Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria of Healthy and Diseased Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Isolation of Endophytic Actinobacteria
2.3. Phenotypic and Molecular Characterization of Actinobacterial Isolates
2.4. Screening for Antagonistic Actinobacteria
2.5. Isolation and Characterization of Antifungal Compounds
2.6. Antifungal Assay of Elucidated Bioactive Compounds
2.7. Culture-Independent Community Analysis
2.8. Statistical Analysis
3. Results
3.1. Isolation and Distribution of Endophytic Actinobacteria
3.2. In Vitro Antagonism of S. sclerotiorum and Identification of Bioactive Strains
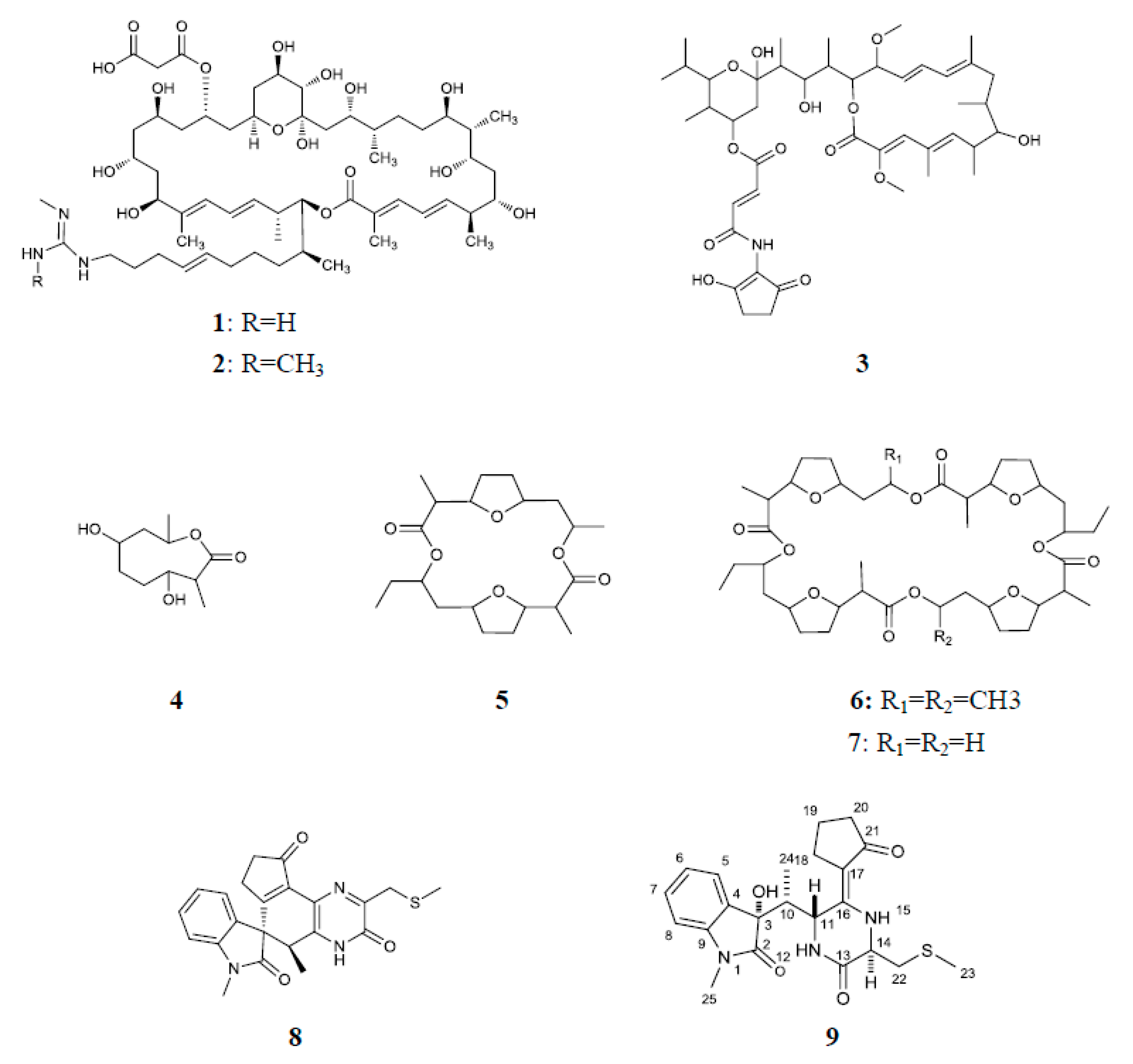
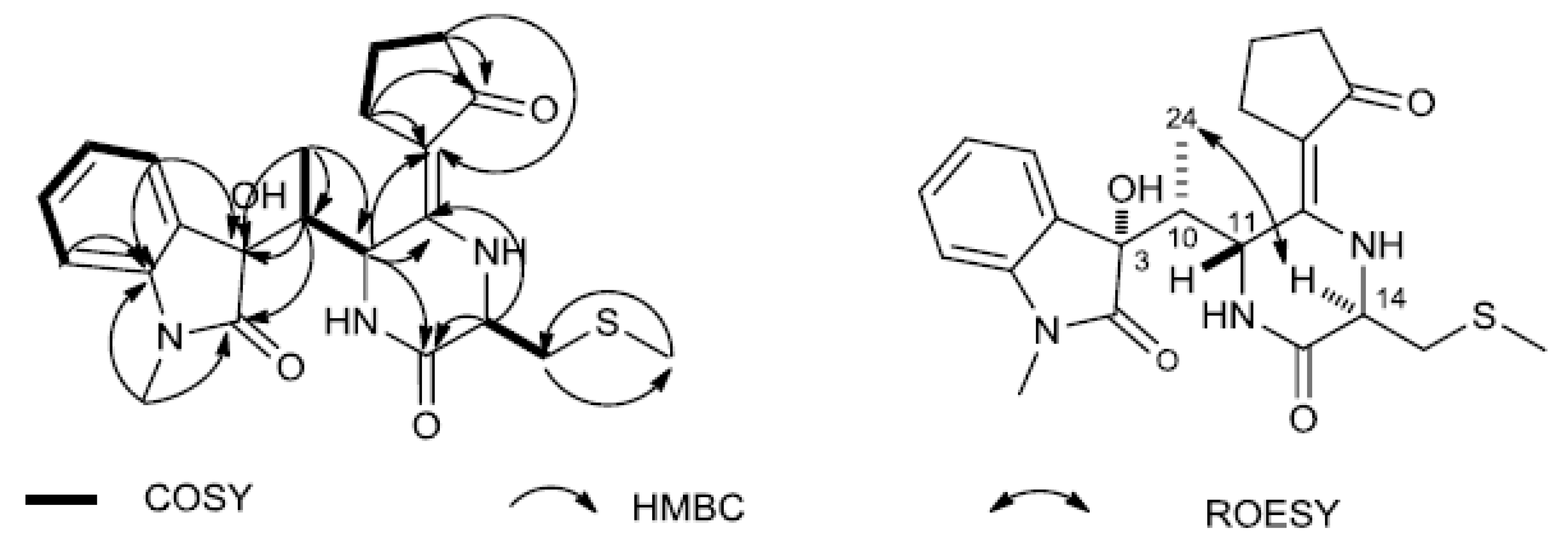
3.3. Identification and Activity Evaluation of the Antifungal Compounds
3.4. Culture-Independent Communities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Boland, G.J.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant. Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Bardin, S.D.; Huang, H.C. Research on biology and control of Sclerotinia diseases in Canada. Can. J. Plant. Pathol. 2001, 23, 88–98. [Google Scholar] [CrossRef]
- Firoz, M.J.; Xiao, X.; Zhu, F.X.; Fu, Y.P.; Jiang, D.H.; Schnabel, G.; Luo, C.X. Exploring mechanisms of resistance to dimethachlone in Sclerotinia sclerotiorum. Pest. Manag. Sci. 2016, 72, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Boland, G.J. Stability analysis for evaluating the influence of environment on chemical and biological control of white mold (Sclerotinia sclerotiorum) of bean. Biol. Control. 1997, 9, 7–14. [Google Scholar] [CrossRef]
- Lee, Y.H.; Cho, Y.S.; Lee, S.W.; Hong, J.K. Chemical and biological controls of balloon flower stem rots caused by Rhizoctonia solani and Sclerotinia sclerotiorum. Plant. Pathol. J. 2012, 28, 156–163. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, J.; Zhang, Y.; He, S.; Zhu, F. Baseline sensitivity and toxic actions of boscalid against Sclerotinia sclerotiorum. Crop. Prot. 2018, 110, 83–90. [Google Scholar] [CrossRef]
- Ma, H.X.; Feng, X.J.; Chen, Y.; Chen, C.J.; Zhou, M.G. Occurrence and characterization of dimethachlon insensitivity in Sclerotinia sclerotiorum in jiangsu province of China. Plant. Dis. 2009, 93, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Hou, Y.P.; Wang, J.X.; Zhou, M.G. Sensitivity of Sclerotinia sclerotiorum to fludioxonil: in vitro determination of baseline sensitivity and resistance risk. Crop. Prot. 2011, 30, 876–882. [Google Scholar] [CrossRef]
- Zhou, F.; Zhang, X.L.; Li, J.L.; Zhu, F.X. Dimethachlon resistance in Sclerotinia sclerotiorum in China. Plant. Dis. 2014, 98, 1221–1226. [Google Scholar] [CrossRef]
- Smith, S.A.; Tank, D.C.; Boulanger, L.A.; Bascom-Slack, C.A.; Eisenman, K.; Kingery, D. Bioactive endophytes warrant intensified exploration and conservation. PLoS ONE 2008, 3, e3052. [Google Scholar] [CrossRef]
- Sturz, A.V.; Christie, B.R.; Nowak, J. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant. Sci. 2000, 19, 1–30. [Google Scholar] [CrossRef]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.B.; Taghavi, S.; Mezgeay, M.; van der Lelie, D. Endophytic bacteria and their potential applications. CRC. Crit. Rev. Plant. Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant. Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. What are endophytes? In Microbial Root Endophytes; Schulz, B.J.E., Boyle, C.J.C., Sieber, T.N., Eds.; Springer: Berlin, Germany; pp. 1–13.
- Sorensen, J.; Sessitsch, A. Plant-associated bacteria—Lifestyle and molecular interactions. In Modern Soil Microbiology; Van Elsas, J.D., Jansson, J.K., Trevors, J.T., Eds.; CRC Press: Boca Raton, FL, USA; pp. 211–236.
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Liu, C.X.; Song, J.; Wang, X.J.; Xiang, W.S. Recruitment of defensive microbs and plant protection. Sci. Sin. Vitae 2016, 46, 1–8. [Google Scholar]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant. Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef]
- Bèrdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Qin, S.; Li, W.J.; Dastager, S.G.; Hozzein, W.N. Editorial: Actinobacteria in special and extreme habitats: Diversity, function roles, and environmental adaptations. Front. Microbiol. 2016, 7, 1415. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef]
- Ek-Ramos, M.J.; Gomez-Flores, R.; Orozco-Flores, A.A.; Rodríguez-Padilla, C.; González-Ochoa, G.; Patricia Tamez-Guerra, P. Bioactive products from plant-endophytic Gram-positive bacteria. Front. Microbiol. 2019, 10, 463. [Google Scholar] [CrossRef]
- Conn, V.M.; Franco, C.M. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 2003, 69, 5603–5608. [Google Scholar]
- Tian, X.; Cao, L.; Tan, H.; Han, W.; Chen, M.; Liu, Y.; Zhou, S. Diversity of cultivated and uncultivated actinobacterial endophytes in the stems and roots of rice. Microb. Ecol. 2007, 53, 700–707. [Google Scholar] [CrossRef]
- Zhao, K.; Penttinen, P.; Guan, T.W.; Xiao, J.; Chen, Q.; Xu, J.; Lindström, K.; Zhang, L. The diversity and antimicrobial activity of endophytic actinomycetes isolated from medicinal plants in Panxi plateau, China. Curr. Microbiol. 2011, 62, 182–190. [Google Scholar] [CrossRef]
- Li, J.; Zhao, G.Z.; Huang, H.Y.; Qin, S.; Zhu, W.Y.; Zhao, L.X.; Xu, L.H.; Zhang, S.; Li, W.J.; Strobel, G. Isolation and characterization of culturable endophytic actinobacteria associated with Artemisia annua L. Antonie. Leeuwenhoek. 2012, 101, 515–527. [Google Scholar] [CrossRef]
- Miao, G.P.; Zhu, C.S.; Feng, J.T.; Han, L.R.; Zhang, X. Effects of plant stress signal molecules on the production of wilforgine in an endophytic actinomycete isolated from Tripterygium wilfordii Hook. f. Curr. Microbiol. 2015, 70, 571–579. [Google Scholar] [CrossRef]
- Wei, W.; Zhou, Y.; Chen, F.; Yan, X.; Lai, Y.; Wei, C.; Chen, X.; Xu, J.; Wang, X. Isolation, diversity, and antimicrobial and immunomodulatory activities of endophytic actinobacteria from tea cultivars Zijuan and Yunkang-10 (Camellia sinensis var assamica). Front. Microbiol. 2018, 9, 1304. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Nahal Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Trivedi, P.; He, Z.; Van Nostrand, J.D.; Albrigo, G.; Zhou, J.; Wang, N. Huanglongbing alters the structure and functional diversity of microbial communities associated with citrus rhizosphere. ISME. J. 2012, 6, 363–383. [Google Scholar] [CrossRef]
- Boland, G.J.; Hall, R. Growth room evaluation of soybean cultivars for resistance to Sclerotinia sclerotiorum. Can. J. Plant. Sci. 1986, 66, 559–564. [Google Scholar] [CrossRef]
- Hayakawa, M.; Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 1987, 65, 501–509. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media; Parks, L.C., Ed.; CRC Press: Boca Raton, FL, USA, 1993. [Google Scholar]
- Guan, X.J.; Liu, C.X.; Fang, B.Z.; Zhao, J.W.; Jin, P.J.; Li, J.M. Baia soyae gen. nov., sp. nov., a mesophilic representative of the family Thermoactinomycetaceae, isolated from soybean root [Glycine max (L.) Merr]. Int. J. Syst. Evol. Microbiol. 2015, 65, 3241–3247. [Google Scholar] [CrossRef]
- Qin, S.; Li, J.; Chen, H.H.; Zhao, G.Z.; Zhu, W.Y.; Jiang, C.L.; Xu, L.H.; Li, W.J. Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 2009, 75, 6176–6186. [Google Scholar] [CrossRef]
- Shirling, E.B.; Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- Maniatis, T.; Fritsch, E.F.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1982. [Google Scholar]
- Kim, S.B.; Brown, R.; Oldfield, C.; Gilbert, S.C.; Iliarionov, S.; Goodfellow, M. Gordonia amicalis sp. nov., a novel dibenzothiophene-desulphurizing actinomycete. Int. J. Syst. Evol. Microbiol. 2000, 50, 2031–2036. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Kimura, M. The Neutral Theory of Molecular Evolution; Cambridge Universiry Press: Cambridge, UK, 1983. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar]
- Hamzah, T.N.T.H.; Lee, S.Y.; Hidayat, A.; Terhem, R.; Faridah-Hanum, I.; Mohamed, R. Diversity and characterization of endophytic fungi isolated from the tropical mangrove species, Rhizophora mucronata, and identification of potential antagonists against the soil-borne fungus, Fusarium solani. Front. Microbiol. 2018, 9, 1707. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.J.; Yan, Y.J.; Jiang, L.; Wang, J.D.; Li, B.J.; Xiang, W.S. Isolation and identification of 5-hydroxyl-5-methyl-2-hexenoic acid from Actinoplanes sp. HBDN08 with antifungal activity. Bioresource. Technol. 2010, 101, 8383–8388. [Google Scholar] [CrossRef]
- Liu, C.X.; Zhang, J.; Wang, X.J.; Qian, P.T.; Wang, J.D.; Gao, Y.M.; Yan, Y.J.; Zhang, S.Z.; Xu, P.F.; Li, W.B.; et al. Antifungal activity of borrelidin produced by a Streptomyces strain isolated from soybean. J. Agric. Food. Chem. 2012, 60, 1251–1257. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Liu, C.X.; Zhang, J.; Shen, Y.; Li, C.; He, H.R.; Wang, X.J.; Xiang, W.S. Actinomycetospora atypica sp. nov., a novel soil actinomycete and emended description of the genus Actinomycetospora. Antonie. Leeuwenhoek. 2014, 105, 891–897. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Yuan, G.; Lin, H.; Wang, C.; Hong, K.; Liu, Y.; Li, J. 1H and 13C assignments of two new macrocyclic lactones isolated from Streptomyces sp. 211726 and revised assignments of azalomycins F3a, F4a and F5a. Magn. Reson. Chem. 2011, 49, 30–37. [Google Scholar] [CrossRef]
- Crevelin, E.J.; Canova, S.P.; Melo, I.S.; Zucchi, T.D.; da Silva, R.E.; Moraes, L.A. Isolation and characterization of phytotoxic compounds produced by Streptomyces sp. AMC 23 from red mangrove (Rhizophora mangle). Appl. Biochem. Biotechnol. 2013, 171, 1602–1616. [Google Scholar] [CrossRef]
- Han, B.; Cui, C.B.; Cai, B.; Ji, X.F.; Yao, X.S. Actinolactomycin, a new 2-oxonanonoidal antitumor antibiotic produced by Streptomyces flavoretus 18522, and its inhibitory effect on the proliferation of human cancer cells. Chin. Chem. Lett. 2005, 4, 471–474. [Google Scholar]
- Zhao, P.J.; Fan, L.M.; Li, G.H.; Zhu, N.; Shen, Y.M. Antibacterial and antitumor macrolides from Streptomyces sp. Is9131. Arch. Pharm. Res. 2005, 28, 1228–1232. [Google Scholar] [CrossRef]
- Shishlyannikova, T.A.; Kuzmin, A.V.; Fedorova, G.A.; Shishlyannikov, S.M.; Lipko, I.A.; Sukhanova, E.V.; Belkova, N.L. Ionofore antibiotic polynactin produced by Streptomyces sp. 156A isolated from Lake Baikal. Nat. Prod. Res. 2017, 31, 639–644. [Google Scholar] [CrossRef]
- Lan, Y.X.; Zou, Y.; Huang, T.T.; Wang, X.Z.; Brock, N.L.; Deng, Z.X.; Lin, S.J. Indole methylation protects diketopiperazine configuration in the maremycin biosynthetic pathway. Sci. Chin. Chem. 2016, 59, 1224–1228. [Google Scholar] [CrossRef]
- Duan, Y.Y.; Liu, Y.Y.; Huang, T.; Zou, Y.; Huang, T.T.; Hu, K.F.; Deng, Z.X.; Lin, S.J. Divergent biosynthesis of indole alkaloids FR900452 and spiro-maremycins. Org. Biomol. Chem. 2018, 16, 5446–5451. [Google Scholar] [CrossRef]
- Reiter, B.; Pfeifer, U.; Schwab, H.; Sessitsch, A. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 2002, 68, 2261–2268. [Google Scholar] [CrossRef]
- Upreti, R.; Thomas, P. Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front. Microbiol. 2015, 6, 255. [Google Scholar] [CrossRef]
- Huang, J.F.; Wei, Z.; Tan, S.Y.; Mei, X.L.; Yin, S.X.; Shen, Q.R.; Xu, Y.C. The rhizosphere soil of diseased tomato plants as a source for novel microorganisms to control bacterial wilt. Appl. Soil. Ecol. 2013, 72, 79–84. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Kitto, S.L.; Caplan, J.L.; Hsueh, Y.H.; Kearns, D.B.; Wu, Y.S.; Bai, H.P. Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant. Physiol. 2012, 160, 1642–1661. [Google Scholar] [CrossRef]
- Barea, J.M.; Pozo, M.J.; Azcón, R.; Azcón-Aguilar, C. Microbial co-operation in the rhizosphere. J. Exp. Bot. 2005, 56, 1761–1778. [Google Scholar] [CrossRef] [Green Version]
- Dennis, P.G.; Miller, A.J.; Hirsch, P.R. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS. Microbiol. Ecol. 2010, 72, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Bull, C.T. Relationship between root colonization and suppression of Gaeumannomyces graminis var tritici by Pseudomonas fluorescens strain 2–79. Phytopathology 1991, 81, 954–959. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Leeman, M.; van Oorschot, M.M.P.; van der Sluis, I.; Schippers, B.; Bakker, P.A.H.M. Dose-response relationships in biological-control of Fusarium-wilt of radish by Pseudomonas spp. Phytopathology 1995, 85, 1075–1081. [Google Scholar] [CrossRef]
- Werner, G.; Hagenmaier, H.; Drautz, H.; Baumgartner, A.; Zähner, H. Metabolic products of microorganisms. 224. Bafilomycins, a new group of macrolide antibiotics. Production, isolation, chemical structure and biological activity. J. Antibiot. 1984, 37, 110–117. [Google Scholar] [CrossRef]
- Frändberg, E.; Petersson, C.; Lundgren, L.N.; Schnürer, J. Streptomyces halstedii K122 produces the antifungal compounds bafilomycin B1 and C1. Can. J. Microbiol. 2000, 46, 753–758. [Google Scholar] [CrossRef]
- Arai, M. Azalomycins B and F, two new antibiotics. II. Properties of azalomycins B and F. J. Antibiot. 1960, 13, 51–56. [Google Scholar]
- Chandra, A.; Nair, M.G. Azalomycin F complex from Streptomyces hygroscopicus, MSU/MN-4-75B. J. Antibiot. 1995, 48, 896–898. [Google Scholar] [CrossRef]
- Copping, L.G.; Menn, J.J. Biopesticides: A review of their action, applications and efficacy. Pest. Manage. Sci. 2000, 56, 651–676. [Google Scholar] [CrossRef]
- Silva, L.J.; Crevelin, E.J.; Souza, W.R.; Moraes, L.A.; Melo, I.S.; Zucchi, T.D. Streptomyces araujoniae produces a multiantibiotic complex with ionophoric properties to control Botrytis cinerea. Phytopathology 2014, 104, 1298–1305. [Google Scholar] [CrossRef]
- Cui, C.B.; Kakeya, H.; Osada, H. Novel mammalian cell cycle inhibitors, spirotryprostatins A and B, produced by Aspergillus fumigatus, which inhibit mammalian cell cycle at G2/M phase. Tetrahedron 1996, 52, 12651–12666. [Google Scholar] [CrossRef]
- Zheng, C.J.; Kim, Y.H.; Kim, W.G. Glioperazine B, as a new antimicrobial agent against Staphylococcus aureus, and glioperazine C: two new dioxopiperazines from Bionectra byssicola. Biosci. Biotechnol. Biochem. 2007, 71, 1979–1983. [Google Scholar] [CrossRef]
- Byun, H.G.; Zhang, H.; Mochizuki, M.; Adachi, K.; Shizuri, Y.; Lee, W.J.; Kim, S.K. Novel antifungal diketopiperazine from marine fungus. J. Antibiot. 2003, 56, 102–106. [Google Scholar] [CrossRef]
- Ma, Y.M.; Liang, X.A.; Kong, Y.; Jia, B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J. Agric. Food. Chem. 2016, 64, 6659–6671. [Google Scholar] [CrossRef]
- Takase, S.; Shigematsu, N.; Shima, I.; Uchida, I.; Hashimoto, M.; Tada, T. Structure of FR900452, a novel platelet-activating factor inhibitor from a Streptomyces. J. Org. Chem. 1987, 52, 3485–3487. [Google Scholar] [CrossRef]
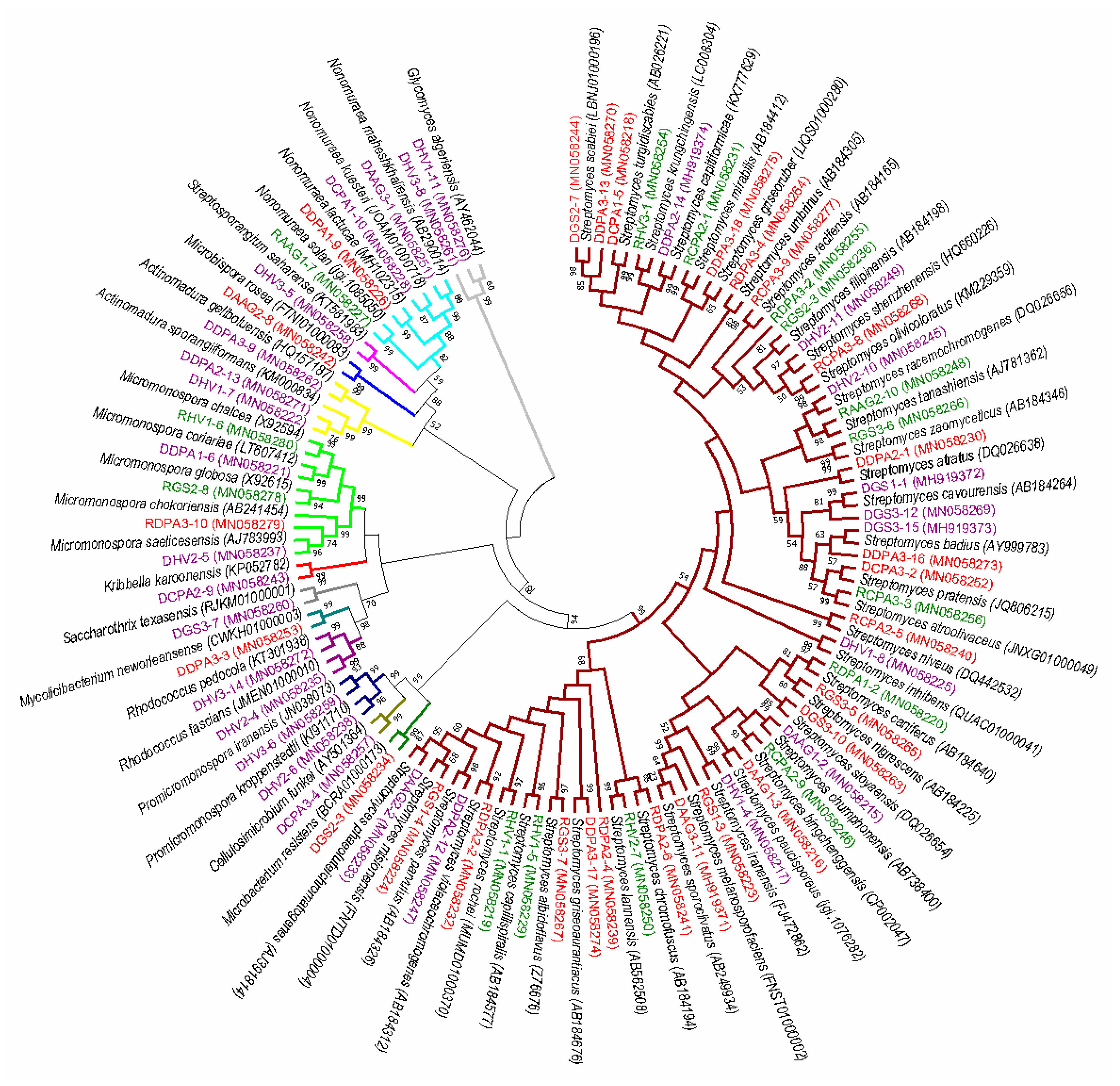
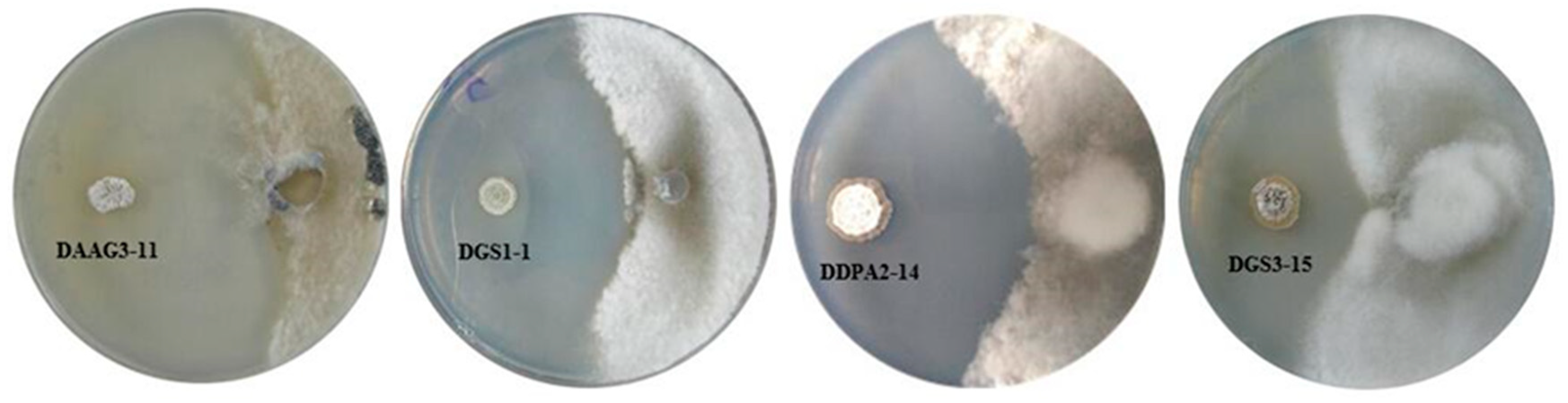

| Isolate No. and NCBI Genbank Accesion No. | Closest Type Strain with Accession Number | Similarity | Isolated From | Colony Number | S. sclerotiorum Mycelial Growth Inhibition (%) * |
|---|---|---|---|---|---|
| DAAG3-11 (MH919371) | Streptomyces sporoclivatus (AB249934) | 100% | Healthy soybean root | 11 | 87.6 ± 1.8 a |
| Diseased soybean root | 176 | ||||
| DGS1-1 (MH919372) | Streptomyces cavourensis (AB184264) | 99.9% | Diseased soybean root | 13 | 78.9 ± 1.9 b |
| DDPA2-14 (MH919374) | Streptomyces capitiformicae (KX777629) | 100% | Diseased soybean root | 9 | 68.6 ± 3.4 c |
| DGS3-15 (MH919373) | Streptomyces pratensis (JQ806215) | 99.9% | Diseased soybean root | 6 | 54.1 ± 2.2 d |
| Position | δC | δH (J in Hz) |
|---|---|---|
| 2 | 178.33 | |
| 3 | 78.69 | |
| 4 | 130.01 | |
| 5 | 125.53 | 7.43 (1H, d, 7.8) |
| 6 | 123.24 | 7.12 (1H, td, 7.6, 1.0) |
| 7 | 130.36 | 7.37 (1H, td, 7.7, 1.2) |
| 8 | 109.08 | 6.89 (1H, d, 7.9) |
| 9 | 142.83 | |
| 10 | 42.84 | 2.06 (1H, m) |
| 11 | 52.94 | 5.36 (1H, s) |
| 12 | 11.01 (brs) | |
| 13 | 168.23 | |
| 14 | 52.8 | 4.14 (1H, dd, 8.2, 3.3) |
| 15 | 7.09 (brs) | |
| 16 | 152.27 | |
| 17 | 100.43 | |
| 18 | 27.04 | 2.24 (1H, s) |
| 2.44 (1H, ddd, 13.9, 8.4, 3.8) | ||
| 19 | 21.04 | 1.83 (2H, m) |
| 20 | 38.79 | 2.28 (2H, m) |
| 21 | 204.84 | |
| 22 | 38.75 | 3.17 (1H, m) |
| 22 | 2.83 (1H, dd, 14.0, 8.2) | |
| 23 | 16.36 | 2.15 (3H, s) |
| 24 | 8.92 | 1.19 (3H, d, 7.0) |
| 25 | 26.65 | 3.23 (3H, s) |
| Compounds | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| EC50 (mg/L) | 4.87 ± 0.16 a | 4.96 ± 0.13 a | 0.21 ± 0.02 b | 49.14 ± 0.82 c | 5.33 ± 0.15 ae | 3.69 ± 0.05 d | 5.60 ± 0.11 e | 3.46 ± 0.12 d | 3.70 ± 0.05 d |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, C.; Zhuang, X.; Yu, Z.; Wang, Z.; Wang, Y.; Guo, X.; Xiang, W.; Huang, S. Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria of Healthy and Diseased Soybean. Microorganisms 2019, 7, 243. https://doi.org/10.3390/microorganisms7080243
Liu C, Zhuang X, Yu Z, Wang Z, Wang Y, Guo X, Xiang W, Huang S. Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria of Healthy and Diseased Soybean. Microorganisms. 2019; 7(8):243. https://doi.org/10.3390/microorganisms7080243
Chicago/Turabian StyleLiu, Chongxi, Xiaoxin Zhuang, Zhiyin Yu, Zhiyan Wang, Yongjiang Wang, Xiaowei Guo, Wensheng Xiang, and Shengxiong Huang. 2019. "Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria of Healthy and Diseased Soybean" Microorganisms 7, no. 8: 243. https://doi.org/10.3390/microorganisms7080243





