1. Introduction
Proteases, also denoted as peptidases, proteinases, or proteolytic enzymes, can be classified according to the nature of the functional group at the active site. Most proteases belong to one of the four major families: Aspartic, cysteine, metallo, and serine peptidases. They are widely used in biotechnology, mainly in the food, leather, and detergent industries, in ecological bioremediation processes, and to produce therapeutic peptides [
1]. They comprise a large number of proteins that account for a significant proportion of an organism’s gene count. Thus, species in the genera,
Aspergillus or
Penicillium, contain more than 200 and 100 annotated genes encoding for putative proteases in the MEROPS database (
https://www.ebi.ac.uk/merops/), respectively. These enzymes play a major role in the physiology, morphogenesis, and metabolism of fungi. Their production is regulated in response to environmental signals, such as extracellular pH and carbon and nitrogen sources [
2]. Proteases secreted into the environment play a crucial role in nutrition because they are needed for external digestion of macromolecular nutrients. In addition to nutrient utilization, microbial proteases are involved in many physiological processes, such as morphogenesis, germination, and conidial discharge [
3]. Proteases play an important role in the mechanism of the virulence of pathogens by participating in the penetration and dissemination within the host, as well as by combating the host’s defense mechanisms [
4,
5,
6,
7]. The role of fungal proteases in plant infection has been less characterized than that of bacterial and animal pathogens. For example,
Sclerotinia sclerotiorum produced aspartyl proteases, non-aspartyl acidic proteases, and serine proteases during infection of sunflower, and the increase of protease production was correlated with intensive colonization and maceration of the host tissues [
8]. A UV-induced mutant of the tomato pathogen
Colletotrichum coccodes defective in extracellular protease activity was unable to infect tomato fruits, although it showed normal vegetative growth and cellulase activity [
9]. In
Fusarium oxysporum f. sp.
lycopersici, the synergistic action of a serine protease, FoSep1, and a metalloprotease, FoMep1, was required for cleavage and removal of the chitin-binding domain (CBD) from two tomato CBD-chitinases [
10]. In addition, mutants of
F. oxysporum f. sp.
lycopersici lacking both FoSep1 and FoMep1 exhibited reduced virulence on tomato, confirming that secreted fungal proteases are important virulence factors by targeting CDB-chitinases to compromise an important component of the plant’s basal defense [
10]. Fungalysins are a conserved family of metalloproteases in fungi and their role as chitinase-degrading enzymes has been demonstrated in
Colletotrichum graminicola. The absence of the fungalisyn metalloprotease-encoding
CgfI gene delayed fungal development during the infection process on maize leaves and, in parallel, maize leaves exhibit increased chitinase activity, suggesting that the fungus employs a CgfI-mediated strategy to control chitin signaling [
11].
Botrytis cinerea is a typical necrotroph that secretes aspartic proteases during infection on various plant tissues. However, single or double deletant mutants in five genes encoding aspartic proteases did not result in any defect in virulence [
12].
PrtT is a fungal-specific transcription activator of extracellular proteases that was first isolated and characterized in
Aspergillus niger [
13]. It is present in several
Aspergilli and
Penicillia, but absent in the genome of
Aspergillus nidulans [
14]. This transcription factor belongs to the fungal-specific Gal
4-like Zn
2Cys
6 binuclear cluster protein family and plays an important role in the production of secreted proteases. Disruption of
prtT in
A. niger resulted in transformants unable to form a protease degradation halo on plates containing skim milk [
13]. Moreover, an
Aspergillus oryzae prtT disruption mutant produced lower levels of the alkaline serine protease S8 (AlpA) and to a lesser extent, the neutral metalloprotease M36 (NpI) compared to the wild type, confirming the role of PrtT in the regulation of the major proteases in this fungus [
13]. Unexpectedly, microarray analysis revealed that the expression of genes involved in iron uptake and ergosterol synthesis was dramatically decreased in the
Aspergillus fumigatus Δ
prtT mutant, together with an upregulation of different secondary metabolite clusters [
15]. However, in two independent works, this transcription factor was found to be not essential for virulence in this human opportunistic fungal pathogen, suggesting that either residual protease activity is sufficient to enable virulence or that proteases are dispensable for pathogenicity in this fungus [
14,
16]. Regarding the genus
Penicillium, PrtT has been only characterized in
Penicillium oxalicum. A transcription profiling analysis using RNA-Seq showed that many putative peptidase-encoding genes were either up- or down-regulated in a
P. oxalicum Δ
prtT mutant, including both secreted and intracellular proteases [
17], confirming that PrtT is a global regulator of proteases. In addition, this transcriptomic study found that PrtT putatively regulates the transcription of specific amylases and major facilitator superfamily (MFS) transporters involved in the transport of nutrients, and of specific transporters and enzymes involved in lignocellulose degradation in response to nutrient limitation.
Penicillium digitatum is the most important postharvest pathogen of citrus fruit grown under Mediterranean conditions. It is a necrotrophic fungus that requires wounds in the fruit peel to penetrate and colonize the fruit tissue mostly through the deployment of maceration enzymes. The genome of this fungus contains 275 putative carbohydrate-active enzymes (CAZymes) assigned mostly to glycoside hydrolases, carbohydrate esterases, and polysaccharide lyases, among others, and to a lesser extent, to enzymes related to the degradation of cellulose and hemicellulose [
18]. In comparison with other
Penicillium spp.,
P. digitatum is enriched in polygalacturonases and pectinesterases, both involved in pectin degradation. This necrotrophic fungus possesses a small secretome compared to
Penicillium expansum or
Penicillium italicum, and proteases constitute a large proportion of its secretome [
19]. The genome of
P. digitatum encodes 119 proteases and 29 non-peptidase homologs (MEROPS peptidase database for
P. digitatum, release 12.1, April 2019) [
20]. The most abundant category corresponds to the superfamily of serine proteases, followed by metallo and cysteine proteases. In a previous study, we observed that genes coding for putative fungal proteases and plant cell wall-degrading enzymes represent the largest categories during the orange–
P. digitatum interaction, with five secreted protease-encoding genes being among the most highly expressed genes during fruit infection [
21]. In this report, we aim to analyze the role of
P. digitatum proteases on virulence. In view of the large number of secreted proteases, we focused on PrtT, which regulates extracellular proteases, with the aim of reducing the production of secreted proteases as much as possible while avoiding the gene compensation effects observed when eliminating a single member of a large gene family. If the major proteases were regulated by PrtT, knocking out the corresponding gene could offer an alternative to study to the role of
P. digitatum proteases in virulence. For this purpose, we followed a functional approach by constructing and characterizing a deletion mutant of the
prtT gene. In addition, we undertook a pharmacological approach by using a set of protease inhibitors during the infection of citrus fruit by
P. digitatum. Our results showed that 1,10-phenanthroline, a metalloprotease inhibitor, is able to control the development of
P. digitatum in citrus fruit.
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
Penicillium digitatum (Pers.:Fr.) Sacc. strain Pd1 (PDIP, deposited at the Spanish Type Culture Collection with accession code CECT20795) was isolated from an infected grapefruit [
18]. To prepare conidial suspensions, the strain was grown on potato-dextrose-agar (PDA) at 24 °C for 7 days. Conidia were scraped off the agar with a sterile spatula, suspended in sterile distilled water, and filtered through a nylon mesh. Conidia concentration was determined with a hemocytometer.
2.2. Generation and Verification of P. digitatum prtT Mutants
A BlastP search with the sequence of PrtT from
A. niger (accession number XM_001402018.2) as the query was performed against the
P. digitatum Pd1 proteome [
18]. To construct the
prtT gene replacement plasmid, 1.8 kb upstream and downstream flanking fragments of the
prtT gene (PDIP_25240) were amplified from genomic DNA of
P. digitatum (Pd1/PDIP), using the specific primers O1, O2, A3, and A4 (
Table 1). These primers include vector-specific 9 bp long overhangs containing a single 2-deoxyuridine nucleoside in the 5’ end, which ensured directionality in the cloning reaction. The two flanking fragments were introduced into pRF-HU2 following the USER (uracil-specific excision reagent) protocol described by Frandsen et al. [
22]. The resulted plasmid (denoted as pDprtT) was introduced into
Escherichia coli DH5α chemical competent cells. Kanamycin-resistant transformants were screened by PCR for the presence of the promoter and the terminator with primer pairs RF1/RF6 and RF2/RF5, respectively (
Table 1). Proper fusions were further confirmed by DNA sequencing and then the plasmid was transferred to
Agrobacterium tumefaciens AGL1 electrocompetent cells. Transformation of
P. digitatum Pd1 was done as previously described [
18]. Equal volumes of induced bacterial culture and conidial suspension of
P. digitatum strain Pd1 (10
5 conidia/mL) were mixed and spread onto filter papers, which were placed on agar plates containing the co-cultivation medium. After co-cultivation at 24 °C for 48 h, the membranes were transferred to PDA plates containing 100 μg/mL of hygromycin B (InvivoGen, San Diego, CA, USA), as the selection agent for fungal transformants, and 200 μg/mL of cefotaxime (Calbiochem, San Diego, CA, USA) to inhibit the growth of
A. tumefaciens cells. Hygromycin-resistant colonies appeared after 4 to 5 days of incubation at 24 °C. To ensure correct deletion of the
prtT gene and the absence of ectopic insertions, conventional PCR and quantitative PCR (qPCR) were used to determine the gene copy number of the T-DNA inserted in
P. digitatum. Firstly, disruption of the
prtT gene was confirmed by PCR analyses of the transformants. Integration of the T-DNA by homologous recombination was examined using primer pairs HPHTER2/1F and HPHRO4/4R (
Table 1) for the promoter and the terminator regions, respectively. Further verification of deletion of the target gene and the insertion of the hygromycin marker was done with primers 5F/6R followed by digestion with
EcoRI. To determine the number of T-DNA molecules that had been integrated into the genome of each selected transformant, a qPCR analysis was carried out following an already demonstrated methodology described by several authors [
21,
23,
24], using Pd1 DNA as the control. A primer pair (7F/8R) was designed within the T-DNA in the terminator region of the target gene, close to the selection marker. The
P. digitatum actin gene (PDIP_18200) was chosen as a reference using the primer pair PdACTFor2/PdACTRev2 (
Table 1). qPCR reactions were performed in a LightCycler480 System (Roche Diagnostics, Basel, Switzerland) using SYBR Green to monitor DNA amplification. For each primer pair and each sample, the PCR efficiencies (E) and the quantification cycle (Cq) were assessed using the LinRegPCR software version 2017.1 [
25]. The number of T-DNA copies that were integrated in the genome of the transformants was calculated according to the formula: Copy number = (E
target gene)^ΔCq
target gene(wild type – transformant)/(E
reference gene) ^ΔCq
reference gene(wild type – transformant) based on Pfaffl [
26], which depends on E and the Cq value of the transformant versus the wild-type strain, and normalized in comparison to a reference gene that is present with the same copy number in both wild-type and transformant strains.
2.3. Characterization of the ΔprtT Knockout Mutants
For growth assessment and sporulation quantification, PDA plates were inoculated centrally with 5 μL of a conidia suspension (105 conidia/mL) of the P. digitatum parental strain Pd1, the ectopic prtT mutant, and two ΔprtT knockout mutants. Cultures were incubated at 24 °C for up to 7 days. Mycelial growth was determined by measuring two perpendicular diameters of the growing colonies at day 7 after inoculation. Sporulation assessment was carried out by scraping the surface of the 7-day-old cultures with a spatula. Conidia concentration was measured by using a haemocytometer.
Proteolytic activity on solid medium was assessed based on Ward [
27]. Spores (10
5 conidia/mL) were inoculated onto filter discs overlaid on solid complete medium plates (PDA) containing a colony restrictor (2 mg/mL dichloran). After 4 days of incubation at 24 °C, the filters were removed and the plates were overlaid with a layer of skim milk agarose (1% agarose, 1% skim milk, 0.45% CaCl
2, 0.6% acetic acid, pH 5.5), and milk clotting was allowed to proceed at 37 °C for 3 days. The extent of clotting was proportional to the number of proteases secreted by the colony that had occupied that position on the plate.
2.4. Chemicals
A protease inhibitor cocktail containing 1.4 mM of trans-Epoxysuccinyl-L-leucylamido(4-guanidino)butane (E-64), 500 mM of 1,10-phenanthroline, 100 mM of 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF), and 2.2 mM of pepstatin A was purchased from Sigma-Aldrich (P8215) (St. Louis, MO, USA). AEBSF, bestatin hydrochloride, E-64, phosphoramidon disodium salt, pepstatin A, ferrozine, diethyldithiocarbamate, and 1,10-phenanthroline hydrochloride monohydrate were also purchased from Sigma-Aldrich. Ethylenediaminetetraacetic acid calcium disodium salt dehydrate (EDTA) and dimethylsulfoxide (DMSO) were obtained from Applichem (Darmstadt, Germany).
2.5. Orange Fruit Infection Assays
To analyze the role of prtT in the pathogenicity of P. digitatum, we artificially inoculated the parental strain Pd1, one ectopic mutant (eprtT3), and two knockout mutants (ΔprtT44 and ΔprtT70) on sweet oranges: ‘Navelate’ and ‘Lane late’ mature oranges that were obtained from a packinghouse in Lliria, Valencia (Spain) the same day of harvesting before receiving any postharvest treatment. They were brought to the laboratory, surface-disinfected with 5% sodium hypochlorite for 5 min, rinsed with tap water, and allowed to dry. The next day, oranges were wounded four times around the equator with a nail (3 mm in depth) and were immediately inoculated by adding 10 μL of a conidial suspension (104 conidia/mL). Three replicates of five infected fruits with four wounds per fruit were placed on plastic boxes and incubated at 20 °C and 90% relative humidity for 7 days. Disease incidence (measured as the percentage of infection) and severity (as maceration diameter, in mm) were determined at day 5 and 7 post inoculation (dpi). Analysis of variance was performed to test the different incidence among strains at 5 dpi. Means were separated using the Tukey test with p < 0.05, using Statgraphics Stratus (Statgraphics Technologies, Inc., The Plains, VA, USA).
To study the effect on virulence of either the protease inhibitor cocktail, its individual components, and other different protease inhibitors and chelators, P. digitatum conidia were artificially co-inoculated with the proteinase inhibitor cocktail, E-64, 1,10-phenanthroline, AEBSF, pepstatin A, the double or triple combination of the different components, and with bestatin, phosphoramidon, EDTA, EGTA, ferrozine, and DETC in mature oranges as described above. The assayed concentration of each compound is indicated in the figure legend. Incidence and severity were measured up to 7 dpi.
The effect of metals and the chelator 1,10-phenanthroline was assayed by co-inoculation of 104 conidia/mL of P. digitatum Pd1 with different metal ions (ZnSO4, CuSO4, MnSO4, and FeSO4) at 0.5 mM either in the presence or absence of 1,10-phenanthroline 0.5 mM in mature oranges as described previously. Disease incidence was determined at 4, 5, and 6 dpi.
2.6. Gene Expression Analysis
For RNA extraction, mature oranges were wounded using a nail and inoculated with 10 μL of a conidial suspension (106 conidia/mL, 16 wounds per fruit) from either the P. digitatum parental strain Pd1, the ectopic mutant (eprtT3), or a knockout mutant (ΔprtT70). Inoculated fruits were stored at 20 °C and high humidity for 24, 48, and 72 h. After each storage time, cylinders of peel containing the flavedo and the albedo of the fruit were removed using a cork borer of 5 mm centered in the inoculation point. Each biological replicate consisted of 80 discs (16 wounds per 5 fruits) and three biological replicates were collected at each sampling time point. All samples were immediately frozen in liquid nitrogen and then ground to a fine powder for subsequent RNA extraction. Spores of the parental strain, the ectopic mutant, and the knockout mutant were also frozen for subsequent RNA extraction.
Total RNA extraction from
P. digitatum spores and from macerated orange peel tissue was done following a previously published protocol [
28] with minor modifications. One gram of frozen tissue was extracted with 10 mL of RNA extraction buffer (100 mM Tris HCl pH 8.0, 100 mM LiCl, 10 mM EDTA pH 8.0, 1% SDS, 1% PVP-40, and 1% β-mercaptoethanol). After phenol extraction, total nucleic acids were precipitated by adding one-tenth volume of 3M sodium acetate, pH 5.2, and two volumes of cold ethanol, and incubating at −20 °C for at least 30 min. For non-macerated orange peel tissue, RNA extraction was done according to López-Pérez et al. [
21]. RNA concentration was measured spectrophotometrically. DNase treatment and first-strand cDNA synthesis were conducted with the Maxima H Minus cDNA synthesis kit with dsDNase (Thermo Scientific, Waltham, MA, USA) using 2 μg of total RNA according to the manufacturer’s instructions. RT-qPCR was conducted following the MIQE (Minimum Information for Publication of Quantitative Real-Time PCR Experiments) guidelines [
29]. Gene-specific primer sets (
Table 1) were designed for gene expression analysis with Primer3Plus [
30]. Real-time qPCR reactions were performed in a LightCycler480 System (Roche Diagnostics, Basel, Switzerland) using SYBR Green to monitor cDNA amplification. Gene expression measurements were derived from three biological replicates and two technical replicates. Relative gene expression (RGE) was calculated using the formula described by Pfaffl [
26]. For each primer pair and each sample, the PCR efficiency (E) and the quantification cycle (Cq) were assessed using LinRegPCR software version 2017.1. Amplicon specificity was examined by analysis of the melting curve. The Cq value for the reference normalization factor (REF) was calculated by taking actin (PDIP_18200) as the reference gene, using primer pairs PdACTFor2/PdACTRev2 [
21].
4. Discussion
This study aimed to characterize the role of secreted proteases in the virulence of
P. digitatum towards citrus fruit. Because most of the protease-encoding genes belong to gene families containing an elevated number of members, it is not technically feasible to delete more than a few of these genes at a time. This methodology has been described in
B. cinerea by constructing single and double knockout mutants of five members from an aspartic proteinase gene family; however, the role of them in the virulence is not completely clear [
12]. In our study, instead of simultaneously deleting several
P. digitatum protease genes at a time, we designed two alternative approaches to determine the contribution of secreted proteolytic activities to the virulence of
P. digitatum: (i) Construction of
prtT knockout mutants and characterization of the mutants during
in vitro and
in vivo growth, and (ii) the application of different protease inhibitors during the infection of
P. digitatum in sweet oranges.
As a first approach, we focused on the
P. digitatum prtT gene, which encodes a putative transcription factor controlling the expression of multiple secreted proteases. The disruption of the
prtT gene has been previously described in
A. niger [
13],
A. fumigatus [
14,
15,
16,
31], and
P. oxalicum [
17]. As far as we are aware, there are no reports on the possible role of secreted proteases in the virulence of fungal pathogens of citrus fruit. Characterization of two independent knockout mutants revealed that PrtT is required for the production of several extracellular proteases by
P. digitatum (
Figure 2B). However, the absence of the regulator has just a small influence on the expression of the genes encoding the major putative extracellular proteases secreted by
P. digitatum during the infection of sweet oranges (
Figure 4) and has no effect in the virulence of the fungus (
Figure 3).
Secreted protease activity depends on the pH of the growth media and the nitrogen or carbon source, among others [
2]. For example, in
A. fumigatus, protease activity was repressed by ammonia, or elevated pH, and activated in the presence of proteins as the sole nitrogen source [
16]. In the present study, conidia production per area of growth of the
P. digitatum Δ
prtT knockout mutants were similar to those of the parental strain under the tested conditions (
Figure 2A). It has been described that proteases constitute the largest group of
P. digitatum genes up-regulated during the infection of oranges and that they might contribute to pathogenicity in different ways, such as degrading plant cell components or inactivating defense proteins [
21]. We have shown that the protease activity of the Δ
prtT knockout mutants grown on PDA and further incubated with skim milk was reduced to almost undetectable levels (
Figure 2B) with respect to the parental strain and the ectopic mutant. This result indicates that PrtT is involved in the regulation of at least some proteases that are required by
P. digitatum to degrade skim milk when the fungus grows on PDA medium. However, whether PrtT is involved in the regulation of additional proteases would require further experiments.
In order to determine the role of PrtT in virulence, the knockout Δ
prtT70 mutant, the ectopic mutant, and the parental strain
P. digitatum were artificially inoculated in sweet oranges (
Figure 3). After 7 days of inoculation, no significant differences were observed in the incidence and the maceration diameter among them, suggesting that PrtT is not involved in the virulence of this postharvest pathogen in citrus fruit. Similar results have been previously observed in
A. fumigatus, in which the
prtT gene appears not to be essential for pathogenicity in animal models [
14,
16,
31]. The
A. fumigatus Δ
prtT mutant showed reduced killing of lung alveolar cells and erythrocyte lysis [
14,
16]; however, the mutant strain showed wild-type virulence in infected neutropenic mice, suggesting that perhaps residual protease activity was sufficient to enable virulence [
14,
16]. Our results suggest that although PrtT regulates a group of secreted proteases (
Figure 2B), it has no role in virulence (
Figure 3). As already pointed out in the work done with
A. fumigatus PrtT, this result could suggest that either residual protease activity is sufficient to enable virulence or that proteases are dispensable for pathogenicity in this fungus [
14,
16]. Another possible explanation is that the major extracellular proteases secreted by the pathogen during the infection process are not regulated by the
prtT gene. To test this hypothesis, we analyzed the expression of the two genes coding for the major putative proteases during the
P. digitatum infection process: The aspartic endopeptidase
pep1 encoding gene (PDIP_25240) and the tripeptidyl peptidase
aor1 encoding gene (PDIP_12220) [
21]. The expression of these two genes during the infection of oranges was barely affected by the loss of the
prtT gene, indicating that the regulation of these genes depends mostly on another factor(s). XprG is another transcription factor that regulates extracellular protease production in
Aspergillus nidulans, a fungus that lacks a PrtT homolog. Deletion of both
A. fumigatus xprG and
prtT genes resulted in the generation of a mutant with almost no ability to degrade proteins; however, it retained wild-type virulence in murine systemic and pulmonary models of infection [
31]. In the case of
P. digitatum, we identified a single XprG ortholog by amino-acid similarity to
A. fumigatus XrpG (data not showed). The possibility that these two major proteinases are relevant for
P. digitatum virulence in sweet oranges and the role of XrpG in the regulation of protease secretion should be further explored. In future experiments, we might generate
P. digitatum deletion mutants in these two genes encoding major extracellular proteases and in the
xrpG putative transcription factor gene.
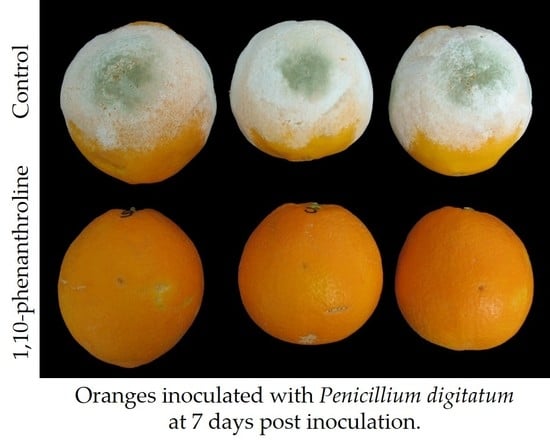
In the second approach, we investigated the effect of the application of different protease inhibitors on the virulence of
P. digitatum. The presence of protease inhibitors has been described in plants and they are part of the pathogenesis-related proteins [
32]. The first protease inhibitor proteins, trypsin and chymotrypsin inhibitors, with antifungal activity were described in
Brassica oleracea by Lorito et al. [
33], and subsequently, other protease-inhibitor proteins, such as cystatin, have been described in plants [
34,
35,
36,
37]. In the present study, we investigated the role of different protease inhibitors on the virulence of
P. digitatum in oranges, and after 6 days post-inoculation, only 1,10-phenanthroline and the combinations containing this metalloprotease inhibitor were effective in controlling the development of
P. digitatum in oranges. We tested other metalloprotease inhibitors, such as bestatin and phosphoramidon, and different metal ion chelators, such as EDTA, EGTA, ferrozine, and DETC (
Figure 6A); although none of them were as effective as 1,10-phenanthroline in reducing the development of
P. digitatum in citrus fruit, we observed some protective effect with some chelators, specially EDTA.
1,10-phenanthroline is a membrane permeable heterocyclic compound with the ability to sequester metal ions in biological systems, forming coordination compounds with them [
38]. It has the capability of inhibiting the biological role of metal-dependent proteins, interfering with metal acquisition, bioavailability, and metabolism for crucial reactions; disturbing the microbial cell homeostasis; and culminating in the blockage of microbial nutrition, growth, development, and playing an important role in the
in vivo infection progression [
39]. The utilization of metal complexes containing 1,10-phenanthroline as antimicrobials against a broad spectrum of bacteria and as a potential alternative to antibiotics has been described previously [
39]. Phenanthroline-based complexes can penetrate the cell membrane and can interact with relevant biomolecules in the microorganisms, leading to inhibition of the cell growth and causing cell death, exhibiting a broad spectrum of both antibacterial (e.g., against
E. coli and
Pseudomonas aeruginosa) and antifungal (e.g., against
A. niger and
Fusarium solani) activities. Metal sequestration is also found in nature as a means to combat microbial infection. The process by which a host organism sequesters trace minerals in an effort to limit pathogenicity during infection has been designated ‘nutritional immunity’ [
40,
41,
42]. Well–studied examples of nutritional immunity include the production of the iron binding lactoferrin or the zinc and manganese binding protein calprotectin [
40,
41,
42]. The antimicrobial activity of these proteins is mostly due to their capability to bind metal ions, as is the case of siderophores secreted by many biocontrol microbial antagonists [
43,
44]. Moreover, it has been hypothesized that the high level of gluconic acid secretion found during pathogenicity of apple fruits by
P. expansum could be involved in the formation of iron chelates, which could favor iron acquisition and pathogenicity [
45].
In the present work, we co-inoculated
P. digitatum with some metal ions (ZnSO
4, CuSO
4, MnSO
4, and FeSO
4) in sweet oranges to further analyze the role of 1,10-phernatroline as a metalloproteinase inhibitor and as a chelator. We chose these four metal ions because 1,10-phenanthroline has a very high affinity for Fe
2+, Zn
2+, and Cu
2+, but very low affinity for Mn
2+. We hypothesized that the effect of these metal ions reverting the inhibition of 1,10-phenanthhroline would be related to their affinity to this chelator. The co-inoculation had no effect in the development of the pathogen, with an incidence of 100% of infected wounds after 7 dpi (
Figure 6B). However, as indicated previously, the application of 1,10-phenanthroline drastically reduced the growth of the fungal pathogen in the fruit. The application of a plant protease inhibitor as an antifungal agent has been evidenced in transgenic rice constitutively expressing a potato carboxypeptidase inhibitor; these plants exhibit resistance against the economically important pathogens,
Magnaporthe oryzae and
Fusarium verticillioides [
46]. The effect of 1,10-phenanthroline preventing the infection of citrus fruit by
P. digitatum was partially reverted by application of CuSO
4, and was completely reverted by the addition of ZnSO
4 and FeSO
4 (
Figure 6B), indicating that the fungus is most susceptible to zinc and iron and, to a lesser extent, copper deprivation during the infection process. The concept of fungal micronutrient scavenging can be used in future studies aimed at developing a product containing a chelator, such as 1,10-phenanthroline, capable of reducing the development of fungal pathogens during postharvest.