Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon
Abstract
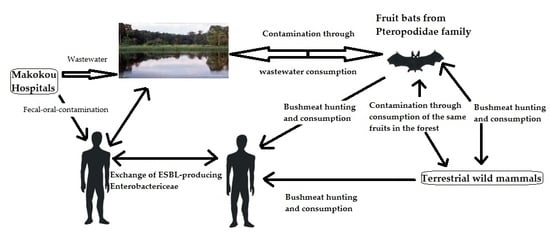
:1. Introduction
2. Materials and Methods
2.1. Research License
2.2. Study Area
2.3. Collection of Fecal Samples
2.4. Culture, Isolation and Identification of Colonies
2.5. Antibiotic Susceptibility Testing
2.6. Phylogenetic Analyses
2.7. Statistical Analyses
2.8. Accession Numbers
3. Results
3.1. Enterobacteria Found in Bat Faecal Samples
3.2. Antibiotic Susceptibility
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cláudio, V.C.; Gonzalez, I.; Barbosa, G.; Rocha, V.; Moratelli, R.; Rassy, F. Bacteria richness and antibiotic-resistance in bats from a protected area in the Atlantic Forest of Southeastern Brazil. PLoS ONE 2018, 13, e0203411. [Google Scholar] [CrossRef] [PubMed]
- Allocati, N.; Petrucci, A.; Di Giovanni, P.; Masulli, M.; Di Ilio, C.; De Laurenzi, V. Bat–man disease transmission: Zoonotic pathogens from wildlife reservoirs to human populations. Cell Death Discov. 2016, 2, 16048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dietrich, M.; Kearney, T.; Seamark, E.C.; Markotter, W. The excreted microbiota of bats: Evidence of niche specialisation based on multiple body habitats. FEMS Microbiol. Lett. 2017, 364, fnw284. [Google Scholar] [CrossRef] [PubMed]
- Mühldorfer, K. Bats and bacterial pathogens: A review. Zoonoses Public Health 2013, 60, 93–103. [Google Scholar] [CrossRef]
- Graves, S.; Kennelly-Merrit, S.; Tidemann, C.; Rawlinson, P.; Harvey, K.; Thornton, I. Antibiotic-resistance patterns of enteric bacteria of wild mammals on the Krakatau Islands and West Java, Indonesia. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1988, 322, 339–353. [Google Scholar] [CrossRef]
- McDougall, F.; Boardman, W.; Gillings, M.; Power, M. Bats as reservoirs of antibiotic resistance determinants: A survey of class 1 integrons in Grey-headed Flying Foxes (Pteropus poliocephalus). Infect. Genet. Evol. 2019, 70, 107–113. [Google Scholar] [CrossRef]
- Streicker, D.G.; Turmelle, A.S.; Vonhof, M.J.; Kuzmin, I.V.; McCracken, G.F.; Rupprecht, C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 2010, 329, 676–679. [Google Scholar] [CrossRef] [Green Version]
- Dolejska, M.; Papagiannitsis, C.C. Papagiannitsis, Plasmid-mediated resistance is going wild. Plasmid 2018, 99, 99–111. [Google Scholar] [CrossRef]
- Adesiyun, A.; Downes, M. Prevalence of antimicrobial resistance and enteropathogenic serogroups in Escherichia coli isolates. Vet. Arhiv 1999, 69, 335–347. [Google Scholar]
- Benavides, J.; Shiva, C.; Virhuez, M.; Tello, C.; Appelgren, A.; Vendrell, J.; Solassol, J.; Godreuil, S.; Streicker, D. Extended-spectrum beta-lactamase-producing Escherichia coli in common vampire bats Desmodus rotundus and livestock in Peru. Zoonoses Public Health 2018, 65, 454–458. [Google Scholar] [CrossRef] [Green Version]
- Djedjiga, A.C.C. Etude de la Flore Bactérienne Résistante aux Antibiotiques chez les Chauves-Souris. Mémoires de Master. 2017. Available online: http://www.univ-bejaia.dz/dspace/handle/123456789/204 (accessed on 28 December 2018).
- Garcês, A.; Correia, S.; Amorim, F.; Pereira, J.; Igrejas, G.; Poeta, P. First report on extended-spectrum beta-lactamase (ESBL) producing Escherichia coli from European free-tailed bats (Tadarida teniotis) in Portugal: A One-Health approach of a hidden contamination problem. J. Hazard. Mater. 2017, 370, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Iroha, I.; Afiukwa, F.; Oji, A.; Ejikeugwu, P.; Nwakeze, E. Occurrence of extended spectrum beta lactamase producing Escherichia coli from human clinical and wild birds (pigeons, bats, parrots and ducks) samples from Ebonyi state, Nigeria. World J. Pharm. Sci. 2015, 4, 20–29. [Google Scholar]
- Obi, T.; Chibana, M.; Taira, C.; Nakayama, A.; Miyazaki, K.; Takase, K.; Nakamura, I.; Miyamoto, A.; Kawamoto, Y. Antimicrobial susceptibility in Enterobacteriaceae recovered from Okinawa least horseshoe bat Rhinolophus pumilus. Wildl. Biol. 2014, 20, 64–66. [Google Scholar] [CrossRef]
- Oluduro, A.O. Antibiotic-resistant commensal Escherichia coli in faecal droplets from bats and poultry in Nigeria. Vet. Ital. 2012, 48, 297–308. [Google Scholar]
- Hassell, J.M.; Ward, M.J.; Muloi, D.; Bettridge, J.M.; Robinson, T.P.; Kariuki, S.; Ogendo, A.; Kiiru, J.; Imboma, T.; Kang’ethe, E.K. Clinically relevant antimicrobial resistance at the wildlife–livestock–human interface in Nairobi: An epidemiological study. Lancet Planet. Health 2019, 3, e259–e269. [Google Scholar] [CrossRef] [Green Version]
- Ambler, R.; Coulson, A.; Frere, J.-M.; Ghuysen, J.-M.; Joris, B.; Forsman, M.; Levesque, R.; Tiraby, G.; Waley, S. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 1991, 276 Pt 1, 269. [Google Scholar] [CrossRef]
- Paterson, D.L.; Bonomo, R.A. Extended-spectrum β-lactamases: A clinical update. Clin. Microbiol. Rev. 2005, 18, 657–686. [Google Scholar] [CrossRef] [Green Version]
- Sosa, A.D.J.; Amábile-Cuevas, C.F.; Byarugaba, D.K.; Hsueh, P.-R.; Kariuki, S.; Okeke, I.N. Antimicrobial Resistance in Developing Countries; Springer: New York, NY, USA, 2010. [Google Scholar]
- Bradford, P.A. Extended-spectrum β-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 2001, 14, 933–951. [Google Scholar] [CrossRef] [Green Version]
- Benavides, J.A.; Godreuil, S.; Bodenham, R.; Ratiarison, S.; Devos, C.; Petretto, M.-O.; Raymond, M.; Escobar-Páramo, P. No evidence for transmission of antibiotic-resistant Escherichia coli strains from humans to wild western lowland gorillas in Lope National Park, Gabon. Appl. Environ. Microbiol. 2012, 78, 4281–4287. [Google Scholar] [CrossRef] [Green Version]
- Blanco, G.; Lemus, J.A.; Grande, J.; Gangoso, L.; Grande, J.M.; Donázar, J.A.; Arroyo, B.; Frías, O.; Hiraldo, F. Retracted Geographical variation in cloacal microflora and bacterial antibiotic resistance in a threatened avian scavenger in relation to diet and livestock farming practices. Environ. Microbiol. 2007, 9, 1738–1749. [Google Scholar] [CrossRef]
- Cantón, R.; González-Alba, J.M.; Galán, J.C. CTX-M enzymes: Origin and diffusion. Front. Microbiol. 2012, 3, 110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A. Plasmids and the spread of resistance. Int. J. Med. Microbiol. 2013, 303, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L. Extended-spectrum β-lactamase-producing and AmpC-producing Escherichia coli from livestock and companion animals, and their putative impact on public health: A global perspective. Clin. Microbiol. Infect. 2012, 18, 646–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, K.H.; Damborg, P.; Andreasen, M.; Nielsen, S.S.; Guardabassi, L. Carriage and fecal counts of cefotaxime M-producing Escherichia coli in pigs: A longitudinal study. Appl. Environ. Microbiol. 2013, 79, 794–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janatova, M.; Albrechtova, K.; Petrzelkova, K.J.; Dolejska, M.; Papousek, I.; Masarikova, M.; Cizek, A.; Todd, A.; Shutt, K.; Kalousova, B. Antimicrobial-resistant Enterobacteriaceae from humans and wildlife in Dzanga-Sangha Protected Area, Central African Republic. Vet. Microbiol. 2014, 171, 422–431. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, A.; Mevius, D.J.; Olsen, B.; Bonnedahl, J. The colistin resistance mcr-1 gene is going wild. J. Antimicrob. Chemother. 2016, 71, 2335–2336. [Google Scholar] [CrossRef] [Green Version]
- Mbehang Nguema, P.P.; Okubo, T.; Tsuchida, S.; Fujita, S.; Yamagiwa, J.; Tamura, Y.; Ushida, K. Isolation of multiple drug-resistant enteric bacteria from feces of wild Western Lowland Gorilla (Gorilla gorilla gorilla) in Gabon. J. Vet. Med. Sci. 2015, 77, 619. [Google Scholar] [CrossRef] [Green Version]
- Mbehang Nguema, P.P.; Tsuchida, S.; Ushida, K. Bacteria culturing and isolation under field conditions of Moukalaba-Doudou National Park, Gabon, and preliminary survey on bacteria carrying antibiotic resistance genes. Tropics 2015, 23, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Schaumburg, F.; Alabi, A.; Kokou, C.; Grobusch, M.P.; Köck, R.; Kaba, H.; Becker, K.; Adegnika, A.A.; Kremsner, P.G.; Peters, G. High burden of extended-spectrum β-lactamase-producing Enterobacteriaceae in Gabon. J. Antimicrob. Chemother. 2013, 68, 2140–2143. [Google Scholar] [CrossRef] [Green Version]
- Schaumburg, F.; Alabi, A.S.; Frielinghaus, L.; Grobusch, M.P.; Köck, R.; Becker, K.; Issifou, S.; Kremsner, P.G.; Peters, G.; Mellmann, A. The risk to import ESBL-producing Enterobacteriaceae and Staphylococcus aureus through chicken meat trade in Gabon. BMC Microbiol. 2014, 14, 286. [Google Scholar] [CrossRef]
- Bauer, A.; Kirby, W.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–497. [Google Scholar] [CrossRef] [PubMed]
- Chaverri, G. Aerobic bacterial flora from the digestive tract of the common vampire bat, Desmodus rotundus (Chiroptera: Phyllostomidae). Rev. Biol. Trop. 2006, 54, 717–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, M.; Aggarwal, A. Aggarwal, Occurrence of the CTX-M, SHV and the TEM genes among the extended spectrum β-lactamase producing isolates of Enterobacteriaceae in a tertiary care hospital of North India. J. Clin. Diagn. Res. 2013, 7, 642–645. [Google Scholar] [PubMed]
- Thabit, A.G.; El-Khamissy, T.R.; Ibrahim, M.A.; Attia, A.E. Detection of extended-spectrum β-lactamase enzymes (ESBLs) produced by Escherichia coli urinary pathogens at Assiut University Hospital. Bull. Pharm. Sci. Assiut Univ. 2011, 34, 93–103. [Google Scholar]
- Peng, X.; Yu, K.-Q.; Deng, G.-H.; Jiang, Y.-X.; Wang, Y.; Zhang, G.-X.; Zhou, H.-W. Comparison of direct boiling method with commercial kits for extracting fecal microbiome DNA by Illumina sequencing of 16S rRNA tags. J. Microbiol. Methods 2013, 95, 455–462. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Adesiyun, A.A.; Stewart-Johnson, A.; Thompson, N.N. Isolation of enteric pathogens from bats in Trinidad. J. Wildl. Dis. 2009, 45, 952–961. [Google Scholar] [CrossRef] [Green Version]
- Islam, A.; Mikolon, A.; Mikoleit, M.; Ahmed, D.; Khan, S.U.; Sharker, M.Y.; Hossain, M.J.; Islam, A.; Epstein, J.H.; Zeidner, N. Isolation of Salmonella virchow from a fruit bat (Pteropus giganteus). EcoHealth 2013, 10, 348–351. [Google Scholar] [CrossRef]
- Klite, P. Intestinal bacterial flora and transit time of three neotropical bat species. J. Bacteriol. 1965, 90, 375–379. [Google Scholar] [CrossRef] [Green Version]
- Moreno, G.; Lopes, C.; Seabra, E.; Pavan, C.; Correa, A. Bacteriological study of the intestinal flora of bats (Desmodus rotundus) (author’s transl). Arq. Inst. Biol. 1975, 42, 229–232. [Google Scholar]
- Nowak, K.; Fahr, J.; Weber, N.; Lübke-Becker, A.; Semmler, T.; Weiss, S.; Mombouli, J.-V.; Wieler, L.H.; Guenther, S.; Leendertz, F.H. Highly diverse and antimicrobial susceptible Escherichia coli display a naïve bacterial population in fruit bats from the Republic of Congo. PLoS ONE 2017, 12, e0178146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sallem, R.B.; Gharsa, H.; Slama, K.B.; Rojo-Bezares, B.; Estepa, V.; Porres-Osante, N.; Jouini, A.; Klibi, N.; Sáenz, Y.; Boudabous, A. First detection of CTX-M-1, CMY-2, and QnrB19 resistance mechanisms in fecal Escherichia coli isolates from healthy pets in Tunisia. Vector Borne Zoonotic Dis. 2013, 13, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Slama, K.B.; Sallem, R.B.; Jouini, A.; Rachid, S.; Moussa, L.; Sáenz, Y.; Estepa, V.; Somalo, S.; Boudabous, A.; Torres, C. Diversity of genetic lineages among CTX-M-15 and CTX-M-14 producing Escherichia coli strains in a Tunisian hospital. Curr. Microbiol. 2011, 62, 1794–1801. [Google Scholar] [CrossRef]
- Gakuya, F.; Kyule, M.; Gathura, P.; Kariuki, S. Antimicrobial resistance of bacterial organisms isolated from rats. East Afr. Med. J. 2001, 78, 646–649. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Ma, Z.-B.; Zeng, Z.-L.; Yang, X.-W.; Huang, Y.; Liu, J.-H. The role of wildlife (wild birds) in the global transmission of antimicrobial resistance genes. Zool. Res. 2017, 38, 55–80. [Google Scholar] [CrossRef] [Green Version]
- Hordijk, J.; Schoormans, A.; Kwakernaak, M.; Duim, B.; Broens, E.; Dierikx, C.; Mevius, D.; Wagenaar, J.A. High prevalence of fecal carriage of extended spectrum β-lactamase/AmpC-producing Enterobacteriaceae in cats and dogs. Front. Microbiol. 2013, 4, 242. [Google Scholar] [CrossRef] [Green Version]
- Wellington, E.M.; Boxall, A.B.; Cross, P.; Feil, E.J.; Gaze, W.H.; Hawkey, P.M.; Johnson-Rollings, A.S.; Jones, D.L.; Lee, N.M.; Otten, W. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect. Dis. 2013, 13, 155–165. [Google Scholar] [CrossRef]
- Liakopoulos, A.; Mevius, D.; Ceccarelli, D. A review of SHV extended-spectrum β-lactamases: Neglected yet ubiquitous. Front. Microbiol. 2016, 7, 1374. [Google Scholar] [CrossRef]
- Rolland, R.; Hausfater, G.; Marshall, B.; Levy, S. Antibiotic-resistant bacteria in wild primates: Increased prevalence in baboons feeding on human refuse. Appl. Environ. Microbiol. 1985, 49, 791–794. [Google Scholar] [CrossRef] [Green Version]
- Sherley, M.; Gordon, D.M.; Collignon, P.J. Variations in antibiotic resistance profile in Enterobacteriaceae isolated from wild Australian mammals. Environ. Microbiol. 2000, 2, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Vlieghe, E.; Phoba, M.; Tamfun, J.M.; Jacobs, J. Antibiotic resistance among bacterial pathogens in Central Africa: A review of the published literature between 1955 and 2008. Int. J. Antimicrob. Agents 2009, 34, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacopello, C.; Foti, M.; Mascetti, A.; Grosso, F.; Ricciardi, D.; Fisichella, V.; Piccolo, F.L. Antimicrobial resistance patterns of Enterobacteriaceaein European wild bird species admitted in a wildlife rescue centre. Vet. Ital. 2016, 52, 139–144. [Google Scholar] [PubMed]
- Aarestrup, F.M.; Wegener, H.C. The effects of antibiotic usage in food animals on the development of antimicrobial resistance of importance for humans in Campylobacter and Escherichia coli. Microbes Infect. 1999, 1, 639–644. [Google Scholar] [CrossRef]
- Dhama, K.; Rajagunalan, S.; Chakraborty, S.; Verma, A.; Kumar, A.; Tiwari, R.; Kapoor, S. Food-borne pathogens of animal origin-diagnosis, prevention, control and their zoonotic significance: A review. Pak. J. Biol. Sci. 2013, 16, 1076. [Google Scholar] [CrossRef] [Green Version]
- Heredia, N.; García, S. Animals as sources of food-borne pathogens: A review. Anim. Nutr. 2018, 4, 250–255. [Google Scholar] [CrossRef]
- Nowak, K. African fruit bats as potential reservoir for zoonotic pathogens-the example of Escherichia coli. Public Health 2018. [Google Scholar] [CrossRef]
- Biyela, P.; Lin, J.; Bezuidenhout, C. The role of aquatic ecosystems as reservoirs of antibiotic resistant bacteria and antibiotic resistance genes. Water Sci. Technol. 2004, 50, 45–50. [Google Scholar] [CrossRef]
- Korine, C.; Adams, R.; Russo, D.; Fisher-Phelps, M.; Jacobs, D. Bats and water: Anthropogenic alterations threaten global bat populations. In Bats in the Anthropocene: Conservation of Bats in a Changing World; Springer: Cham, Switzerland, 2016; pp. 215–241. [Google Scholar]
- Guenther, S.; Aschenbrenner, K.; Stamm, I.; Bethe, A.; Semmler, T.; Stubbe, A.; Stubbe, M.; Batsajkhan, N.; Glupczynski, Y.; Wieler, L.H. Comparable high rates of extended-spectrum-beta-lactamase-producing Escherichia coli in birds of prey from Germany and Mongolia. PLoS ONE 2012, 7, e53039. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, E.; Hong, P.-Y.; Bedon, L.C.; Mackie, R.I. Carriage of antibiotic-resistant enteric bacteria varies among sites in Galapagos reptiles. J. Wildl. Dis. 2012, 48, 56–67. [Google Scholar] [CrossRef]
- Islam, M.N. Prevalence and Antibiogram of E. coli and Salmonella spp. Isolates in Small Fruits Bat (Rousettus leschenaulti) and Associated Public Health Risk in Bangladesh. Ph.D. Thesis, Chittagong Veterinary and Animal Sciences University, Chittagong, Bangladesh, 2014. [Google Scholar]
- Campbell, S. So long as it’s near water: Variable roosting behaviour of the large-footed myotis (Myotis macropus). Aust. J. Zool. 2009, 57, 89–98. [Google Scholar] [CrossRef]
- Ciechanowski, M. Community structure and activity of bats (Chiroptera) over different water bodies. Mamm. Biol. Z. Säugetierkund. 2002, 67, 276–285. [Google Scholar] [CrossRef]
- Grindal, S.; Morissette, J.; Brigham, R. Concentration of bat activity in riparian habitats over an elevational gradient. Can. J. Zool. 1999, 77, 972–977. [Google Scholar] [CrossRef]
- Vaughan, N.; Jones, G.; Harris, S. Habitat use by bats (Chiroptera) assessed by means of a broad-band acoustic method. J. Appl. Ecol. 1997, 34, 716–730. [Google Scholar] [CrossRef]
- Jackrel, S.L.; Matlack, R.S. Influence of surface area, water level and adjacent vegetation on bat use of artificial water sources. Am. Midl. Nat. 2010, 164, 74–80. [Google Scholar] [CrossRef]
- Sirami, C.; Jacobs, D.S.; Cumming, G.S. Artificial wetlands and surrounding habitats provide important foraging habitat for bats in agricultural landscapes in the Western Cape, South Africa. Biol. Conserv. 2013, 164, 30–38. [Google Scholar] [CrossRef]
- Amos, G.; Hawkey, P.; Gaze, W.; Wellington, E. Waste water effluent contributes to the dissemination of CTX-M-15 in the natural environment. J. Antimicrob. Chemother. 2014, 69, 1785–1791. [Google Scholar] [CrossRef]
- Conte, D.; Palmeiro, J.K.; da Silva Nogueira, K.; de Lima, T.M.R.; Cardoso, M.A.; Pontarolo, R.; Pontes, F.L.D.; Dalla-Costa, L.M. Characterization of CTX-M enzymes, quinolone resistance determinants, and antimicrobial residues from hospital sewage, wastewater treatment plant, and river water. Ecotoxicol. Environ. Saf. 2017, 136, 62–69. [Google Scholar] [CrossRef]
- Moremi, N.; Manda, E.V.; Falgenhauer, L.; Ghosh, H.; Imirzalioglu, C.; Matee, M.; Chakraborty, T.; Mshana, S.E. Predominance of CTX-M-15 among ESBL producers from environment and fish gut from the shores of Lake Victoria in Mwanza, Tanzania. Front. Microbiol. 2016, 7, 1862. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.K.; Racey, P.A. Bats as bushmeat in Madagascar. Madag. Conserv. Dev. 2008, 3. [Google Scholar] [CrossRef] [Green Version]
- Kuzmin, I.V.; Bozick, B.; Guagliardo, S.A.; Kunkel, R.; Shak, J.R.; Tong, S.; Rupprecht, C.E. Bats, emerging infectious diseases, and the rabies paradigm revisited. Emerg. Health Threat. J. 2011, 4, 7159. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Primers | Sequences | Basic Pair Length | Hybridization Temperature | References |
|---|---|---|---|---|
| SHV-F | 5′-GATGAACGCTTTCCCATGATG-3′ | 214 bp | 59 °C | [36] |
| SHV-R | 5′-CGCTGTTATCGCTCATGGTAA-3′ | |||
| TEM-F | 5′-AGTGCTGCCATAACCATGAGTG-3′ | 550 bp | 63 °C | [36] |
| TEM-R | 5′-CTGACTCCCCGTCGTGTAGATG-3′ | |||
| CTX UNIV-F | 5′-TCTTCCAGAATAAGGAATCCC-3′ | 909 bp | 57 °C | [36] |
| CTX UNIV-R | 5′-CCGTTTCCGCTATTACAAAC-3′ |
| Enterobacteria Strains | Isolates n = 29 | ESBL Detected n (%) |
|---|---|---|
| Citrobacter freundii | 1 (3.45) | 0 |
| Enterobacter aerogenes | 3 (10.34) | 0 |
| Enterobacter cloacae | 2 (6.89) | 1 (3.45) |
| Enterobacter hormaechei | 1 (3.45) | 0 |
| Escherichia coli | 11 (37.93) | 6 (20.69) |
| Ewingella americana | 1 (3.45) | 0 |
| Klebsiella pneumoniae | 5 (17.24) | 4 (13.79) |
| Morganella morganii | 1 (3.45) | 0 |
| Pantoea sp. | 1 (3.45) | 0 |
| Proteus vulgaris | 1 (3.45) | 0 |
| Serratia plymuthica | 2 (6.89) | 0 |
| Antibiotic Agent | Number and Percentage (%) of ESBL-Producing Enterobacteria Strains by Species | |||
|---|---|---|---|---|
| E. cloacae (n = 1) | E. coli (n = 6) | K. pneumoniae (n = 4) | Total (n = 11) | |
| Amoxicillin | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Ampicillin | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Amoxicillin/clavulanic acid | 1 (100) | 1 (16.16) | 4 (100) | 6 (54.54) |
| Ticarcillin | 1 (100) | 6(100) | 4 (100) | 11 (100) |
| Ticarcillin/clavulanic acid | 1 (100) | 5 (83.33) | 4 (100) | 10 (90.90) |
| Piperacillin | 1 (100) | 5 (83.33) | 4 (100) | 10 (90.90) |
| Piperacillin/tazobactam | 1 (100) | 0 | 2 (50) | 3 (27.27) |
| Cephalexin | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Cefoxitin | 1 (100) | 2 (33.33) | 3 (75) | 6 (54.54) |
| Cefotaxime | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Cefpodoxime | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Ceftazidime | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Cefepime | 1 (100) | 4 (66.67) | 4 (100) | 9 (81.81) |
| Aztreonam | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Imipenem | 0 | 0 | 0 | 0 |
| Ertapenem | 1 (100) | 2 (33.33) | 1 (25) | 4 (36.36) |
| Amikacin | 1 (100) | 2 (33.33) | 0 | 3 (27.27) |
| Gentamycin | 1 (100) | 2 (33.33) | 4 (80) | 7 (63.63) |
| Kanamycin | 1 (100) | 4 (66.67) | 4 (100) | 9 (81.81) |
| Netilmicin | 0 | 2 (33.33) | 2 (50) | 4 (36.36) |
| Streptomycin | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Tobramycin | 1 (100) | 3 (50) | 3 (60) | 7 (63.63) |
| Erythromycin | 1 (100) | 6 (100) | 4 (100) | 11 (100) |
| Fosfomycin | 1 (100) | 1 (16.16) | 2 (40) | 4 (36.36) |
| Tetracycline | 0 | 5 (83.33) | 4 (100) | 9 (81.81) |
| Colistin | 1 (100) | 1 (9.09) | 4 (80) | 6 (54.54) |
| Trimethoprim/sulfamethoxazole | 0 | 4 (66.67) | 4 (100) | 8 (72.72) |
| Chloramphenicol | 0 | 0 | 0 | 0 |
| Nalidixic acid | 1 (100) | 2 (33.33) | 4 (100) | 7 (63.63) |
| Ciprofloxacin | 1 (100) | 5 (83.33) | 4 (100) | 10 (90.90) |
| Ofloxacin | 0 | 3 (50) | 3 (60) | 6 (54.54) |
| Levofloxacin | 0 | 2 (33.33) | 3 (60) | 5 (45.45) |
| Nitrofurantoin | 0 | 0 | 2 (50) | 2 (18.18) |
| Colony | Species of Bat | Bacterial Strain | Profiles of ESBL-Producing Enterobacteriae | ESBL Gene |
|---|---|---|---|---|
| CH 82 (1) | Epomops franqueti | E. coli | AX-TIC-PRL-CL-CTX-CAZ-CPD-ATM-AMP-TE-STR-SXT-ERY | blaCTX-M-15 |
| CH 71 (1) | E. franqueti | E. coli | AX-TIC-TIM-PRL-CL-CTX-CAZ-CPD-ATM-AMP-ERT-CIP-OFX-STR-ERY-SXT-TE | blaCTX-M-15 |
| CH 41 (1) | E. franqueti | E. coli | AX-TIC-TIM-PRL-CL-FOX-CTX-CAZ-CPD-FEP-ATM-AMP-CIP-KAN-CT-E-STR-TE | blaCTX-M-15 |
| CH 42 (1) | Megaloglossus woermanni | E. coli | AX-TIC-TIM-PRL-CL-CTX-CAZ-CPD-FEP-ATM-AMP-CIP-OFX-LEV-AK-CN-KAN-STR-ERY-TOB-SXT-TE | blaCTX-M-15 |
| CH 18 (3) | M. woermanni | E. coli | AX-TIC-TIM-CL-FOX-CTX-CAZ-CPD-FEP-ATM-AMP-ERT-NA-CIP-AK-CN-KAN-NET-STR-E-TOB-CT-FOS | blaCTX-M-15 |
| CH 41 (2) | M. woermanni | E. coli | AX-AMC-TIC-TIM-PRL-CL-CTX-CAZ-CPD-FEP-ATM-AMP-NA-CIP-OFX-LEV-KAN-NET-STR-ERY-TOB-SXT-TE | blaCTX-M-15 |
| CH 8 (2) | E. franqueti | E. cloacae | AX-AMC-TIC-TIM-PRL-TPZ-CL-FOX-CTX-CAZ-CPD-FEP-ATM-ERT-NA-CIP-AK-CN-KAN-STR-TOB-CT-FOS | blaCTX-M-15 |
| CH 17 (2) | E. franqueti | K. pneumoniae | AX-AMC-TIC-TIM-PRL-CTX-CAZ-CPD-FEP-ATM-CIP-OFX-KAN-CT | blaCTX-M-15 |
| CH 43 (1) | E. franqueti | K. pneumoniae | AX-AMC-TIC-TIM-PRL-TPZ-CL-FOX-CTX-CAZ-CPD-FEP-ATM-CIP-CN-KAN-STR-CT-FTN-SXT-TE- | blaCTX-M-15, blaSHV-11 |
| CH 42 (2) | M. woermanni | K. pneumoniae | AX-AMC-TIC-TIM-PRL-CL-FOX-CTX-CAZ-CPD-FEP-ATM-NA-CIP-CN-KAN-NET-S-TOB-CT-TE-SXT | blaCTX-M-15, blaSHV-11 |
| CH 38 (2) | E. franqueti | K. pneumoniae | AX-AMC-TIC-TIM-PRL-TPZ-CL-FOX-CTX-CAZ-CPD-FEP-ATM-ERT-CIP-OFX-LEV-TOB-CN-KAN-NET-STR-FOS-TE-SXT | blaCTX-M-15, blaSHV-11 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mbehang Nguema, P.P.; Onanga, R.; Ndong Atome, G.R.; Obague Mbeang, J.C.; Mabika Mabika, A.; Yaro, M.; Lounnas, M.; Dumont, Y.; Zohra, Z.F.; Godreuil, S.; et al. Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon. Microorganisms 2020, 8, 138. https://doi.org/10.3390/microorganisms8010138
Mbehang Nguema PP, Onanga R, Ndong Atome GR, Obague Mbeang JC, Mabika Mabika A, Yaro M, Lounnas M, Dumont Y, Zohra ZF, Godreuil S, et al. Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon. Microorganisms. 2020; 8(1):138. https://doi.org/10.3390/microorganisms8010138
Chicago/Turabian StyleMbehang Nguema, Pierre Philippe, Richard Onanga, Guy Roger Ndong Atome, Jean Constant Obague Mbeang, Arsène Mabika Mabika, Moussa Yaro, Manon Lounnas, Yann Dumont, Zaidi Fatma Zohra, Sylvain Godreuil, and et al. 2020. "Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon" Microorganisms 8, no. 1: 138. https://doi.org/10.3390/microorganisms8010138
APA StyleMbehang Nguema, P. P., Onanga, R., Ndong Atome, G. R., Obague Mbeang, J. C., Mabika Mabika, A., Yaro, M., Lounnas, M., Dumont, Y., Zohra, Z. F., Godreuil, S., & Bretagnolle, F. (2020). Characterization of ESBL-Producing Enterobacteria from Fruit Bats in an Unprotected Area of Makokou, Gabon. Microorganisms, 8(1), 138. https://doi.org/10.3390/microorganisms8010138