecular diversity and ecology with particular reference to the Helotiales. Environ. Microbiol. 2009, 11, 3166–3178. [Google Scholar] [CrossRef] [PubMed]
- You, Y.-H.; Yoon, H.-J.; Woo, J.-R.; Rim, S.-O.; Lee, J.-H.; Kong, W.-S.; Kim, J.-G. Diversity of endophytic fungi isolated from the rootlet of Pinus densiflora colonized by Tricholoma matsutake. Korean J. Mycol. 2011, 39, 223–226. [Google Scholar] [CrossRef]
- Kluber, L.A.; Smith, J.E.; Myrold, D.D. Distinctive fungal and bacterial communities are associated with mats formed by ectomycorrhizal fungi. Soil Biol. Biochem. 2011, 43, 1042–1050. [Google Scholar] [CrossRef]
- Li, Q.; Chen, Ch.; Penttinen, P.; Xiong, Ch.; Zheng, L.; Huang, W. Microbial diversity associated with Tricholoma matsutake fruiting bodies. Microbiology 2016, 85, 531–539. [Google Scholar] [CrossRef]
- Pacioni, G.; Leonardi, M.; Aimola, P.; Ragnelli, A.M.; Rubini, A.; Paolocci, F. Isolation and characterization of some mycelia inhabiting Tuber ascomata. Mycol. Res. 2007, 111, 1450–1460. [Google Scholar] [CrossRef] [PubMed]
- Baar, J.; Stanton, N.L. Ectomycorrhizal fungi challenged by saprotrophic basidiomycetes and soil microfungi under different ammonium regimes in vitro. Mycol. Res. 2000, 104, 691–697. [Google Scholar] [CrossRef]
- Leake, J.R.; Donnelly, D.P.; Boddy, L. Interactions between ecto-mycorrhizal and saprotrophic fungi. In Mycorrhizal ecology; van der Heijden, M.G.A., Sanders, I.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 345–372. [Google Scholar]
- Summerbell, R.C. From Lamarckian fertilizers to fungal castles: Recapturing the pre-1985 literature on endophytic and saprotrophic fungi associated with ectomycorrhizal root systems. Stud. Mycol. 2005, 53, 191–256. [Google Scholar] [CrossRef]
- Mucha, J.; Zadworny, M.; Werner, A.; Napierala-Filipiak, A.; Lakomy, P. Antagonistic activity of the ectomycorrhizal fungus Suillus bovinus challenged by saprotrophic fungi from different soils. Nova Hedwig. 2008, 87, 373–385. [Google Scholar] [CrossRef]
- Kope, H.H.; Fortin, J.A. Inhibition of phytopathogenic fungi in vitro by cell free culture media of ectomycorrhizal fungi. New Phytol. 1989, 113, 57–63. [Google Scholar] [CrossRef]
- Whipps, J.M. Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can. J. Bot. 2004, 82, 1198–1227. [Google Scholar] [CrossRef]
- Malyshkin, P.E. Stimulation of tree growth by microorganisms. In Mycotrophy in Plants; Academy of Sciences of the USSR: Moscow, Russia, 1955; pp. 211–220. [Google Scholar]
- Voznyakovskaya, Y.M.; Ryzhkova, A. Microflora accompanying mycorrhizas. In Mycotrophy of Woody Plants; Academy of Sciences of the USSR: Moscow, Russia, 1955; pp. 320–323. [Google Scholar]
- Yun, W.; Hall, I.R.; Evans, L.A. Ectomycorrhizal fungi with edible fruiting bodies 1. Tricholoma matsutake and Related Fungi. Econ. Bot. 1997, 51, 311–327. [Google Scholar]
- Yamanaka, T.; Ota, Y.; Konno, M.; Kawai, M.; Ohta, A.; Neda, H.; Terashima, Y.; Yamada, A. The host ranges of conifer-associated Tricholoma matsutake, Fagaceae-associated T. bakamatsutake and T. fulvocastaneum are wider in vitro than in nature. Mycologia 2014, 106, 397–406. [Google Scholar] [CrossRef]
- Kataoka, R.; Siddiqui, Z.A.; Kikuchi, J.; Ando, M.; Sriwati, R.; Nozaki, A.; Futai, K. Detecting nonculturable bacteria in the active mycorrhizal zone of the pine mushroom Tricholoma matsutake. J. Microbiol. 2012, 50, 199–206. [Google Scholar] [CrossRef]
- Ohara, H.; Hamada, M. Disappearance of bacteria from the zone of active mycorrhizas in Tricholoma matsutake (S. Ito et Imai) Singer. Nature 1967, 213, 528. [Google Scholar] [CrossRef]
- Park, M.S.; Oh, S.-Y.; Cho, H.J.; Fong, J.J.; Cheon, W.-J.; Lim, Y.W. Trichoderma songyi sp. nov., a new species associated with the pine mushroom (Tricholoma matsutake). Antonie Van Leeuwenhoek 2014, 106, 593–603. [Google Scholar] [CrossRef] [PubMed]
- Song, H.-S.; Min, K.-H. Microfungal flora of Tricholoma matsutake producing and nonproducing sites in the forest of Pinus densiflora. Korean J. Mycol. 1991, 19, 109–119. [Google Scholar]
- Kim, M.; Yoon, H.J.; You, Y.H.; Kim, Y.E.; Woo, J.R.; Seo, Y.G.; Lee, G.M.; Kim, Y.J.; Kong, W.S.; Kim, J.G. Metagenomic analysis of fungal communities inhabiting the fairy ring zone of Tricholoma matsutake. J. Microbiol. Biotechnol. 2013, 23, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-Y.; Fong, J.J.; Park, M.S.; Lim, Y.W. Distinctive feature of microbial communities and bacterial functional profiles in Tricholoma matsutake dominant soil. PLoS ONE 2016, 11, e0168573. [Google Scholar] [CrossRef] [PubMed]
- Vaario, L.-M.; Fritze, H.; Spetz, P.; Heinonsalo, J.; Hanajík, P.; Pennanen, T. Tricholoma matsutake dominates diverse microbial communities in different forest soils. Appl. Environ. Microbiol. 2011, 77, 8523–8531. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-Y.; Lim, Y.W. Effect of fairy ring bacteria on the growth of Tricholoma matsutake in vitro culture. Mycorrhiza 2018, 28, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.-Y.; Lim, Y.W. Root-associated bacteria influencing mycelial growth of Tricholoma matsutake (pine mushroom). J. Microbiol. 2018, 56, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, M.; Kawai, M. Studies on the artificial reproduction of Tricholoma matsutake (S. Ito et Imai) Sing. III. Effects of growth promotion of natural products on the vegetative growth of T. matsutake. Trans. Mycol. Soc. Jpn. 1976, 17, 492–498. [Google Scholar]
- Oh, S.-Y.; Park, M.S.; Cho, H.J.; Lim, Y.W. Diversity and effect of Trichoderma isolated from the roots of Pinus densiflora within the fairy ring of pine mushroom (Tricholoma matsutake). PLoS ONE 2018, 13, e0205900. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W.; Consortium, F.B. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Rogers, S.O.; Bendich, A.J. Extraction of total cellular DNA from plants, algae and fungi. In Plant Molecular Biology Manual; Gelvin, S.B., Schilperoort, R.A., Eds.; Springer: Dordrecht, The Netherlands, 1994; pp. 183–190. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J.L. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR protocols: A guide to methods and applications; Academic Press: New York, NY, USA, 1990. [Google Scholar]
- Glass, N.L.; Donaldson, G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995, 61, 1323–1330. [Google Scholar] [PubMed]
- Carbone, I.; Kohn, L.M. A Method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Samuels, G.J.; Dodd, S.L.; Gams, W.; Castlebury, L.A.; Petrini, O. Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 2002, 94, 146–170. [Google Scholar] [CrossRef] [PubMed]
- Park, M.S.; Fong, J.J.; Oh, S.-Y.; Houbraken, J.; Sohn, J.H.; Hong, S.-B.; Lim, Y.W. Penicillium jejuense sp. nov., isolated from the marine environments of Jeju Island, Korea. Mycologia 2015, 107, 209–216. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Park, M.S.; Fong, J.J.; Lee, H.; Oh, S.-Y.; Jung, P.E.; Min, Y.J.; Seok, S.J.; Lim, Y.W. Delimitation of Russula Subgenus Amoenula in Korea Using Three Molecular Markers. Mycobiology 2013, 41, 191–201. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and gaphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. Available online: https://www.R-project.org/ (accessed on 6 June 2019).
- Kim, I.-Y.; Jung, G.-R.; Han, S.-K.; Cha, J.-Y.; Sung, J.-M. Favorable condition for mycelial growth of Tricholoma matsutake. Korean J. Mycol. 2005, 33, 22–29. [Google Scholar]
- Rueden, C.T.; Schindelin, J.; Hiner, M.C.; DeZonia, B.E.; Walter, A.E.; Arena, E.T.; Eliceiri, K.W. ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinform. 2017, 18, 529. [Google Scholar] [CrossRef]
- Wang, F. SIOX plugin in ImageJ: Area measurement made easy. UV4Plants Bull. 2017, 2016, 37–44. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Kubicek, C.P. The first 100 Trichoderma species characterized by molecular data. Mycoscience 2006, 47, 55. [Google Scholar] [CrossRef]
- Visagie, C.M.; Houbraken, J.; Frisvad, J.C.; Hong, S.-B.; Klaassen, C.H.W.; Perrone, G.; Seifert, K.A.; Varga, J.; Yaguchi, T.; Samson, R.A. Identification and nomenclature of the genus Penicillium. Stud. Mycol. 2014, 78, 343–371. [Google Scholar] [CrossRef] [PubMed]
- Chaverri, P.; Castlebury, L.A.; Samuels, G.J.; Geiser, D.M. Multilocus phylogenetic structure within the Trichoderma harzianum/Hypocrea lixii complex. Mol. Phylogenet. Evol. 2003, 27, 302–313. [Google Scholar] [CrossRef]
- Perrone, G.; Stea, G.; Epifani, F.; Varga, J.; Frisvad, J.C.; Samson, R.A. Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biol. 2011, 115, 1138–1150. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.W.; Jacobson, D.J.; Kroken, S.; Kasuga, T.; Geiser, D.M.; Hibbett, D.S.; Fisher, M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000, 31, 21–32. [Google Scholar] [CrossRef]
- Anastasiadis, I.A.; Giannakou, I.O.; Prophetou-Athanasiadou, D.A.; Gowen, S.R. The combined effect of the application of a biocontrol agent Paecilomyces lilacinus, with various practices for the control of root-knot nematodes. Crop Prot. 2008, 27, 352–361. [Google Scholar] [CrossRef]
- Liu, J.; Sun, J.; Qiu, J.; Liu, X.; Xiang, M. Integrated management of root-knot nematodes on tomato in glasshouse production using nematicides and a biocontrol agent, and their effect on soil microbial communities. Nematology 2014, 16, 463–473. [Google Scholar] [CrossRef]
- Cromack, K.; Fichter, B.L.; Moldenke, A.M.; Entry, J.A.; Ingham, E.R. Interactions between soil animals and ectomycorrhizal fungal mats. Agric. Ecosyst. Environ. 1988, 24, 161–168. [Google Scholar] [CrossRef]
- Oh, S.-Y.; Kim, M.; Eimes, J.A.; Lim, Y.W. Effect of fruiting body bacteria on the growth of Tricholoma matsutake and its related molds. PLoS ONE 2018, 13, e0190948. [Google Scholar] [CrossRef] [PubMed]
- Riedlinger, J.; Schrey, S.D.; Tarkka, M.T.; Hampp, R.; Kapur, M.; Fiedler, H.-P. Auxofuran, a novel metabolite that stimulates the growth of fly agaric, is produced by the mycorrhiza helper bacterium Streptomyces strain AcH 505. Appl. Environ. Microbiol 2006, 72, 3550–3557. [Google Scholar] [CrossRef] [PubMed]
- Brulé, C.; Frey-Klett, P.; Pierrat, J.C.; Courrier, S.; Gérard, F.; Lemoine, M.C.; Rousselet, J.L.; Sommer, G.; Garbaye, J. Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol. Biochem. 2001, 33, 1683–1694. [Google Scholar] [CrossRef]
- Sabella, E.; Nutricati, E.; Aprile, A.; Miceli, A.; Sorce, C.; Lorenzi, R.; de Bellis, L. Arthrinium phaeospermum isolated from Tuber borchii ascomata: The first evidence for a “Mycorrhization Helper Fungus”? Mycol. Prog. 2015, 14, 59. [Google Scholar] [CrossRef]
- Labbé, J.L.; Weston, D.J.; Dunkirk, N.; Pelletier, D.A.; Tuskan, G.A. Newly identified helper bacteria stimulate ectomycorrhizal formation in Populus. Front. Plant. Sci. 2014, 5, 579. [Google Scholar] [CrossRef] [PubMed]
- Obase, K. Extending the hyphal area of the ectomycorrhizal fungus Laccaria parva co-cultured with ectomycorrhizosphere bacteria on nutrient agar plate. Mycoscience 2019, 60, 95–101. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, S.-Y.; Park, M.S.; Lim, Y.W. The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake. Microorganisms 2019, 7, 169. https://doi.org/10.3390/microorganisms7060169
Oh S-Y, Park MS, Lim YW. The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake. Microorganisms. 2019; 7(6):169. https://doi.org/10.3390/microorganisms7060169
Chicago/Turabian StyleOh, Seung-Yoon, Myung Soo Park, and Young Woon Lim. 2019. "The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake" Microorganisms 7, no. 6: 169. https://doi.org/10.3390/microorganisms7060169
APA StyleOh, S.-Y., Park, M. S., & Lim, Y. W. (2019). The Influence of Microfungi on the Mycelial Growth of Ectomycorrhizal Fungus Tricholoma matsutake. Microorganisms, 7(6), 169. https://doi.org/10.3390/microorganisms7060169





