On the Ability of Perfluorohexane Sulfonate (PFHxS) Bioaccumulation by Two Pseudomonas sp. Strains Isolated from PFAS-Contaminated Environmental Matrices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Microbial Enrichment Cultures
2.2. Microbial Isolation
2.3. Bacterial Strains Identification
2.4. Culture Conditions of PS27 and PFMF10 as Either Axenic Cultures (i.e., Growing or Resting Cells) or Mixed Culture
2.5. Liquid Chromatography Mass Spectrometry (LC-MS/MS)
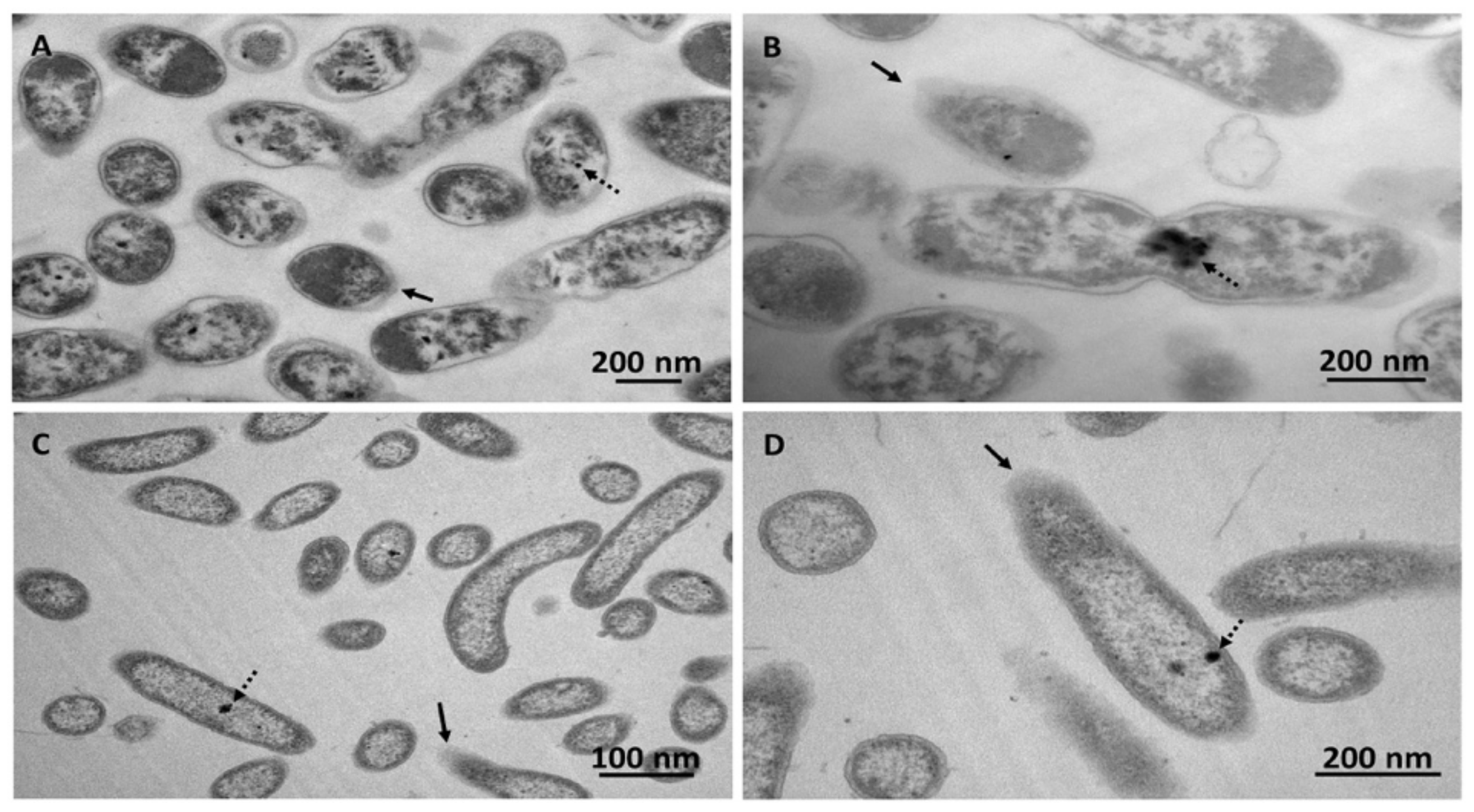
2.6. Transmission Electron Microscopy (TEM)
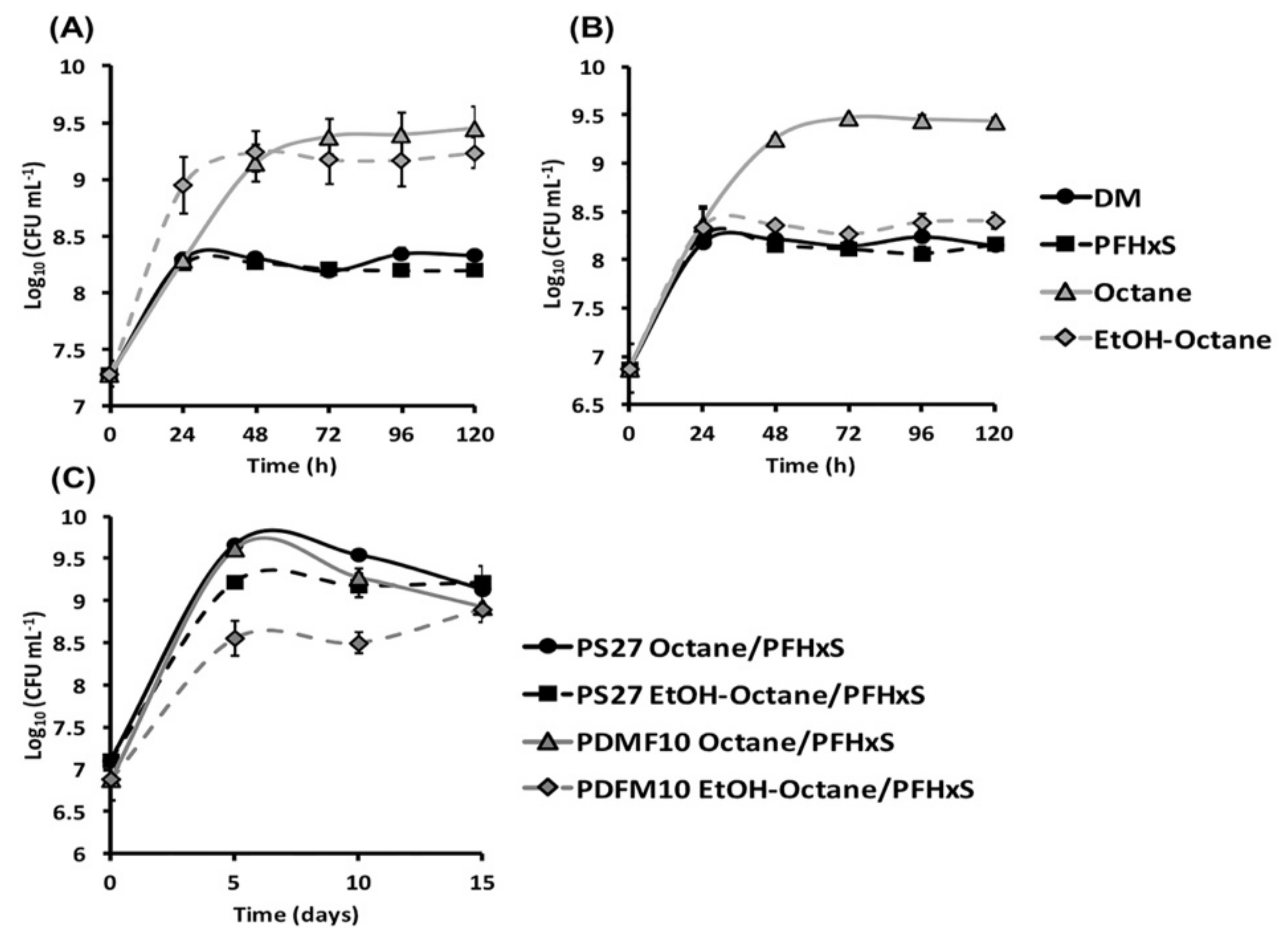
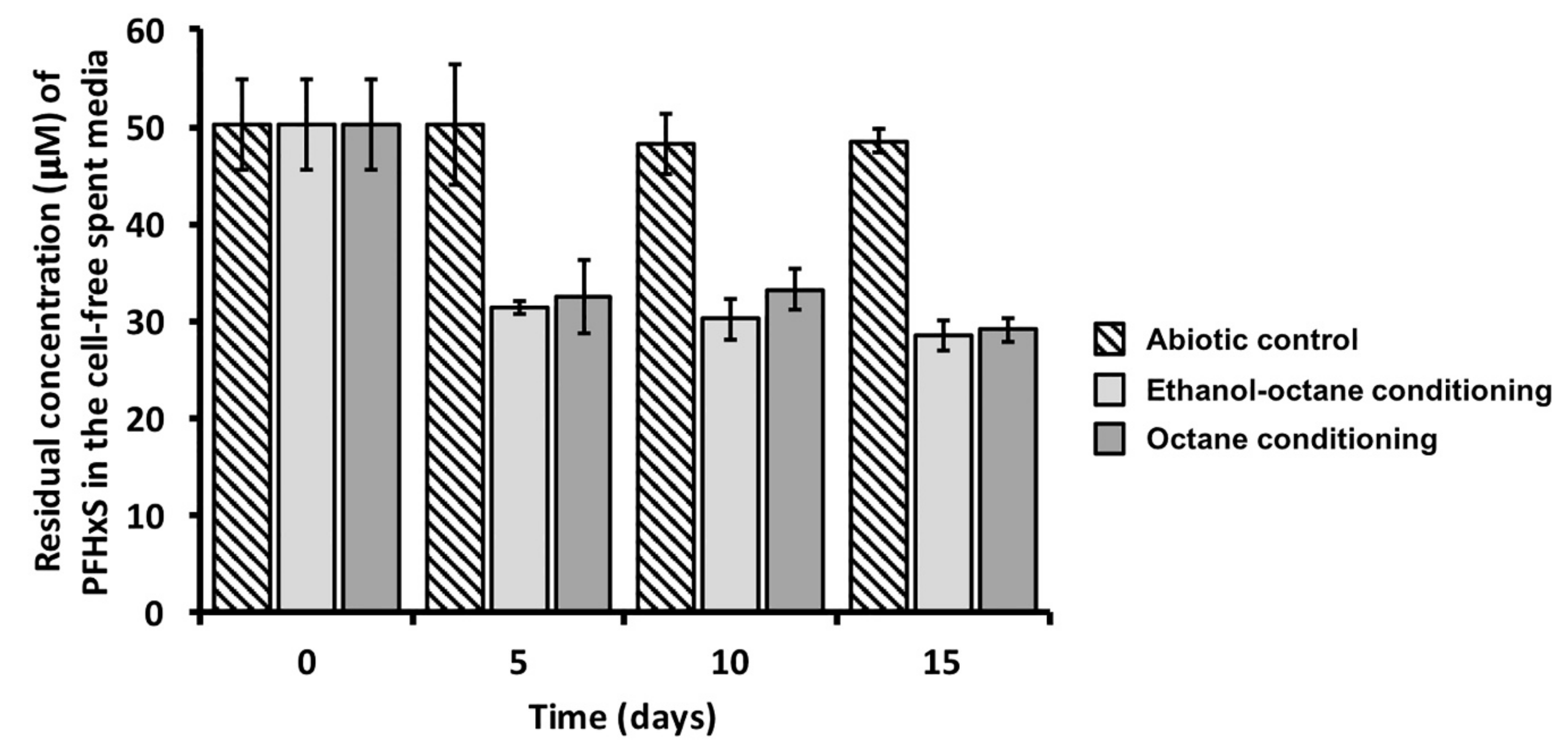
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Buck, R.C.; Franklin, J.; Berger, U.; Conder, J.M.; Cousins, I.T.; de Voogt, P.; Jensen, A.A.; Kannan, K.; Marbury, S.A.; van Leeuwen, S.P.J. Perfluoroalkyl and polyfluoroalkyl substances in the environment: Terminology, classification, and origins. IEAM 2011, 7, 513–541. [Google Scholar] [CrossRef] [PubMed]
- Kissa, E. Fluorinated Surfactants, Synthesis, Properties, Applications; Marcel Dekker, Inc.: New York, NY, USA, 1994. [Google Scholar] [CrossRef]
- Kissa, E. Fluorinated Surfactants and Repellents, 2nd ed.; Marcel Dekker, Inc.: New York, NY, USA, 2001. [Google Scholar] [CrossRef]
- Prevedouros, K.; Cousins, I.T.; Buck, R.C.; Korzeniowski, S.H. Sources, fate and transport of perfluorocarboxylates. Environ. Sci. Technol. 2006, 40, 32–44. [Google Scholar] [CrossRef] [PubMed]
- Lindstrom, A.B.; Strynar, M.J.; Libelo, E.L. Polyfluorinated compounds: Past, present, and future. Environ. Sci. Technol. 2011, 45, 7954–7961. [Google Scholar] [CrossRef] [PubMed]
- Jian, J.M.; Guo, Y.; Zeng, L.; Liu, L.-Y.; Lu, X.; Wang, F.; Zeng, E.Y. Global distribution of perfluorochemicals (PFCs) in potential human exposure source—A review. Environ. Int. 2017, 108, 51–62. [Google Scholar] [CrossRef]
- Wang, Z.; Boucher, J.M.; Scheringer, M.; Cousins, I.T.; Hungerbühler, K. Toward a comprehensive global emission inventory of C4-C10 perfluoroalkanesulfonic acids (PFSAs) and related precursors: Focus on the life cycle of C8-based products and ongoing industrial transition. Environ. Sci. Technol. 2017, 51, 4482–4493. [Google Scholar] [CrossRef]
- Wang, Z.; DeWitt, J.C.; Higgins, C.P.; Cousins, I.T. A never-ending story of perand polyfluoroalkyl substances (PFASs)? Environ. Sci. Technol. 2017, 51, 2508–2518. [Google Scholar] [CrossRef]
- Giesy, J.P.; Kannan, K. Peer Reviewed: Perfluorochemical Surfactants in the Environment. Environ. Sci. Technol. 2002, 36, 146A–152A. [Google Scholar] [CrossRef] [Green Version]
- Hansen, K.J.; Clemen, L.A.; Ellefson, M.E.; Johnson, H.O. Compound-Specific, Quantitative Characterization of Organic Fluorochemicals in Biological Matrices. Environ. Sci. Technol. 2001, 35, 766–770. [Google Scholar] [CrossRef]
- Kannan, K. Perfluoroalkyl and polyfluoroalkyl substances: Current and future perspectives. Environ. Chem. 2011, 8, 333–338. [Google Scholar] [CrossRef] [Green Version]
- United Nations Environment Programme. Stockholm Convention on Persistent Organic Pollutants; United Nations Environment Programm: Nairobi, Kenya, 2014. [Google Scholar]
- Kabore, H.A.; Vo Duy, S.; Munoz, G.; Meite, L.; Desrosiers, M.; Liu, J.; Sory, T.K.; Sauve, S. Worldwide drinking water occurrence and levels of newly identified perfluoroalkyl and polyfluoroalkyl substances. Sci. Total. Environ. 2018, 616, 1089–1100. [Google Scholar] [CrossRef]
- Ateia, M.; Maroli, A.; Tharayil, N.; Karanfil, T. The overlooked short- and ultrashort-chain poly- and perfluorinated substances: A review. Chemosphere 2019, 220, 866–882. [Google Scholar] [CrossRef]
- Pawel, R.; Nobuyoshi, Y.; Iris Man Ka, S.; Sachi, T. Perfluorinated compounds in streams of the shihwa industrial zone and Lake Shihwa, South Korea. Environ. Toxicol. Chem. 2006, 25, 2374–2380. [Google Scholar] [CrossRef]
- Olsen, G.W.; Chang, S.C.; Noker, P.E.; Gorman, G.S.; Ehresman, D.J.; Lieder, P.H.; Butenhoff, J.L. A comparison of the pharmacokinetics of perfluorobutanesulfonate (PFBS) in rats, monkeys, and humans. Toxicology 2009, 256, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Barmentlo, S.H.; Stel, J.M.; van Doorn, M.; Eschauzier, C.; de Voogt, P.; Kraak, M.H.S. Acute and chronic toxicity of short chained perfluoroalkyl substances to Daphnia magna. Environ. Pollut. 2015, 198, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cousins, I.T.; Scheringer, M.; Hungerbuehler, K. Hazard assessment of fluorinated alternatives to long-chain perfluoroalkyl acids (PFAAs) and their precursors: Status quo, ongoing challenges and possible solutions. Environ. Int. 2015, 75, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wang, Q.; Shan, G.; Zhu, L.; Yang, L.; Liu, M. Occurrence, partitioning and bioaccumulation of emerging and legacy per- and polyfluoroalkyl substances in Taihu Lake, China. Sci. Total. Environ. 2018, 5, 251–259. [Google Scholar] [CrossRef]
- Ma, X.; Shan, G.; Chen, M.; Zhao, J.; Zhu, L. Riverine inputs and source tracing of perfluoroalkyl substances (PFASs) in Taihu Lake, China. Sci. Total Environ. 2018, 15, 18–25. [Google Scholar] [CrossRef]
- ECHA. Member State Committee Support Document for the Identification of Perfluorohexane-1-Sulphonic Acid and Its Salts as Substances of Very High Concern Because of Their Vpvb (Article 57 E) Properties. 2017. Available online: https://echa.europa.eu/documents/10162/40a82ea7-dcd2-5e6f-9bff-6504c7a226c5) (accessed on 12 September 2019).
- Valsecchi, S.; Rusconi, M.; Mazzoni, M.; Viviano, G.; Pagnotta, R.; Zaghi, C.; Serrini, G.; Polesello, S. Occurrence and sources of perfluoroalkyl acids in Italian river basins. Chemosphere 2015, 129, 126–134. [Google Scholar] [CrossRef]
- Frassinetti, S.; Setti, L.; Corti, A.; Farrinelli, P.; Montevecchi, P.; Vallini, G. Biodegradation of dibenzothiophene by a nodulating isolate of Rhizobium meliloti. Can. J. Microbiol. 1998, 44, 289–297. [Google Scholar] [CrossRef]
- Coy, M.R.; Hoffmann, M.; Kingdom Gibbard, H.N.; Kuhns, E.H.; Pelz-Stelinski, K.S.; Stelinski, L.L. Nested-quantitative PCR approach with improved sensitivity for the detection of low titer levels of Candidatus Liberibacter asiaticus in the Asian citrus psyllid, Diaphorina citri Kuwayama. J. Microbiol. Methods 2014, 102, 15–22. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBio-Cloud: A taxonomically united database of 16s rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2016, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Piacenza, E.; Presentato, A.; Ambrosi, E.; Speghini, A.; Turner, R.J.; Vallini, G.; Lampis, S. Physical-chemical properties of biogenic selenium nanoparticles produced by Stenotrophomonas maltophilia SeITE02 and Ochrobactrum sp. MPV1. Front. Microbiol. 2018, 9, 3178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Presentato, A.; Cappelletti, M.; Sansone, A.; Ferreri, C.; Piacenza, E.; Demeter, M.C.; Crognale, S.; Petruccioli, M.; Milazzo, G.; Fedi, S.; et al. Aerobic Growth of Rhodococcus aetherivorans BCP1 Using Selected Naphthenic Acids as the Sole Carbon and Energy Sources. Front. Microbiol. 2018, 9, 672. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.A.; Marbury, S.A.; Martin, J.W.; Muir, D.C.G. Thermolysis of fluoropolymers as a potential source of halogenated organic acids in the environment. Nature 2001, 412, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Dinglasan, M.J.A.; Ye, Y.; Edwards, E.; Mabury, S. Fluorotelomer alcohol biodegradation yields poly- and perfluorinated acids. Environ. Sci. Technol. 2004, 38, 2857–2864. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Szostek, B.; Buck, R.C.; Folsom, P.W.; Sulecki, L.M.; Capka, V.; Berti, W.R.; Gannon, J.T. Fluorotelomer alcohol biodegradation-direct evidence that perfluorinated carbon chains breakdown. Environ. Sci. Technol. 2005, 39, 7516–7528. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Szostek, B.; Folsom, P. Aerobic biotransformation of 14C-labeled 8:2 telomer B alcohol by activated sludge from a domestic sewage treatment plant. Environ. Sci. Technol. 2005, 39, 531–538. [Google Scholar] [CrossRef]
- Wang, N.; Szostek, B.; Buck, R.C.; Folsom, P.W.; Sulecki, L.M.; Gannon, J.T. 8:2 Fluorotelomer alcohol aerobic soil biodegradation: Pathways, metabolites, and metabolite yields. Chemosphere 2009, 75, 1089–1096. [Google Scholar] [CrossRef]
- Key, B.D.; Howell, R.D.; Criddle, C.S. Defluorination of organofluorine sulfur compounds by Pseudomonas sp. strain D2. Environ. Sci. Technol. 1998, 32, 2283–2287. [Google Scholar] [CrossRef]
- Liu, J.; Lee, L.S.; Nies, L.F.; Nakatsu, C.H.; Turco, R.F. Biotransformation of 8:2 Fluorotelomer Alcohol in Soil and by Soil Bacteria Isolates. Environ. Sci. Technol. 2007, 41, 8024–8030. [Google Scholar] [CrossRef]
- Kim, M.H.; Wang, N.; McDonald, T.; Chu, K.H. Biodefluorination and Biotransformation of Fluorotelomer Alcohols by Two Alkane-Degrading Pseudomonas Strains. Biotechnol. Bioeng. 2012, 109, 3041–3049. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, J.; Liang, R.; Liu, J. Characterization of the Medium- and Long-Chain n-Alkanes Degrading Pseudomonas aeruginosa Strain SJTD-1 and Its Alkane Hydroxylase Genes. PLoS ONE 2014, 9, 105506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, J.; Ichikawa, Y.; Sagae, H.; Komura, I.; Kanou, H.; Yamada, K. Isolation and identification of n-butane-assimilating bacterium. Agric. Biol. Chem. 1980, 44, 1835–1840. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, J.; Perlman, D. Production of intracellular and extracellular protein from n-butane by Pseudomonas butanovora sp. nov. Adv. Appl. Microbiol. 1980, 26, 117–127. [Google Scholar] [CrossRef]
- Witholt, B.; de Smet, M.J.; Kingma, J.; van Beilen, J.B.; Kok, M.; Lageveen, R.G.; Eggink, G. Bioconversions of aliphatic compounds by Pseudomonas oleovorans in multiphase bioreactors: Background and economic potential. Trends Biotechnol. 1990, 8, 46–52. [Google Scholar] [CrossRef]
- Arp, D.J. Butane metabolism by butane-grown Pseudomonas butanovora. Microbiology 1999, 145, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Doughty, D.M.; Sayavedra-Soto, L.A.; Arp, D.J.; Bottomley, P.J. Effects of dichloroethene isomers on the induction and activity of butane monooxygenase in the alkane-oxidizing bacterium Pseudomonas butanovora. Appl. Environ. Microbiol. 2005, 71, 6054–6059. [Google Scholar] [CrossRef] [Green Version]
- Halsey, K.H.; Sayavedra-Soto, L.A.; Bottomley, P.J.; Arp, D.J. Trichloroethylene degradation by butane-oxidizing bacteria causes a spectrum of toxic effects. Appl. Microbiol. Biotechnol. 2005, 68, 794–801. [Google Scholar] [CrossRef]
- Hamamura, N.; Page, C.; Long, T.; Semprini, L.; Arp, D.J. Chloroform cometabolism by butane-grown CF8, Pseudomonas butanovora, and Mycobacterium vaccae JOB5 and methane-grown Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 1997, 63, 3607–3613. [Google Scholar] [CrossRef] [Green Version]
- Heipieper, H.J.; de Bont, J.N. Adaptation of Pseudomonas putida S12 to Ethanol and Toluene at the Level of Fatty Acid Composition of Membranes. Appl. Environ. Microbiol. 1994, 60, 4440–4444. [Google Scholar] [CrossRef] [Green Version]
- Heipieper, H.J.; Meulenbeld, G.; van Oirschot, Q.; de Bont, J.N. Effect of Environmental Factors on the trans/cis Ratio of Unsaturated Fatty Acids in Pseudomonas putida S12. Appl. Environ. Microbiol. 1996, 62, 2773–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heipieper, H.J.; de Waard, P.; van der Meer, P.; Killian, J.A.; Isken, S.; de Bont, J.A.M.; Eggink, G.; de Wolf, F. Regiospecific effect of 1-octanol on cis-trans isomerisation of unsaturated fatty acids in the solvent-tolerant strain Pseudomonas putida S12. Appl. Microbiol. Biotechnol. 2001, 57, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Heipieper, H.J.; Neumann, G.; Kabelitz, N.; Kaestner, M.; Richnow, H. Carbon isotope fractionation during cis-trans isomerization of unsaturated fatty acids in Pseudomonas putida. Appl. Microbiol. Biotechnol. 2004, 66, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Mulakhudair, A.R.; Al-Mashhadani, M.; Hanotu, J.; Zimmerman, W. Inactivation combined with cell lysis of Pseudomonas putida using a low pressure carbon dioxidemicrobubble technology. J. Chem. Technol. Biotechnol. 2017, 92, 1961–1969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frank, R.A.; Kavanagh, R.; Kent Burnison, B.; Arsenault, G.; Headley, J.V.; Peru, K.M.; Van Der Kraak, G.; Solomon, K.R. Toxicity assessment of collected fractions from an extracted naphthenic acid mixture. Chemosphere 2008, 72, 1309–1314. [Google Scholar] [CrossRef] [PubMed]
- Sikkema, J.; de Bont, J.A.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [CrossRef]
- Anderson, A.J.; Dawes, E.A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoate. Microbiol. Rev. 1990, 4, 450–472. [Google Scholar] [CrossRef] [Green Version]
- Steinbüchel, A. Polyhydroxyalkanoic acids. In Biomaterials; Byrom, D., Ed.; MacMillan: London, UK, 1991; pp. 123–213. [Google Scholar]
- Dalal, J.; Lal, B. Microbial polyhydroxyalkanoates: Current status and future prospects. In High Value Fermentation Products: Human Welfare II; Saran, S., Babu, V., Chaubey, A., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2019; pp. 351–387. [Google Scholar] [CrossRef]
- Kwon, B.G.; Lim, H.J.; Na, S.H.; Choi, B.I.; Shin, D.S.; Chung, S.Y. Biodegradation of perfluorooctanesulfonate (PFOS) as an emerging contaminant. Chemosphere 2014, 109, 221–225. [Google Scholar] [CrossRef]
- Yi, L.B.; Chai, L.Y.; Xie, Y.; Peng, Q.J.; Peng, Q.Z. Isolation, identification, and degradation performance of a PFOA-degrading strain. Genet. Mol. Res. 2016, 15, 235. [Google Scholar] [CrossRef]
- Chetverikov, S.P.; Sharipov, D.A.; Korshunova, T.Y.; Loginov, O.N. Degradation of Perfluorooctanyl Sulfonate by Strain Pseudomonas plecoglossicida 2.4-D. Appl. Biochem. Microbiol. 2017, 53, 533–538. [Google Scholar] [CrossRef]
- Huang, S.; Jaffé, P.R. Defluorination of Perfluorooctanoic Acid (PFOA) and Perfluorooctane Sulfonate (PFOS) by Acidimicrobium sp. Strain A6. Environ. Sci. Technol. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, J.R.; Sáez, M.; Dolfing, J.; de Voogt, P. Biodegradation of perfluorinated compounds. In Reviews of Environmental Contamination and Toxicology; Whitacre, D., Ed.; Springer: New York, NY, USA, 2008; pp. 53–71. [Google Scholar] [CrossRef]
- Hua, F.; Wang, H. Uptake modes of octadecane by Pseudomonas sp. DG17 and synthesis of biosurfactant. J. Appl. Microbiol. 2011, 112, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Q.; Zhang, W.L.; Liang, Y.N. Adsorption of perfluoroalkyl and polyfluoroalkyl substances (PFASs) from aqueous solution—A review. Sci. Total Environ. 2019, 694, 133606. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Presentato, A.; Lampis, S.; Vantini, A.; Manea, F.; Daprà, F.; Zuccoli, S.; Vallini, G. On the Ability of Perfluorohexane Sulfonate (PFHxS) Bioaccumulation by Two Pseudomonas sp. Strains Isolated from PFAS-Contaminated Environmental Matrices. Microorganisms 2020, 8, 92. https://doi.org/10.3390/microorganisms8010092
Presentato A, Lampis S, Vantini A, Manea F, Daprà F, Zuccoli S, Vallini G. On the Ability of Perfluorohexane Sulfonate (PFHxS) Bioaccumulation by Two Pseudomonas sp. Strains Isolated from PFAS-Contaminated Environmental Matrices. Microorganisms. 2020; 8(1):92. https://doi.org/10.3390/microorganisms8010092
Chicago/Turabian StylePresentato, Alessandro, Silvia Lampis, Andrea Vantini, Flavio Manea, Francesca Daprà, Stefano Zuccoli, and Giovanni Vallini. 2020. "On the Ability of Perfluorohexane Sulfonate (PFHxS) Bioaccumulation by Two Pseudomonas sp. Strains Isolated from PFAS-Contaminated Environmental Matrices" Microorganisms 8, no. 1: 92. https://doi.org/10.3390/microorganisms8010092
APA StylePresentato, A., Lampis, S., Vantini, A., Manea, F., Daprà, F., Zuccoli, S., & Vallini, G. (2020). On the Ability of Perfluorohexane Sulfonate (PFHxS) Bioaccumulation by Two Pseudomonas sp. Strains Isolated from PFAS-Contaminated Environmental Matrices. Microorganisms, 8(1), 92. https://doi.org/10.3390/microorganisms8010092






