Simulating Extracellular Glucose Signals Enhances Xylose Metabolism in Recombinant Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Plasmids
2.2. Cultivation Conditions and Batch Fermentation
2.3. Determination of cAMP
2.4. Assay of Trehalase Activity
2.5. Analysis of Metabolites
2.6. Calculation of Physiological Parameters
2.7. Quantitative PCR
3. Results
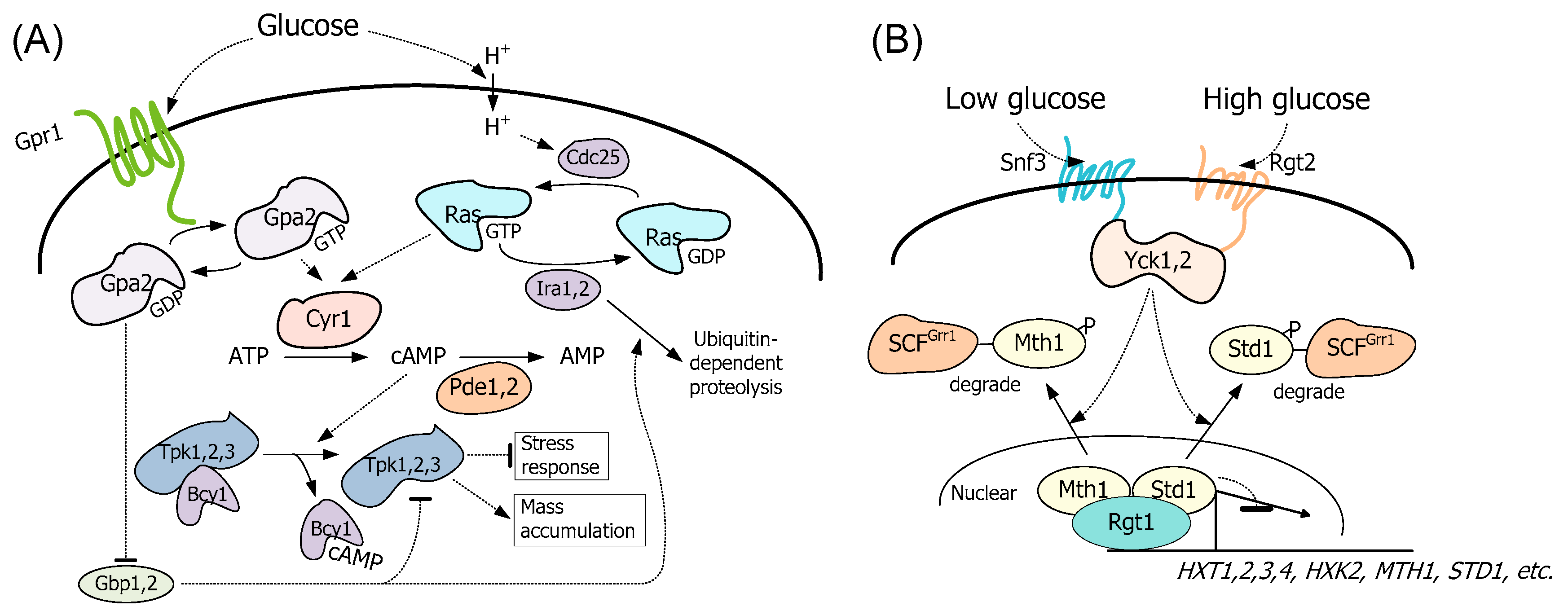
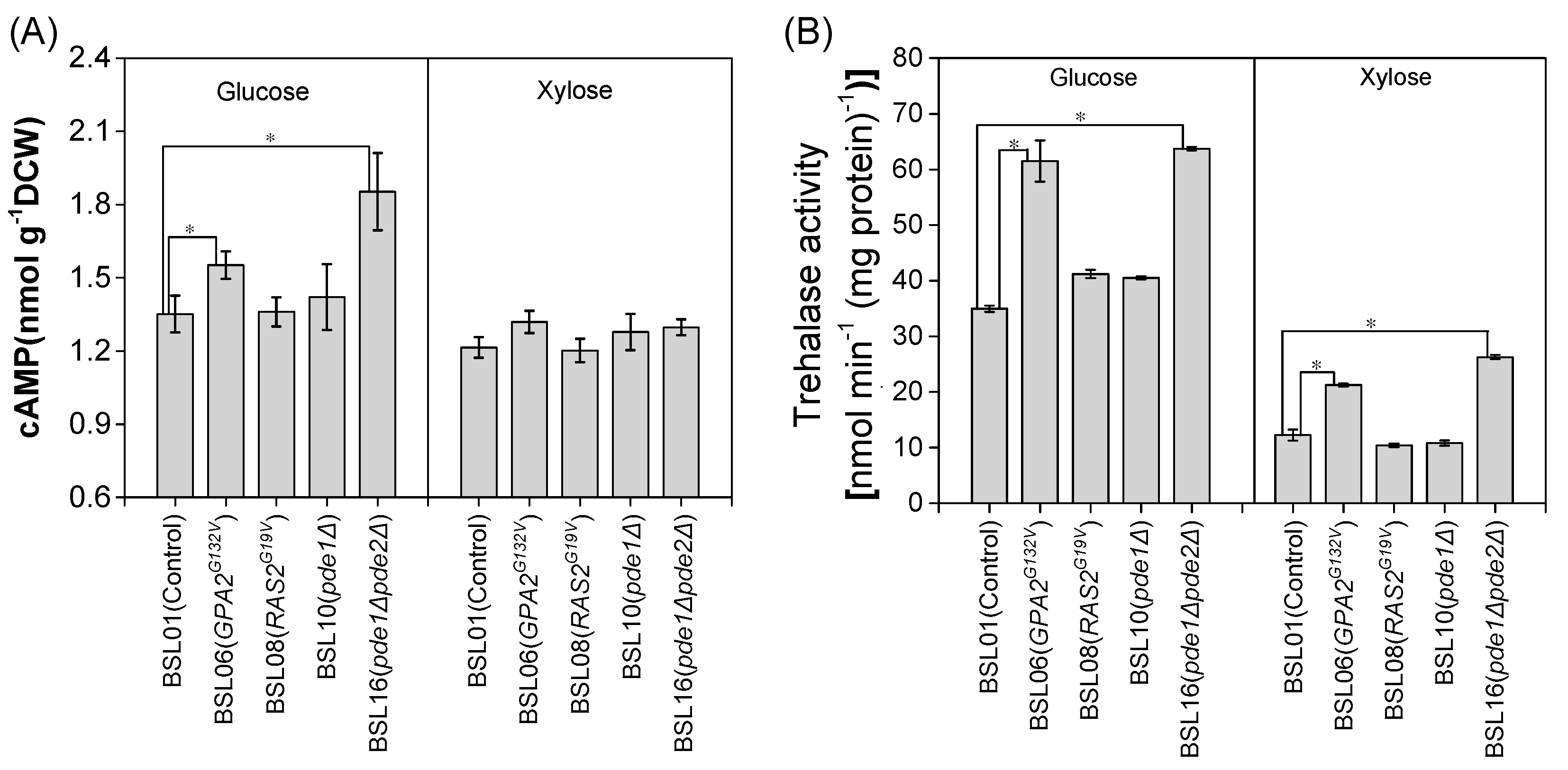
3.1. Expression of the GPA2G132V Allele and Deletion of Both PDE1 and PDE2 Increased PKA Activity in S. cerevisiae
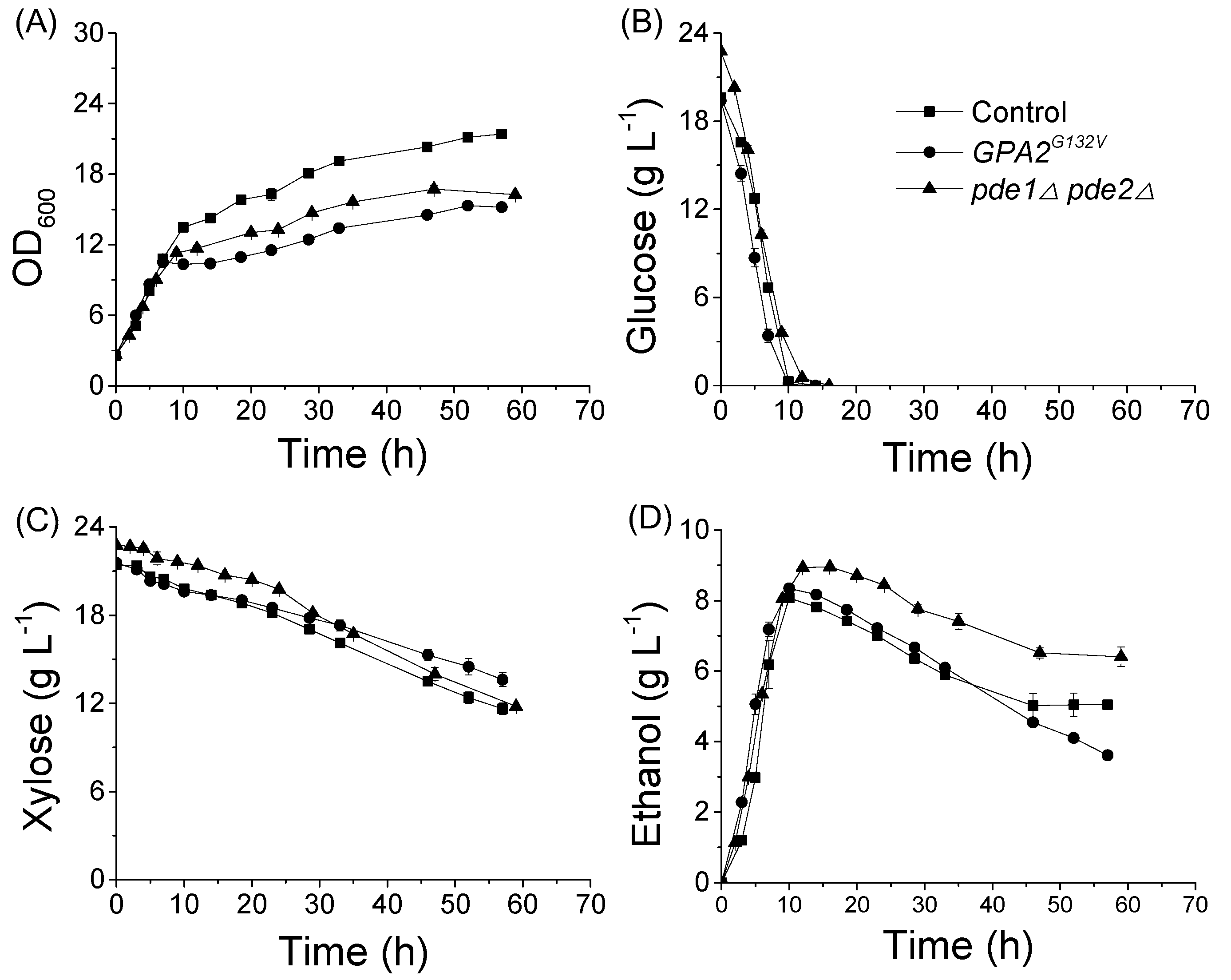
3.2. Expression of GPA2G132V Allele and Deletion of Both PDE1 and PDE2 Affected Glucose and Xylose Metabolism
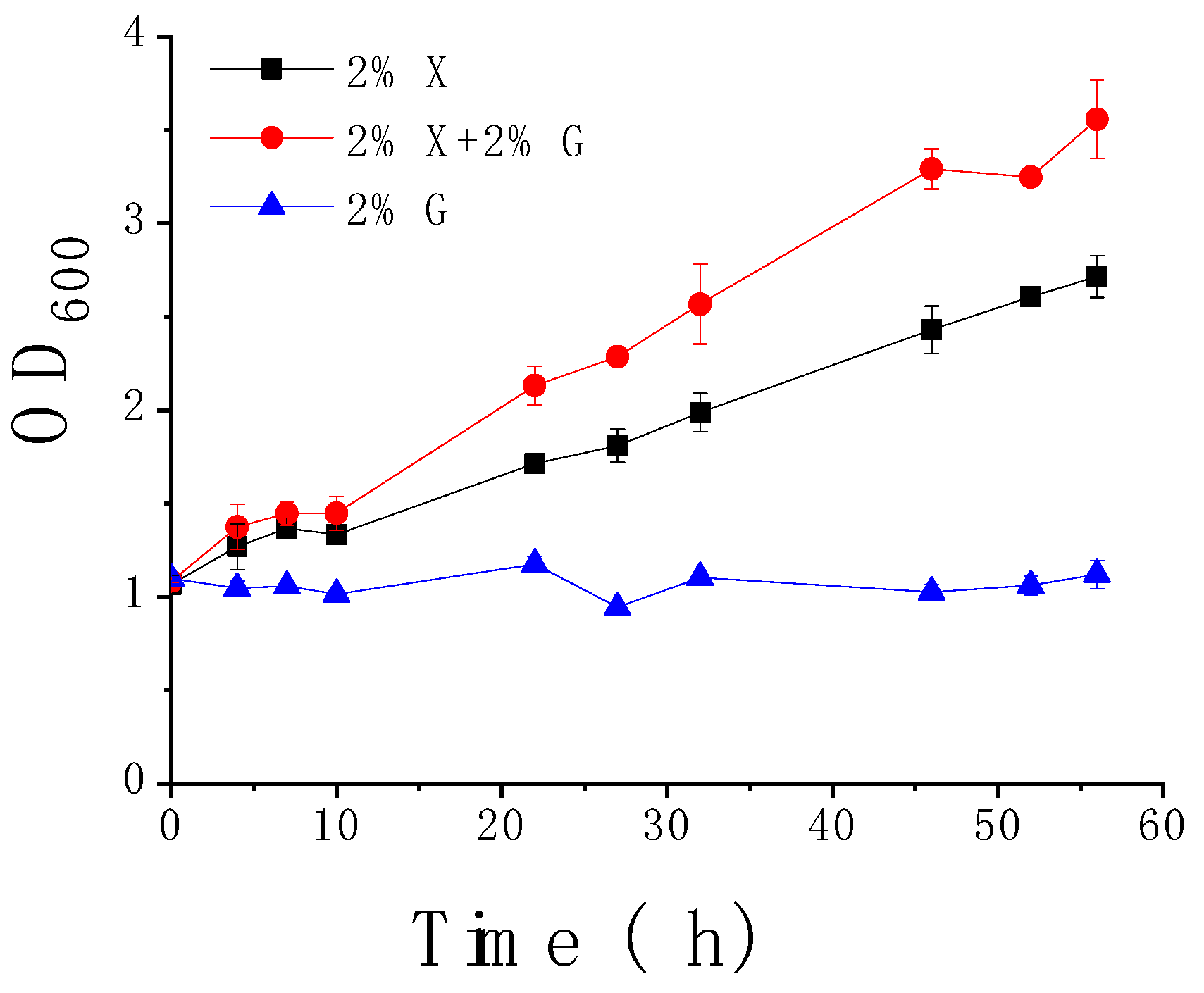
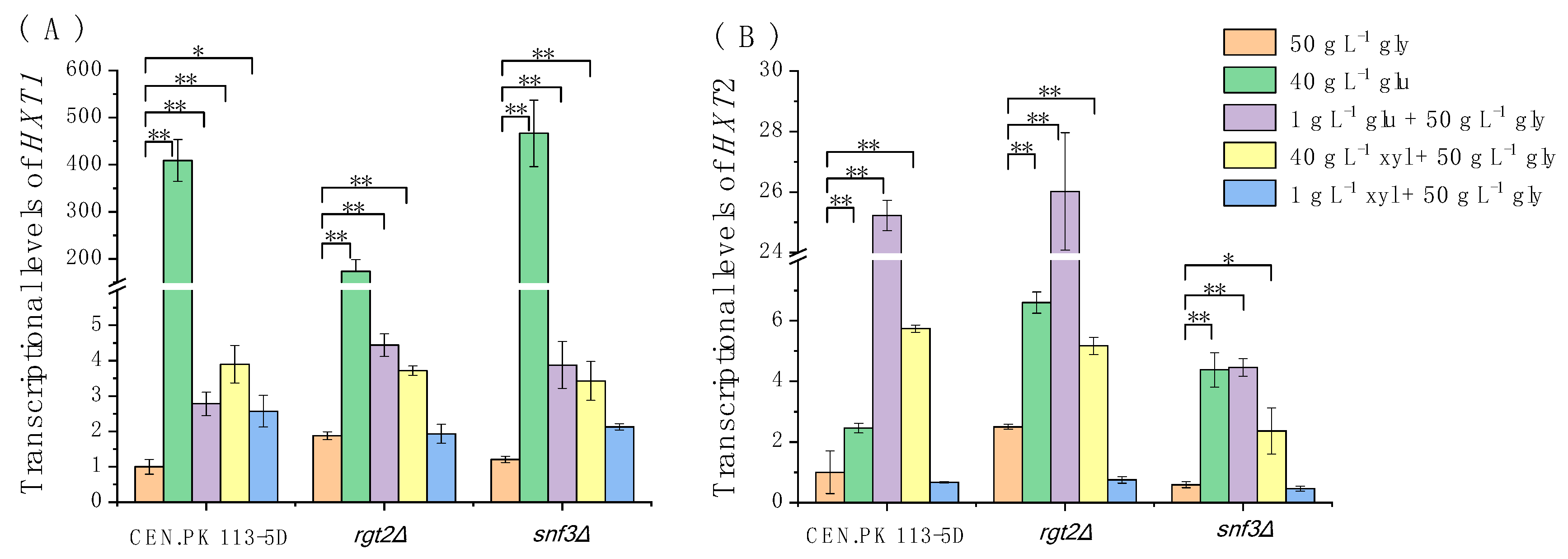
3.3. Weak Responses of Rgt2 and Snf3 to High Concentrations of Extracellular Xylose
3.4. Deleting RGT1 Enhanced Xylose Utilization in S. cerevisiae by Upregulating the Expression of Hexose Transporter Genes
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| Primers | Sequence 5′→3′ |
|---|---|
| For gene expression | |
| BamHI-GPA2-1up | GCGCGGATCCATGGGTCTCTGCGCATCTTC |
| GPA2-1dn | ACTTACCACTTTCAACGGCACCCAG |
| GPA2-2up | CTGCTGGGTGCCGTTGAAAGTGGTAAG |
| SbfI-GPA2-2dn | CGCGCCTGCAGGTCATTGTAACACTCCAGAGTCTTTC |
| GRE3-ty1up | CAGATGACGAGAAGAAAGGTCACATC |
| GRE3-ty1dn | CCGAAGGAGGAGTAAGCAACTAC |
| KanMX-up0505 | ATATCCAAGTAGTTGCTTACTCCTCCTTCGGTAGGTCTAGAGATCTGTTTAGCTTGCC |
| KanMX-dn0505 | CATTTTGAAGCTATGGTGTGTGGGCCACTAGTGGATCTGATATCACC |
| GPA2-up0505 | GTGATATCAGATCCACTAGTGGCCCACACACCATAGCTTCAAAATGTTTC |
| GPA2-dn0505 | GCCCATCTAAGCAATACTTGGGAAGCATGATTACGCCAAGCTTTAAC |
| GRE3-ty2up | CGTAATCATGCTTCCCAAGTATTGCTTAGATGGGC |
| GRE3-ty2dn | TCAGGCAAAAGTGGGGAATTTACC |
| G-int-up | CCATTTGAAGAGAAATACCCTCCAG |
| RAS-ty1up | GAAAACGGGAAACAAGGTTCACATCAG |
| RAS-ty1dn | CGACTTGTTCAAAGGCATTTTTTTTTCTGTATATCTCCTTTCAATTCGAAAACGG |
| Rasup1 | AAGAGAGTACAAGCTAGTCGTCGTTGGTGGTGTTGGTGTTGGTAAATCTGC |
| Rasup2 | ATGCCTTTGAACAAGTCGAACATAAGAGAGTACAAGCTAGTCGTCG |
| RAS-up | GAATTGAAAGGAGATATACAGAAAAAAAAATGCCTTTGAACAAGTCGAAC |
| RAS-dn | AGCGTACGAAGCTTCAGCTTTAACTTATAATACAACAGCCACCCGATC |
| K-RAS-up | AGTTAAAGCTGAAGCTTCGTACGCTG |
| K-RAS-dn | GCGTTTCTACAACTATTTCCTTTTTAGCATAGGCCACTAGTGGATCTG |
| RAS-ty2up | CCACTAGTGGCCTATGCTAAAAAGGAAATAGTTGTAGAAACGCTAAGACG |
| RAS-ty2dn | CTCTGGAACGTCCTCATATTCACC |
| seq-rasR | GTATTCTGGGCCTCCATGTC |
| int-Y | CAGAAGGACCTTTTCATTCACC |
| RuXI-up | GGATCCATGGCAAAAGAATATTTTCCGTTTAC |
| RuXI-dn | CCTGCAGGTTATTTGCAGTGGAGGGCG |
| For gene knockout | |
| PDE1-ty1up | GAATGTACCAGCTACGGGAGATG |
| PDE1-ty1dn | CGTACGAAGCTTCAGCTCAGCCCACTAATATGGTCCAGATG |
| K-PDE1-up | CATCTGGACCATATTAGTGGGCTGAGCTGAAGCTTCGTACGCTG |
| K-PDE1-dn | CTAATATACCCTTGAGATGCACAAGCGCATAGGCCACTAGTGGATCTG |
| PDE1-ty2up | GATCCACTAGTGGCCTATGCGCTTGTGCATCTCAAGGGTATATTAG |
| PDE1-ty2dn | CTCAAGTCTCCTAAGTTCCTCTCC |
| PDE2-ty1up | ATGTCCACCCTTTTTCTGATTGG |
| PDE2-ty1dn | AGCGTACGAAGCTTCAGCTCTCTCGTGTATGATCTATCGCAG |
| K-PDE2-up | TCTGCGATAGATCATACACGAGAGAGCTGAAGCTTCGTACGCTG |
| K-PDE2-dn | CTATCCTCATACTGAGAATGCAATGCGCATAGGCCACTAGTGGATCTG |
| PDE2-ty2up | GATCCACTAGTGGCCTATGCGCATTGCATTCTCAGTATGAGGATAG |
| PDE2-ty2dn | CGTTTGGAATATGCGGATGGTC |
| KanMXhomoup-F | TGTGACGTGGCTTATGAGCACCAG |
| KanMXhomoup-R | AGTACAAGGGAGAGGCGATTTCCAACG |
| KanMXhomodn-F | ATGCTCCCTTCTCCTGTAGGTCAGG |
| KanMXhomodn-R | TGGCAATGGCAATAGTGATGGCACC |
| KanMX-F | GGAAATCGCCTCTCCCTTGTACTGACATGGAGGCCCAGAATAC |
| KanMX-R | CTGACCTACAGGAGAAGGGAGCATCAGTATAGCGACCAGC |
| SNF3-1 | AGTGGCGTATTGGCCTTGTT |
| SNF3-2 | GCCGAGGATAGGACTATTGTCTAG |
| SNF3-3 | CAATAGTCCTATCCTCGGCACGCGGCCGCCAGCTGAAGCTTCG |
| SNF3-4 | GTAACCCAGAATCATGGACTGGACCGGCAGATCCGCGGCCGCA |
| SNF3-5 | CCAGTCCATGATTCTGGGTTACTG |
| SNF3-6 | CAACAACCAGCACCTTACGTCTAC |
| RGT2-1 | CTGTTCCTGCATCGTCCATCGGC |
| RGT2-2 | CGGCAGGTAGTCACCGTTGAGTC |
| RGT2-3 | TCAACGGTGACTACCTGCCGACGCGGCCGCCAGCTGAAGCTTCG |
| RGT2-4 | CTTGGATCTTCGATCGGGAGGCCACCGGCAGATCCGCGGCCGCATAG |
| RGT2-5 | GGCCTCCCGATCGAAGATCCAAG |
| RGT2-6 | ACTGAAGGCGACGTTGACGGC |
| For qPCR | |
| Actin-F | CAAACCGCTGCTCAATCTTC |
| Actin-R | AGTTTGGTCAATACCGGCAG |
| HXT1-qPCR-F | GTCGGTATGGTCTGCTGTTATG |
| HXT1-qPCR-R | ACAGTTACCAGCACCCTTTG |
| HXT2-qPCR-F | GGCTCTCAACAAACTTCTATCCAC |
| HXT2-qPCR-R | GGGAGTTCAGCGTTAGTGTATTC |
References
- Kwak, S.; Jo, J.H.; Yun, E.J.; Jin, Y.S.; Seo, J.H. Production of biofuels and chemicals from xylose using native and engineered yeast strains. Biotechnol. Adv. 2019, 37, 271–283. [Google Scholar] [CrossRef]
- Jansen, M.L.A.; Bracher, J.M.; Papapetridis, I.; Verhoeven, M.D.; de Bruijn, H.; de Waal, P.P.; van Maris, A.J.A.; Klaassen, P.; Pronk, J.T. Saccharomyces cerevisiae strains for second-generation ethanol production: From academic exploration to industrial implementation. FEMS Yeast Res. 2017, 17, fox044. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Qiu, C.; Shen, Y.; Li, H.; Bao, X. Engineering of Saccharomyces cerevisiae for the efficient co-utilization of glucose and xylose. FEMS Yeast Res. 2017, 17, fox034. [Google Scholar] [CrossRef] [PubMed]
- Walfridsson, M.; Anderlund, M.; Bao, X.; Hahn-Hagerdal, B. Expression of different levels of enzymes from the Pichia stipitis XYL1 and XYL2 genes in Saccharomyces cerevisiae and its effects on product formation during xylose utilisation. Appl. Microbiol. Biotechnol. 1997, 48, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Kuyper, M.; Harhangi, H.R.; Stave, A.K.; Winkler, A.A.; Jetten, M.S.; de Laat, W.T.; den Ridder, J.J.; Op den Camp, H.J.; van Dijken, J.P.; Pronk, J.T. High-level functional expression of a fungal xylose isomerase: The key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res. 2003, 4, 69–78. [Google Scholar] [CrossRef]
- Toivari, M.H.; Aristidou, A.; Ruohonen, L.; Penttila, M. Conversion of xylose to ethanol by recombinant Saccharomyces cerevisiae: Importance of xylulokinase (XKS1) and oxygen availability. Metab. Eng. 2001, 3, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Johansson, B.; Hahn-Hagerdal, B. The non-oxidative pentose phosphate pathway controls the fermentation rate of xylulose but not of xylose in Saccharomyces cerevisiae TMB3001. FEMS Yeast Res. 2002, 2, 277–282. [Google Scholar] [PubMed]
- Zhou, H.; Cheng, J.S.; Wang, B.L.; Fink, G.R.; Stephanopoulos, G. Xylose isomerase overexpression along with engineering of the pentose phosphate pathway and evolutionary engineering enable rapid xylose utilization and ethanol production by Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 611–622. [Google Scholar] [CrossRef]
- Kuyper, M.; Toirkens, M.J.; Diderich, J.A.; Winkler, A.A.; van Dijken, J.P.; Pronk, J.T. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 2005, 5, 925–934. [Google Scholar] [CrossRef]
- Qi, X.; Zha, J.; Liu, G.G.; Zhang, W.; Li, B.Z.; Yuan, Y.J. Heterologous xylose isomerase pathway and evolutionary engineering improve xylose utilization in Saccharomyces cerevisiae. Front. Microbiol. 2015, 6, 1165. [Google Scholar] [CrossRef]
- Shen, Y.; Chen, X.; Peng, B.; Chen, L.; Hou, J.; Bao, X. An efficient xylose-fermenting recombinant Saccharomyces cerevisiae strain obtained through adaptive evolution and its global transcription profile. Appl. Microbiol. Biotechnol. 2012, 96, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Shen, Y.; Li, X.; Chen, X.; Hou, J.; Bao, X. Improvement of xylose fermentation in respiratory-deficient xylose-fermenting Saccharomyces cerevisiae. Metab. Eng. 2012, 14, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Jiao, C.; Peng, B.; Shen, Y.; Bao, X. Mutation of a regulator Ask10p improves xylose isomerase activity through up-regulation of molecular chaperones in Saccharomyces cerevisiae. Metab. Eng. 2016, 38, 241–250. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.K.; Tremaine, M.; Parreiras, L.S.; Hebert, A.S.; Myers, K.S.; Higbee, A.J.; Sardi, M.; McIlwain, S.J.; Ong, I.M.; Breuer, R.J.; et al. Directed Evolution Reveals Unexpected Epistatic Interactions That Alter Metabolic Regulation and Enable Anaerobic Xylose Use by Saccharomyces cerevisiae. PLoS Genet. 2016, 12, e1006372. [Google Scholar]
- Myers, K.S.; Riley, N.M.; MacGilvray, M.E.; Sato, T.K.; McGee, M.; Heilberger, J.; Coon, J.J.; Gasch, A.P. Rewired cellular signaling coordinates sugar and hypoxic responses for anaerobic xylose fermentation in yeast. PLoS Genet. 2019, 15, e1008037. [Google Scholar] [CrossRef]
- Wei, S.; Liu, Y.; Wu, M.; Ma, T.; Bai, X.; Hou, J.; Shen, Y.; Bao, X. Disruption of the transcription factors Thi2p and Nrm1p alleviates the post-glucose effect on xylose utilization in Saccharomyces cerevisiae. Biotechnol. Biofuels 2018, 11, 112. [Google Scholar] [CrossRef]
- Salusjarvi, L.; Kankainen, M.; Soliymani, R.; Pitkanen, J.P.; Penttila, M.; Ruohonen, L. Regulation of xylose metabolism in recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2008, 7, 18. [Google Scholar] [CrossRef]
- Wang, C.; Bao, X.; Li, Y.; Jiao, C.; Hou, J.; Zhang, Q.; Zhang, W.; Liu, W.; Shen, Y. Cloning and characterization of heterologous transporters in Saccharomyces cerevisiae and identification of important amino acids for xylose utilization. Metab. Eng. 2015, 30, 79–88. [Google Scholar] [CrossRef]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef]
- Kim, J.H.; Roy, A.; Jouandot, D., II; Cho, K.H. The glucose signaling network in yeast. Biochim. Biophys. Acta Gen. Subj. 2013, 1830, 5204–5210. [Google Scholar] [CrossRef]
- Thevelein, J.M.; de Winde, J.H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 1999, 33, 904–918. [Google Scholar] [CrossRef] [PubMed]
- Ozcan, S.; Dover, J.; Rosenwald, A.G.; Wolfl, S.; Johnston, M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 1996, 93, 12428–12432. [Google Scholar] [CrossRef] [PubMed]
- Busti, S.; Coccetti, P.; Alberghina, L.; Vanoni, M. Glucose signaling-mediated coordination of cell growth and cell cycle in Saccharomyces cerevisiae. Sensors (Basel) 2010, 10, 6195–6240. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.K.; Behera, S.; Arora, R.; Kumar, S.; Sani, R.K. Xylose transport in yeast for lignocellulosic ethanol production: Current status. J. Biosci. Bioeng. 2018, 125, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Brink, D.P.; Borgstrom, C.; Tueros, F.G.; Gorwa-Grauslund, M.F. Real-time monitoring of the sugar sensing in Saccharomyces cerevisiae indicates endogenous mechanisms for xylose signaling. Microb. Cell Fact. 2016, 15, 183. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Shen, Y.; Jiao, C.; Ge, R.; Zhang, X.; Bao, X. Characterization and evolution of xylose isomerase screened from the bovine rumen metagenome in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2016, 121, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Entian, K.-D.; Kötter, P. 25 Yeast Genetic Strain and Plasmid Collections. Methods Microbiol. 2007, 36, 629–666. [Google Scholar]
- Guldener, U.; Heck, S.; Fiedler, T.; Beinhauer, J.; Hegemann, J.H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996, 24, 2519–2524. [Google Scholar] [CrossRef]
- Pernambuco, M.B.; Winderickx, J.; Crauwels, M.; Griffioen, G.; Mager, W.H.; Thevelein, J.M. Glucose-triggered signalling in Saccharomyces cerevisiae: Different requirements for sugar phosphorylation between cells grown on glucose and those grown on non-fermentable carbon sources. Microbiology 1996, 142, 1775–1782. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zahringer, H.; Thevelein, J.M.; Nwaka, S. Induction of neutral trehalase Nth1 by heat and osmotic stress is controlled by STRE elements and Msn2/Msn4 transcription factors: Variations of PKA effect during stress and growth. Mol. Microbiol. 2000, 35, 397–406. [Google Scholar] [CrossRef]
- Rolland, F.; De Winde, J.H.; Lemaire, K.; Boles, E.; Thevelein, J.M.; Winderickx, J. Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 2000, 38, 348–358. [Google Scholar] [CrossRef]
- Colombo, S.; Ma, P.; Cauwenberg, L.; Winderickx, J.; Crauwels, M.; Teunissen, A.; Nauwelaers, D.; de Winde, J.H.; Gorwa, M.F.; Colavizza, D. Involvement of distinct G-proteins, Gpa2 and Ras, in glucose-and intracellular acidification-induced cAMP signalling in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 3326–3341. [Google Scholar] [CrossRef]
- Osiro, K.O.; Brink, D.P.; Borgstrom, C.; Wasserstrom, L.; Carlquist, M.; Gorwa-Grauslund, M.F. Assessing the effect of d-xylose on the sugar signaling pathways of Saccharomyces cerevisiae in strains engineered for xylose transport and assimilation. FEMS Yeast Res. 2018, 18, fox096. [Google Scholar] [CrossRef]
- Ozcan, S.; Dover, J.; Johnston, M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998, 17, 2566–2573. [Google Scholar] [CrossRef]
- Vanhalewyn, M.; Dumortier, F.; Debast, G.; Colombo, S.; Ma, P.; Winderickx, J.; Van Dijck, P.; Thevelein, J.M. A mutation in Saccharomyces cerevisiae adenylate cyclase, Cyr1K1876M, specifically affects glucose- and acidification-induced cAMP signalling and not the basal cAMP level. Mol. Microbiol. 1999, 33, 363–376. [Google Scholar] [CrossRef]
- Peeters, T.; Louwet, W.; Gelade, R.; Nauwelaers, D.; Thevelein, J.M.; Versele, M. Kelch-repeat proteins interacting with the Galpha protein Gpa2 bypass adenylate cyclase for direct regulation of protein kinase A in yeast. Proc. Natl. Acad. Sci. USA 2006, 103, 13034–13039. [Google Scholar] [CrossRef]
- Osiro, K.O.; Borgstrom, C.; Brink, D.P.; Fjolnisdottir, B.L.; Gorwa-Grauslund, M.F. Exploring the xylose paradox in Saccharomyces cerevisiae through in vivo sugar signalomics of targeted deletants. Microb. Cell Fact. 2019, 18, 88. [Google Scholar] [CrossRef]
- Longo, V.D.; Fabrizio, P. Chronological aging in Saccharomyces cerevisiae. Subcell. Biochem. 2012, 57, 101–121. [Google Scholar]
- Liu, Y.; Yang, F.; Li, S.; Dai, J.; Deng, H. Glutaredoxin Deletion Shortens Chronological Life Span in Saccharomyces cerevisiae via ROS-Mediated Ras/PKA Activation. J. Proteome Res. 2018, 17, 2318–2327. [Google Scholar] [CrossRef]


| S. cerevisiae Strains and Plasmids | Description | Sources |
|---|---|---|
| Plasmids | ||
| pUG6 | E. coli plasmid with segment LoxP-KanMX44-LoxP | [28] |
| pJFE1 | YCplac33; CEN, Ampr, URA3, TEF1p-PGK1t | [26] |
| pJX7 | ΥΕplac195; 2μ, Ampr, URA3, TEF1p-Ru-xylA-PGK1t | [26] |
| YEp-CH | YEp; containing hygromycin B resistant gene and Cre gene under GAL2 regulative regulation promoter | [26] |
| S. cerevisiae Strains | ||
| CEN.PK 113-5D | MATa; ura3-53, belongs to CEN.PK strain family | [27] |
| BSPC039 | CEN.PK113-5D derivative; XKS1(-194,-1)::loxP-PTEF1, gre3(-241, +338)::PTPI1-RKI1-TRKI1-PPGK1-TAL1-TTAL1-PFBA1-TKL1-TTKL1-PADH1-RPE1-TRPE1-loxP | [12] |
| BSL01 | BSPC039 derivative; pJX7 | This work |
| BSL06 | BSL01 derivative; GPA2G132V | This work |
| BSL08 | BSL01 derivative; RAS2G19V | This work |
| BSL10 | BSL01 derivative; pde1Δ | This work |
| BSL16 | BSL01 derivative; pde1Δ; pde2Δ | This work |
| BSL20 | BSL01 derivative; rgt1Δ | This work |
| BSWW1 | Derivative of CEN.PK 113-5D: snf3Δ | This work |
| BSWW2 | Derivative of CEN.PK 113-5D: rgt2Δ | This work |
| Strains | Xylose Fermentation | Glucose–Xylose Co-Fermentation | |||
|---|---|---|---|---|---|
| Specific Xylose Consumption Rates (g L−1 h−1 g−1 DCW) | Specific Ethanol Production Rates (g L−1 h−1 g−1 DCW) | Specific Glucose Consumption Rates (g L−1 h−1 g−1 DCW) | Specific Xylose Consumption Rates (g L−1 h−1 g−1 DCW) | Specific Ethanol Production Rates (g L−1 h−1 g−1 DCW) | |
| BSL01(Control) | 0.103 ± 0.003 | 0.047 ± 0.002 | 1.218 ± 0.004 | 0.046 ± 0.002 | 0.428 ± 0.005 |
| BSL06(GPA2G132V) | 0.123 ± 0.003 | 0.082 ± 0.001 *** | 1.354 ± 0.019 ** | 0.0481 ± 0.002 | 0.489 ± 0.002 *** |
| BSL16(pde1Δ pde2Δ) | 0.156 ± 0.001 ** | 0.081 ± 0.003 ** | 1.260 ± 0.008 * | 0.068 ± 0.004 * | 0.523 ± 0.005 *** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, M.; Li, H.; Wei, S.; Wu, H.; Wu, X.; Bao, X.; Hou, J.; Liu, W.; Shen, Y. Simulating Extracellular Glucose Signals Enhances Xylose Metabolism in Recombinant Saccharomyces cerevisiae. Microorganisms 2020, 8, 100. https://doi.org/10.3390/microorganisms8010100
Wu M, Li H, Wei S, Wu H, Wu X, Bao X, Hou J, Liu W, Shen Y. Simulating Extracellular Glucose Signals Enhances Xylose Metabolism in Recombinant Saccharomyces cerevisiae. Microorganisms. 2020; 8(1):100. https://doi.org/10.3390/microorganisms8010100
Chicago/Turabian StyleWu, Meiling, Hongxing Li, Shan Wei, Hongyu Wu, Xianwei Wu, Xiaoming Bao, Jin Hou, Weifeng Liu, and Yu Shen. 2020. "Simulating Extracellular Glucose Signals Enhances Xylose Metabolism in Recombinant Saccharomyces cerevisiae" Microorganisms 8, no. 1: 100. https://doi.org/10.3390/microorganisms8010100
APA StyleWu, M., Li, H., Wei, S., Wu, H., Wu, X., Bao, X., Hou, J., Liu, W., & Shen, Y. (2020). Simulating Extracellular Glucose Signals Enhances Xylose Metabolism in Recombinant Saccharomyces cerevisiae. Microorganisms, 8(1), 100. https://doi.org/10.3390/microorganisms8010100





