Enzyme Activity Profiles Produced on Wood and Straw by Four Fungi of Different Decay Strategies
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Cultivation and Sampling
2.3. Enzyme Activity Assays
2.4. Fungal-Produced Organic Acids and Culture Acidity
2.5. Statistical Analyses
3. Results
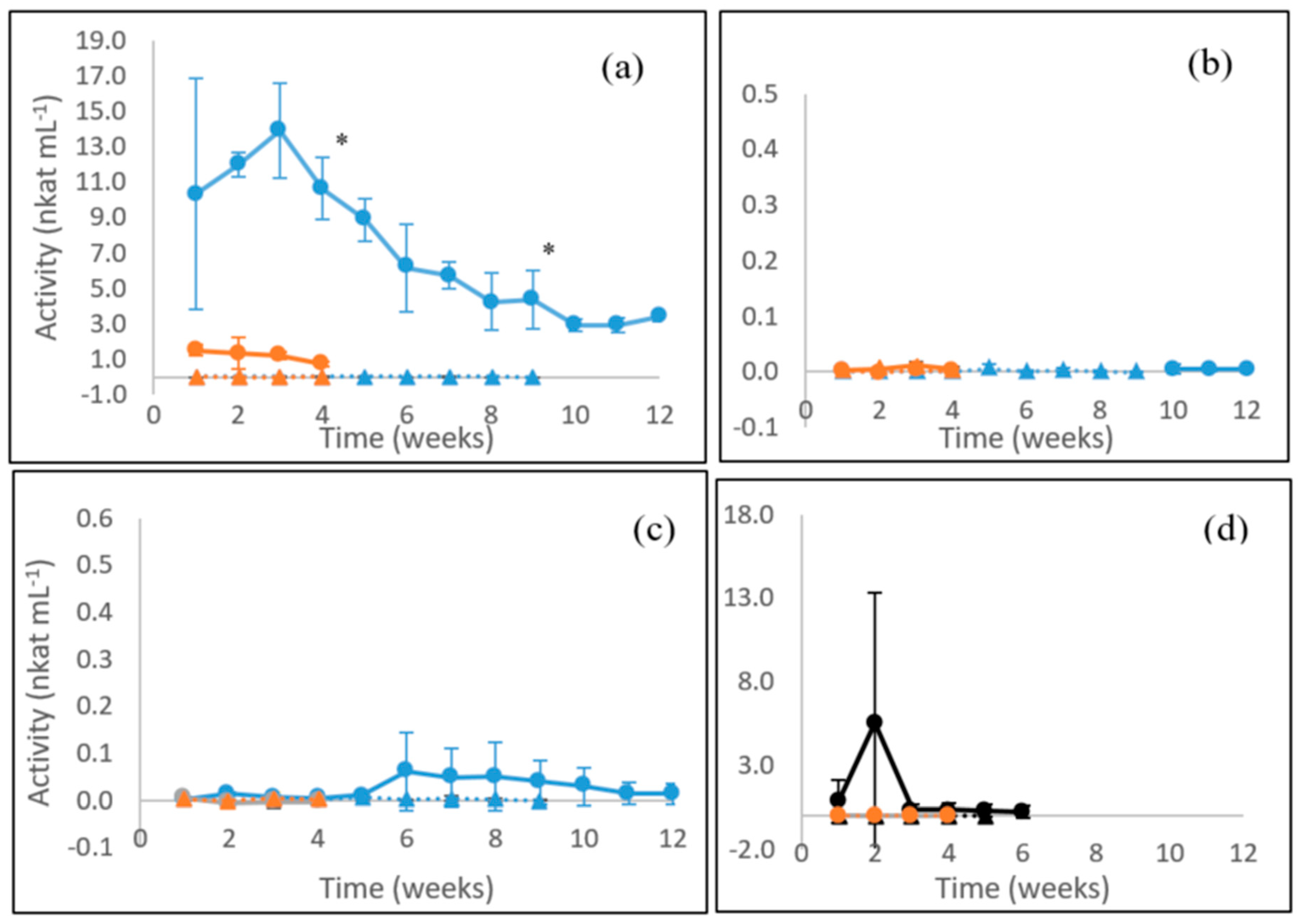
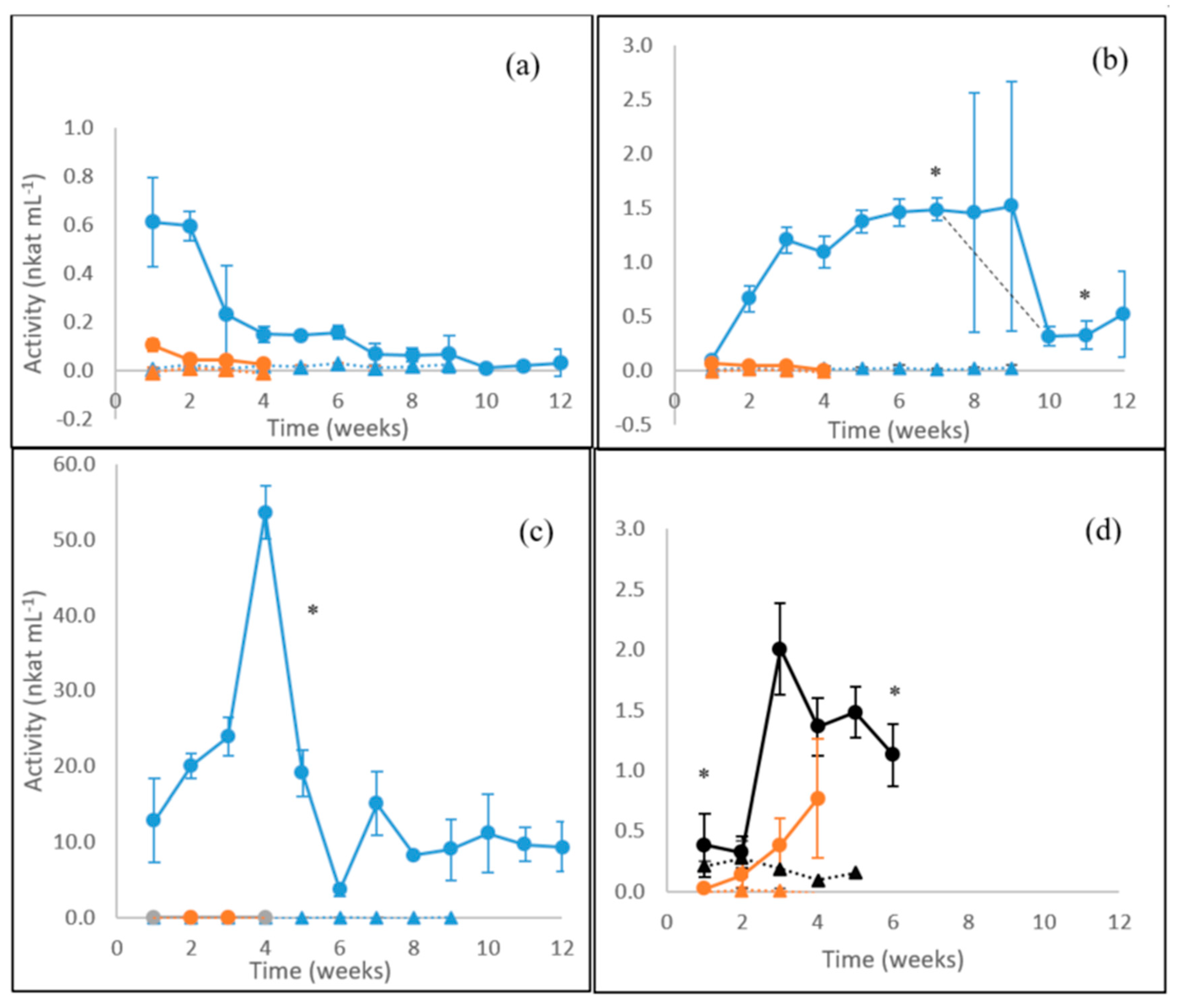
3.1. Laccase Activity
3.2. MnP Activity
3.3. β-glucosidase Activity
3.4. Endoglucanase Activity
3.5. Xylanase Activity
3.6. Summary of Enzyme Production in Fungal Cultures
3.7. Extracellular pH and Concentration of Organic Acids
4. Discussion
4.1. WR Fungus Enzyme Activity Profile
4.2. BR Fungus Enzyme Activity Profile
4.3. Intermediate “GR” Fungus Enzyme Activity Production Profile
4.4. LDF Enzyme Activity Production Profile
4.5. Extracellular pH and Production of Organic Acids
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lundell, T.K.; Mäkelä, M.R.; de Vries, R.P.; Hildén, K.S. Genomics, lifestyles and future prospects of wood-decay and litter-decomposing Basidiomycota. Adv. Bot. Res. 2014, 70, 329–370. [Google Scholar] [CrossRef]
- Hatakka, A.; Hammel, K.E. Fungal biodegradation of lignocelluloses. In The Mycota. Industrial Applications; Esser, K., Hofrichter, M., Eds.; Springer: Berlin, Germany, 2010; Volume 10, pp. 319–340. [Google Scholar]
- Ruiz-Dueñas, F.J.; Martínez, Á.T. Microbial degradation of lignin: How a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2009, 2, 164–177. [Google Scholar]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martinez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef] [PubMed]
- Lundell, T.K.; Mäkelä, M.R.; Hildén, K. Lignin-modifying enzymes in filamentous basidiomycetes-ecological, functional and phylogenetic review. J. Basic Microbiol. 2010, 50, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Dueñas, F.J.; Lundell, T.; Floudas, D.; Nagy, L.G.; Barrasa, J.M.; Hibbett, D.S.; Martínez, A.T. Lignin-degrading peroxidases in Polyporales: An evolutionary survey based on 10 sequenced genomes. Mycologia 2013, 105, 1428–1444. [Google Scholar] [CrossRef]
- Rytioja, J.; Hildén, K.; Yuzon, J.; Hatakka, A.; de Vries, R.P.; Mäkelä, M.R. Plant-polysaccharide-degrading enzymes from Basidiomycetes. Microbiol. Mol. Biol. Rev. 2014, 78, 614–649. [Google Scholar] [CrossRef]
- Riley, R.; Salamov, A.A.; Brown, D.W.; Nagy, L.G.; Floudas, D.; Held, B.W.; Levasseur, A.; Lombard, V.; Morin, E.; Otillar, R.; et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 2014, 111, 9923–9928. [Google Scholar] [CrossRef]
- Nagy, L.G.; Riley, R.; Bergmann, P.J.; Krizsán, K.; Martin, F.M.; Grigoriev, I.V.; Cullen, D.; Hibbett, D. Genetic bases of fungal white rot wood decay predicted by phylogenomic analysis of correlated gene-phenotype evolution. Mol. Biol. Evol. 2017, 34, 35–44. [Google Scholar] [CrossRef]
- Eastwood, D.C.; Floudas, D.; Binder, M.; Majcherczyk, A.; Schneider, P.; Aerts, A.; Asiegbu, F.O.; Baker, S.E.; Barry, K.; Bendiksby, M.; et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science 2011, 333, 762–765. [Google Scholar] [CrossRef]
- Floudas, D.; Held, B.W.; Riley, R.; Nagy, L.G.; Koehler, G.; Ransdell, A.S.; Younus, H.; Chow, J.; Chiniquy, J.; Lipzen, A.; et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fung Gen. Biol. 2015, 76, 78–92. [Google Scholar] [CrossRef]
- Kuuskeri, J.; Häkkinen, M.; Laine, P.; Smolander, O.P.; Tamene, F.; Miettinen, S.; Nousiainen, P.; Kemell, M.; Auvinen, P.; Lundell, T. Time-scale dynamics of proteome and transcriptome of the white-rot fungus Phlebia radiata: Growth on spruce wood and decay effect on lignocellulose. Biotechnol. Biofuel 2016, 9, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.I.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009, 37, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Hofrichter, M.; Ullrich, R.; Pecyna, M.J.; Liers, C.; Lundell, T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010, 87, 871–897. [Google Scholar] [CrossRef] [PubMed]
- Hildén, K.; Hakala, T.K.; Lundell, T. Thermotolerant and thermostable laccases. Biotechnol. Lett. 2009, 31, 1117–1128. [Google Scholar] [CrossRef] [PubMed]
- Cragg, S.M.; Beckham, G.T.; Bruce, N.C.; Bugg, T.D.H.; Distel, D.L.; Dupree, P.; Etxabe, A.; Goodell, B.; Jellison, J.; McGeehan, J.; et al. Lignocellulose degradation mechanisms across the Tree of Life. Curr. Opin. Chem. Biol. 2015, 29, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Shah, F.; Mali, T.; Lundell, T.K. Polyporales brown rot species Fomitopsis pinicola: Enzyme activity profiles, oxalic acid production, and Fe3+-reducing metabolite secretion. Appl. Environ. Microbiol. 2018, 84, e02662-17. [Google Scholar] [CrossRef]
- Morin, E.; Kohler, A.; Baker, A.; Foulongne-Oriol, M.; Lombard, V.; Nagye, L.; Ohm, R.; Patyshakuliyeva, A.; Brun, A.; Aerts, A.; et al. Genome sequence of the button mushroom Agaricus bisporus reveals mechanisms governing adaptation to a humic-rich ecological niche. Proc. Natl. Acad. Sci. USA 2012, 109, 17501–17506. [Google Scholar] [CrossRef]
- Hildén, K.; Mäkelä, M.R.; Steffen, K.T.; Hofrichter, M.; Hatakka, A.; Archer, D.B.; Lundell, T.K. Biochemical and molecular characterization of an atypical manganese peroxidase of the litter-decomposing fungus Agrocybe praecox. Fung Gen. Biol. 2014, 72, 131–136. [Google Scholar] [CrossRef]
- Ohm, R.A.; De Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; De Vries, R.; Record, E.; Levasseur, A.; Baker, S.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef]
- Stajich, J.E.; Wilke, S.K.; Ahren, D.; Au, C.H.; Birren, B.W.; Borodovsky, M.; Burns, C.; Canbäck, B.; Casselton, L.; Cheng, C.; et al. Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc. Natl. Acad. Sci. USA 2010, 107, 11889–11894. [Google Scholar] [CrossRef]
- Mali, T.; Kuuskeri, J.; Shah, F.; Lundell, T.K. Interactions affect hyphal growth and enzyme profiles in combinations of coniferous wood-decaying fungi of Agaricomycetes. PLoS ONE 2017, 12, e0185171. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.V.; Nikitin, R.; Haridas, S.; Kuo, A.; Ohm, R.; Otillar, R.; Riley, R.; Salamov, A.; Zhao, X.; Korzeniewski, F.; et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014, 42, D699–D704. [Google Scholar] [CrossRef] [PubMed]
- Kuuskeri, J.; Mäkelä, M.R.; Isotalo, J.; Oksanen, I.; Lundell, T. Lignocellulose-converting enzyme activity profiles correlate with molecular systematics and phylogeny grouping in the incoherent genus Phlebia (Polyporales, Basidiomycota). BMC Microbiol. 2015, 15, 217. [Google Scholar] [CrossRef] [PubMed]
- Mattila, H.; Kuuskeri, J.; Lundell, T. Single-step, single-organism bioethanol production and bioconversion of lignocellulose waste materials by phlebioid fungal species. Bioresour. Technol. 2017, 225, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Zhu, N.; Liu, J.; Yang, J.; Lin, Y.; Yang, Y.; Ji, L.; Li, M.; Yuan, H. Comparative analysis of the secretomes of Schizophyllum commune and other wood-decay basidiomycetes during solid-state fermentation reveals its unique lignocellulose-degrading enzyme system. Biotechnol. Biofuel 2016, 9, 1–22. [Google Scholar] [CrossRef]
- Mali, T.; Mäki, M.; Hellén, H.; Heinonsalo, J.; Bäck, J.; Lundell, T. Decomposition of spruce wood and release of volatile organic compounds depend on decay type, fungal interactions and enzyme production patterns. FEMS Microbiol. Ecol. 2019, 95, 135. [Google Scholar] [CrossRef]
- Kuuskeri, J. Genomics and Systematics of the White-Rot Fungus Phlebia radiata: Special Emphasis on Wood-Promoted Transcriptome and Proteome. Ph.D. Dissertation, University of Helsinki, Helsinki, Finland, 2016. [Google Scholar]
- Eichlerová, I.; Homolka, L.; Žifčáková, L.; Lisá, L.; Dobiášová, P.; Baldrian, P. Enzymatic systems involved in decomposition reflects the ecology and taxonomy of saprotrophic fungi. Fungal Ecol. 2015, 13, 10–22. [Google Scholar] [CrossRef]
- Valášková, V.; Baldrian, P. Degradation of cellulose and hemicelluloses by the brown rot fungus Piptoporus betulinus-Production of extracellular enzymes and characterization of the major cellulases. Microbiol. SGM 2006, 152, 3613–3622. [Google Scholar] [CrossRef]
- Martinez, D.; Challacombe, J.; Morgenstern, I.; Hibbett, D.; Schmoll, M.; Kubicek, C.P.; Ferreira, P.; Ruiz-Duenas, F.; Martinez, A.; Kersten, P.; et al. Genome, transcriptome, and secretome analysis of wood decay fungus Postia placenta supports unique mechanisms of lignocellulose conversion. Proc. Natl. Acad. Sci. USA 2009, 106, 1954–1959. [Google Scholar] [CrossRef]
- Almási, É.; Sahu, N.; Krizsán, K.; Bálint, B.; Kovács, G.; Kiss, B.; Cseklye, J.; Drula, E.; Henrissat, B.; Nagy, I.; et al. Comparative genomics reveals unique wood-decay strategies and fruiting body development in the Schizophyllaceae. New Phytol. 2019, 224, 902–915. [Google Scholar] [CrossRef]
- Kilaru, S.; Hoegger, P.J.; Kües, U. The laccase multi-gene gene family in Coprinopsis cinerea has seventeen different members that divide into two distinct families. Curr. Gen. 2006, 50, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Wu, S.; Cai, L.; Liu, X.; Wu, H.; Xin, F.; Zhang, M.; Jiang, M. Improved treatment and utilization of rice straw by Coprinopsis cinerea. Appl. Biochem. Biotechnol. 2018, 184, 616–629. [Google Scholar] [CrossRef] [PubMed]
- Nuchdang, S.; Vatanyoopaisarn, S.; Phalakornkule, C. Effectiveness of fungal treatment by Coprinopsis cinerea and Polyporus tricholoma on degradation and methane yields of lignocellulosic grass. Int. J. Biodeter. Biodegrad. 2015, 104, 38–45. [Google Scholar] [CrossRef]
- Mäkelä, M.; Galkin, S.; Hatakka, A.; Lundell, T. Production of organic acids and oxalate decarboxylase in lignin-degrading white rot fungi. Enzym. Microb. Technol. 2002, 30, 542–549. [Google Scholar] [CrossRef]
- Liaud, N.; Giniés, C.; Navarro, D.; Fabre, N.; Crapart, S.; Gimbert, I.; Levasseur, A.; Raouche, S.; Sigoillot, J. Exploring fungal biodiversity: Organic acid production by 66 strains of filamentous fungi. Fungal Biol. Biotechnol. 2014, 1, 1. [Google Scholar] [CrossRef]





| Fungal Species | Isolate Code | Decay Strategy 1 | Preferred Substrate 2 | Reference |
|---|---|---|---|---|
| Phlebia radiata | FBCC0043 | WR | D wood | [12,22] |
| Fomitopsis pinicola | FBCC1181 | BR | D&C wood | [17,22] |
| Schizophyllum commune | H4-8 | “GR” | D wood | [20] |
| Coprinopsis cinerea | AmutBmut P+B− | LDF | Grass plant LC | [21] |
| Highest Enzyme Activity (nkat mL−1) | Week | WR Phlebia radiata | Week | BR Fomitopsis pinicola | Week | “GR” Schizophyllum commune | Week | LDF * Coprinopsis cinerea |
|---|---|---|---|---|---|---|---|---|
| Laccase | 3 | 13.9 ± 2.7 | 10 | 0 1 | 6 | 0 1 | 2 | 5.5 ± 7.8 |
| MnP | 2 | 2.4 + 0.0 | NA 2 | NA 2 | 11 | 0 1 | 5 | 2.9 ± 2.5 |
| β-glucosidase | 1 | 0.6 ± 0.2 | 9 | 1.5 ± 1.1 | 4 | 53.6 ± 3.5 | 3 | 2.0 ± 0.4 |
| Endoglucase | 2 | 3.6 ± 0.4 | 3 | 5.5 ± 1.0 | 5 | 4.4 ± 1.3 | 5 | 3.2 ± 0.8 |
| Xylanase | 1 | 7.0 ± 2.9 | 5 | 42.8 ± 7.1 | 4 | 621 ± 138 | 6 | 8.9 ± 2.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veloz Villavicencio, E.; Mali, T.; Mattila, H.K.; Lundell, T. Enzyme Activity Profiles Produced on Wood and Straw by Four Fungi of Different Decay Strategies. Microorganisms 2020, 8, 73. https://doi.org/10.3390/microorganisms8010073
Veloz Villavicencio E, Mali T, Mattila HK, Lundell T. Enzyme Activity Profiles Produced on Wood and Straw by Four Fungi of Different Decay Strategies. Microorganisms. 2020; 8(1):73. https://doi.org/10.3390/microorganisms8010073
Chicago/Turabian StyleVeloz Villavicencio, Eliana, Tuulia Mali, Hans K. Mattila, and Taina Lundell. 2020. "Enzyme Activity Profiles Produced on Wood and Straw by Four Fungi of Different Decay Strategies" Microorganisms 8, no. 1: 73. https://doi.org/10.3390/microorganisms8010073
APA StyleVeloz Villavicencio, E., Mali, T., Mattila, H. K., & Lundell, T. (2020). Enzyme Activity Profiles Produced on Wood and Straw by Four Fungi of Different Decay Strategies. Microorganisms, 8(1), 73. https://doi.org/10.3390/microorganisms8010073





