Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Plants
2.2. Strains and Cultural Conditions
2.3. Isolation and Screening of DSF Degradation Strains
2.4. Morphological and Physio-Biochemistry Characterization of Isolated Strain HN-8
2.5. Molecular Identification and Phylogenetic Analysis
2.6. Antibiotic Sensitivity Test
2.7. DSF Degradation Capacity Test
2.8. Identification of DSF Degradation Products
2.9. Biocontrol Assay of Strain HN-8 against Xcc
3. Results
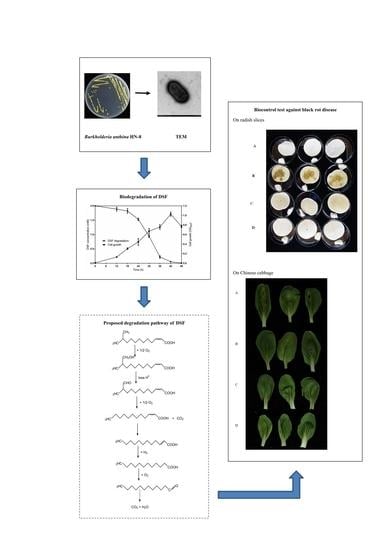
3.1. Isolation and Selection of DSF-Degrading Strain HN-8
3.2. Physio-Biochemistry and Morphological Characterization of Isolate HN-8
3.3. Molecular Identification of the Isolate HN-8
3.4. Antibiotic Sensitivity
3.5. DSF Degradation Kinetics
3.6. Degradation Products of DSF
3.7. Biocontrol Efficacy of Strain HN-8 against Black Rot Caused by Xcc
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ryan, R.P.; An, S.Q.; Allan, J.H.; Mccarthy, Y.; Dow, J.M. The DSF family of cell-cell signals: An expanding class of bacterial virulence regulators. PLoS Pathog. 2015, 11, e1004986. [Google Scholar] [CrossRef] [PubMed]
- Barber, C.E.; Tang, J.L.; Feng, J.X.; Pan, M.Q.; Daniels, M.J. A novel regulatory system required for pathogenicity of Xanthomonas campestris is mediated by a small diffusible signal molecule. Mol. Microbiol. 1997, 24, 555–566. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Arora, N.K. Management of black rot in cabbage by rhizospheric Pseudomonas species and analysis of 2,4-diacetylphloroglucinol by qRT-PCR. Biol. Control 2012, 61, 32–39. [Google Scholar] [CrossRef]
- Williams, P.H. Black rot: A continuing threat to world crucifers. Plant Dis. 1980, 64, 736–742. [Google Scholar] [CrossRef]
- Rosseto, F.R.; Manzine, L.R.; Mario, D.O.N.; Polikarpov, I. Biophysical and biochemical studies of a major endoglucanase secreted by Xanthomonas campestris pv. campestris. Enzyme Microb. Tech. 2016, 91, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Derie, M.L.; Gabrielson, R.L. Black rot of crucifers in a cabbage seed field in Western Washington. Plant Dis. 1988, 72, 45–47. [Google Scholar] [CrossRef]
- Massomo, S.M.S.; Mabagala, R.B.; Swai, I.S.; Hockenhull, J.; Mortensen, C.N. Evaluation of varietal resistance in cabbage against the black rot pathogen, Xanthomonas campestris pv. campestris in Tanzania. Crop Prot. 2004, 23, 315–325. [Google Scholar] [CrossRef]
- Alvarez, A.M. Black Rot of Crucifers. In Mechanisms of Resistance to Plant Diseases; Slusarenko, A.J., Fraser, R.S.S., van Loon, L.C., Eds.; Springer: Dordrecht, The Netherlands, 2000. [Google Scholar]
- Islam, M.T.; Lee, B.R.; Das, P.R.; La, V.H.; Jung, H.I.; Kim, T.H. Characterization of p-coumaric acid-induced soluble and cell wall-bound phenolic metabolites in relation to disease resistance to Xanthomonas campestris pv. campestris in Chinese cabbage. Plant Physiol. Biochem. 2018, 125, 172–177. [Google Scholar] [CrossRef]
- Mandiriza, G.; Kritzinger, Q.; Aveling, T.A.S. The evaluation of plant extracts, biocontrol agents and hot water as seed treatments to control black rot of rape in south africa. Crop Prot. 2018, 114, 129–136. [Google Scholar] [CrossRef]
- Agers, Y.; Bruun, M.S.; Dalsgaard, I.; Larsen, J.L. The tetracycline resistance gene tet E is frequently occurring and present on large horizontally transferable plasmids in Aeromonas spp. from fish farms. Aquaculture 2007, 266, 47–52. [Google Scholar] [CrossRef]
- Alsan, M.; Schoemaker, L.; Eggleston, K.; Kammili, N.; Kolli, P.; Bhattacharya, J. Out-of-pocket health expenditures and antimicrobial resistance in low-income and middle-income countries: An economic analysis. Lancet Infect. Dis. 2015, 15, 1203–1210. [Google Scholar] [CrossRef]
- George, A. Antimicrobial resistance, trade, food safety and security. One Health Amst. Neth. 2018, 5, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T. Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 2018, 26, 313–328. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Ye, T.; Li, Q.; Bhatt, P.; Zhang, L.; Chen, S. Potential of a quorum quenching bacteria isolate Ochrobactrum intermedium D-2 against soft rot pathogen Pectobacterium carotovorum subsp. carotovorum. Front. Microbiol. 2020, 11, 898. [Google Scholar] [CrossRef]
- Wang, H.; Liao, L.; Chen, S.; Zhang, L. A quorum quenching bacterial isolate contains multiple substrate-inducible genes conferring degradation of diffusible signal factor. Appl. Environ. Microbiol. 2020, 86, e2930-19. [Google Scholar] [CrossRef]
- Nealson, K. Autoinduction of bacterial luciferase. Arch. Microbiol. 1977, 112, 73–79. [Google Scholar] [CrossRef]
- Fuqua, C.E.; Greenberg, P. Listening in on bacteria: Acyl-homoserine lactone signaling. Nat. Rev. Mol. Cell Biol. 2002, 3, 685–695. [Google Scholar] [CrossRef]
- Sikdar, R.; Elias, M. Quorum quenching enzymes and their effects on virulence, biofilm, and microbiomes: A review of recent advances. Expert Rev. Anti Infect. Ther. 2020, 4, 1–13. [Google Scholar] [CrossRef]
- Natrah, F.M.I.; Defoirdt, T.; Sorgeloos, P.; Bossier, P. Disruption of bacterial cell-to-cell communication by marine organisms and its relevance to aquaculture. Mar. Biotechnol. 2011, 13, 109–126. [Google Scholar] [CrossRef]
- Fong, J.; Zhang, C.; Yang, R.; Boo, Z.Z.; Tan, S.K.; Nielsen, T.E. Combination therapy strategy of quorum quenching enzyme and quorum sensing inhibitor in suppressing multiple quorum sensing pathways of P. aeruginosa. Sci. Rep. 2018, 8, 1155. [Google Scholar] [CrossRef] [PubMed]
- Givskov, M.; De Nys, R.; Manefield, M.; Gram, L.; Maximilien, R.; Eberl, L.; Molin, S.; Steinberg, P.D.; Kjelleberg, S. Eukaryotic interference with homoserine lactone-mediated prokaryotic signalling. J. Bacteriol. 1996, 178, 6618–6622. [Google Scholar] [CrossRef] [PubMed]
- Rasch, M.; Buch, C.; Austin, B.; Slierendrecht, W.J.; Ekmann, K.S.; Larsen, J.L. An inhibitor of bacterial quorum sensing reduces mortalities caused by vibriosis in rainbow trout Oncorhynchus mykiss, Walbaum. Syst. Appl. Microbiol. 2004, 27, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Defoirdt, T.; Crab, R.; Wood, T.K.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Quorum sensing-disrupting brominated furanones protect the gnotobiotic brine shrimp Artemia franciscana from pathogenic Vibrio harveyi, Vibrio campbellii, and Vibrio parahaemolyticus isolates. Appl. Environ. Microbiol. 2006, 72, 6419–6423. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.G.; Mukherjee, A.; Stacy, D.M.; Lazar, S.; Ané, J.M.; Blackwell, H.E. Interkingdom responses to bacterial quorum sensing signals regulate frequency and rate of nodulation in legume-rhizobia symbiosis. Chembiochem 2016, 17, 2199–2205. [Google Scholar] [CrossRef]
- Palmer, A.G.; Senechal, A.C.; Haire, T.C.; Mehta, N.P.; Valiquette, S.D.; Blackwell, H.E. Selection of appropriate autoinducer analogues for the modulation of quorum sensing at the host-bacterium interface. ACS Chem. Biol. 2018, 13, 3115–3122. [Google Scholar] [CrossRef] [PubMed]
- Azimi, S.; Klementiev, A.D.; Whiteley, M.; Diggle, S.P. Bacterial quorum sensing during infection. Annu. Rev. Microbiol. 2020. [Google Scholar] [CrossRef]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef]
- Corral-Lugo, A.; Daddaoua, A.; Ortega, A.; Espinosa-Urgel, M.; Krell, T. Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 2016, 10, 1126. [Google Scholar] [CrossRef]
- Kim, C.; Kim, J.; Park, H.Y.; Mclean, R.J.C.; Kim, C.K.; Jeon, J.; Yi, S.S.; Kim, Y.G.; Lee, Y.S.; Yoon, J. Molecular modeling, synthesis, and screening of new bacterial quorum sensing antagonists. J. Microbiol. Biotechnol. 2007, 17, 1598–1606. [Google Scholar]
- Galloway, W.R.J.D.; Hodgkinson, J.T.; Bowden, S.D.; Welch, M.; Spring, D.R. Quorum sensing in gram-negative bacteria: Small-molecule modulation of AHL and Al-2 quorum sensing pathways. Chem. Rev. 2011, 111, 28–67. [Google Scholar] [CrossRef] [PubMed]
- Fetzner, S. Quorum quenching enzymes. J. Biotechnol. 2015, 201, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, L.; Cámara, M.; He, Y. The DSF Family of Quorum Sensing Signals: Diversity, Biosynthesis, and Turnover. Trends Microbiol. 2017, 25, 293–303. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Zhou, T.; Xu, X.; Zhang, W.; Fan, X.; Mishra, S.; Zhang, L.; Zhou, X.; Chen, S. Whole-genome sequencing analysis of quorum quenching bacterial strain Acinetobacter lactucae QL-1 identifies the FadY enzyme for degradation of the diffusible signal factor. Int. J. Mol. Sci. 2020, 21, 6729. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Liu, X.; Wu, J.; Lee, J.; Chen, S.; Cheng, Y.; Zhang, C.; Zhang, L. The host-plant metabolite glucose is the precursor of diffusible signal factor (DSF) family signals in Xanthomonas campestris. Appl. Environ. Microbiol. 2015, 81, 2861–2868. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, J.; Yin, W.; Li, P.; Zhou, J.; Chen, S.; He, F.; Cai, J.; Zhang, L. Diffusible signal factor family signals provide a fitness advantage to Xanthomonas campestris pv. campestris in interspecies competition. Environ. Microbiol. 2016, 18, 1534–1545. [Google Scholar] [CrossRef]
- Deng, Y.; Lim, A.; Lee, J.; Chen, S.; An, S.; Dong, Y.; Zhang, L. Diffusible signal factor (DSF) quorum sensing signal and structurally related molecules enhance the antimicrobial efficacy of antibiotics against some bacterial pathogens. BMC Microbiol. 2014, 14, 51. [Google Scholar] [CrossRef]
- Newman, K.L.; Chatterjee, S.; Ho, K.A.; Lindow, S.E. Virulence of plant pathogenic bacteria attenuated by degradation of fatty acid cell-to-cell signaling factors. Mol. Plant Microbe Interact. 2008, 21, 326–334. [Google Scholar] [CrossRef]
- Bhatt, P.; Rene, E.R.; Kumar, A.J.; Zhang, W.; Chen, S. Binding interaction of allethrin with esterase: Bioremediation potential and mechanism. Bioresour. Technol. 2020, 315, 123845. [Google Scholar] [CrossRef]
- Zhan, H.; Wang, H.; Liao, L.; Feng, Y.; Fan, X.; Zhang, L.; Chen, S. Kinetics and novel degradation pathway of permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 2018, 9, 98. [Google Scholar] [CrossRef]
- Bhatt, P.; Huang, Y.; Rene, E.R.; Kumar, A.J.; Chen, S. Mechanism of allethrin biodegradation by a newly isolated Sphingomonas trueperi strain CW3 from wastewater sludge. Bioresour. Technol. 2020, 305, 123074. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Feng, Y.; Zhan, H.; Liu, J.; Yang, F.; Zhang, K.; Zhang, L.; Chen, S. Characterization of a pyrethroid-degrading Pseudomonas fulva strain P31 and biochemical degradation pathway of D-phenothrin. Front. Microbiol. 2018, 9, 1003. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lin, Z.; Zhang, W.; Pang, S.; Bhatt, P.; Rene, E.R.; Kumar, A.J.; Chen, S. New insights into the microbial degradation of D-cyphenothrin in contaminated water/soil environments. Microorganisms 2020, 8, 473. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Zhang, W.; Pang, S.; Lin, Z.; Zhang, Y.; Huang, Y.; Bhatt, P.; Chen, S. Kinetics and new mechanism of azoxystrobin biodegradation by an Ochrobactrum anthropi strain SH14. Microorganisms 2020, 8, 625. [Google Scholar] [CrossRef] [PubMed]
- Bhatt, P.; Zhang, W.; Lin, Z.; Pang, S.; Huang, Y.; Chen, S. Biodegradation of allethrin by a novel fungus Fusarium proliferatum strain CF2 isolated from contaminated soil. Microorganisms 2020, 8, 593. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Zhou, T.; Li, Q.; Xu, X.; Fan, X.; Zhang, L.; Chen, S. Cupriavidus sp. HN-2, a novel quorum quenching bacterial isolate, is a potent biocontrol agent against Xanthomonas campestris pv. campestris. Microorganisms 2020, 8, 45. [Google Scholar] [CrossRef]
- Ye, T.; Zhou, T.; Fan, X.; Pankaj, B.; Zhang, L.; Chen, S. Acinetobacter lactucae strain QL-1, a novel quorum quenching candidate against bacterial pathogen Xanthomonas campestris pv. campestris. Front. Microbiol. 2019, 10, 2867. [Google Scholar] [CrossRef]
- Zhang, W.; Luo, Q.; Zhang, Y.; Fan, X.; Ye, T.; Mishra, S.; Bhatt, P.; Zhang, L.; Chen, S. Quorum quenching in a novel Acinetobacter sp. XN-10 bacterial strain against Pectobacterium carotovorum subsp. carotovorum. Microorganisms 2020, 8, 1100. [Google Scholar] [CrossRef]
- Tsuda, K.; Tsuji, G.; Higashiyama, M.; Ogiyama, H.; Umemura, K.; Mitomi, M.; Kubo, Y. Biological control of bacterial soft rot in Chinese cabbage by Lactobacillus plantarum strain BY under field conditions. Biol. Control 2016, 100, 63–69. [Google Scholar] [CrossRef]
- Fouhy, Y.; Scanlon, K.; Schouest, K.; Spillane, C.; Crossman, L.; Avison, M.B.; Ryan, R.P.; Dow, J.M. Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J. Bacteriol. 2007, 189, 4964–4968. [Google Scholar] [CrossRef]
- Boon, C.; Deng, Y.; Wang, L.H.; He, Y.; Xu, J.L.; Fan, Y.; Pan, S.Q.; Zhang, L.H. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008, 2, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Marques, C.N. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Dow, J.M. Communication with a growing family: Diffusible signal factor (DSF) signaling in bacteria. Trends Microbiol. 2011, 19, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Qian, G.; Yin, F.; Fan, J.; Zhai, Z.; Liu, C. Proteomic analysis of the regulatory function of DSF-dependent quorum sensing in Xanthomonas oryzae pv. oryzicola. Microb. Pathogenesis 2011, 50, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zou, L.; Ji, Z.; Cai, L.; Chen, G. Glucose 6-phosphate isomerase (Pgi) is required for extracellular polysaccharide biosynthesis, DSF signals production and full virulence of Xanthomonas oryzae pv. oryzicola in rice. Physiol. Mol. Plant. Pathol. 2017, 100, 209–219. [Google Scholar] [CrossRef]
- Rossi, B.P.; García, C.; Alcaraz, E.; Franco, M. Stenotrophomonas maltophilia interferes via the DSF-mediated quorum sensing system withCandida albicans filamentation and its planktonic and biofilm modes of growth. Rev. Argent. Microbiol. 2014, 46, 288–297. [Google Scholar]
- Yánez-Mendizábal, V.; Usall, J.; Viñas, I.; Casals, C.; Marín, S.; Solsona, C.; Teixidó, N. Potential of a new strain of Bacillus subtilis CPA-8 to control the major postharvest diseases of fruit. Biocontrol. Sci. Technol. 2011, 21, 409–426. [Google Scholar] [CrossRef]
- Ferraz, L.P.; da Cunha, T.; da Silva, A.C.; Kupper, K.C. Biocontrol ability and putative mode of action ofyeasts against Geotrichum citri-aurantii in citrus fruit. Microbiol. Res. 2016, 188, 72–79. [Google Scholar] [CrossRef]







| Characteristics | Results | Characteristics | Results |
|---|---|---|---|
| Gram staining | − | Amylolysis | − |
| Oxidase | + | Casein hydrolysis | + |
| Gelatin liquefaction | + | Arginine dihydrolase | − |
| Ornithine decarboxylase | + | d-glucose | − |
| d-lactose | − | Raffinose | − |
| Sucrose | − | Cellobiose | − |
| Arabinose | − | Rhamnose | − |
| Maltose | − | Hemolysis | − |
| O-nitrophenyl-β-d-galactoside | + | Lysine decarboxylase | + |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, T.; Zhang, W.; Feng, Z.; Fan, X.; Xu, X.; Mishra, S.; Zhang, L.; Chen, S. Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris. Microorganisms 2020, 8, 1485. https://doi.org/10.3390/microorganisms8101485
Ye T, Zhang W, Feng Z, Fan X, Xu X, Mishra S, Zhang L, Chen S. Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris. Microorganisms. 2020; 8(10):1485. https://doi.org/10.3390/microorganisms8101485
Chicago/Turabian StyleYe, Tian, Wenping Zhang, Zhixuan Feng, Xinghui Fan, Xudan Xu, Sandhya Mishra, Lianhui Zhang, and Shaohua Chen. 2020. "Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris" Microorganisms 8, no. 10: 1485. https://doi.org/10.3390/microorganisms8101485
APA StyleYe, T., Zhang, W., Feng, Z., Fan, X., Xu, X., Mishra, S., Zhang, L., & Chen, S. (2020). Characterization of a Novel Quorum-Quenching Bacterial Strain, Burkholderia anthina HN-8, and Its Biocontrol Potential against Black Rot Disease Caused by Xanthomonas campestris pv. campestris. Microorganisms, 8(10), 1485. https://doi.org/10.3390/microorganisms8101485