Longer Ubiquinone Side Chains Contribute to Enhanced Farnesol Resistance in Yeasts
Abstract
1. Introduction
2. Materials and Method
2.1. Yeast Strains and Media
2.2. Inoculum Preparation
2.3. Ubiquinone Analysis in C. albicans
2.4. Farnesol Sensitivity Assays—High Aeration
2.5. Farnesol Sensitivity Assays—Low Aeration
2.6. Measurement of Intracellular Reactive Oxygen Species (ROS) via DHR123 Staining
2.7. Measurement of Oxygen Consumption Rate in Yeasts
2.8. Salt and Oxidative Stress Assays
2.9. Catalase Activity Induced by Sub-Lethal (20 µM) Farnesol
3. Results
3.1. UQ9 is the Major Ubiquinone in C. albicans
3.2. Farnesol Resistance in S. cerevisiae Depends on the UQ Isoprenologue, UQ6 vs. UQ9
3.3. Intracellular Reactive Oxygen Species (ROS) Accumulation Decreased with UQ9
3.4. Respiratory Rate Remains Unchanged Regardless of UQ Isoprenologue
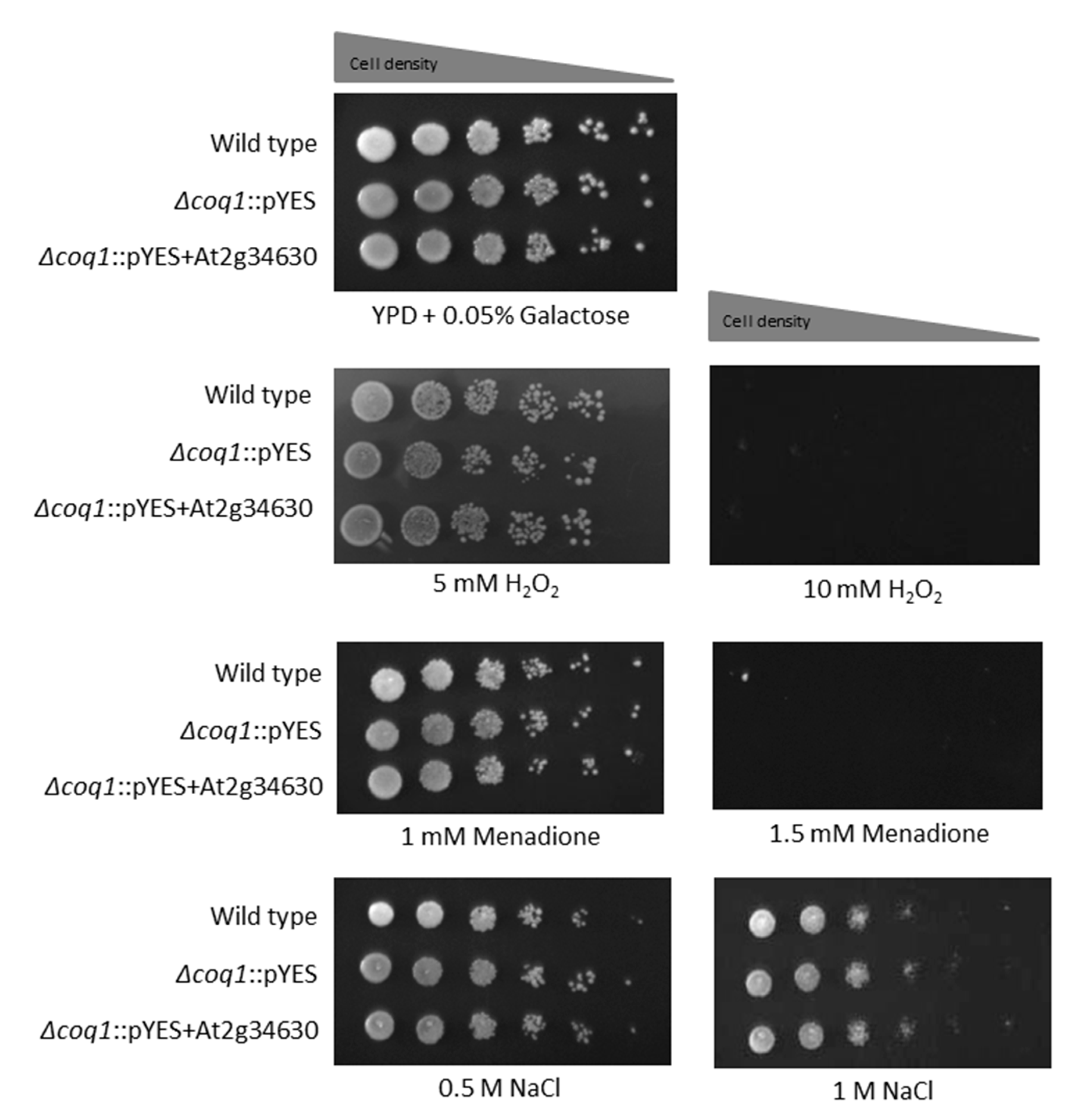
3.5. Oxidative Sensitivity and Osmotolerance in UQ6 vs. UQ9
3.6. Catalase Assays
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Magill, S.S.; Edwards, J.R.; Bamberg, W.; Beldavs, Z.G.; Dumyati, G.; Kainer, M.A.; Lynfield, R.; Maloney, M.; McAllister-Hollod, L.; Nadle, J.; et al. Multistate point-prevalence survey of health care–associated infections. New Engl. J. Med. 2014, 370, 1198–1208. [Google Scholar] [CrossRef] [PubMed]
- Hornby, J.M.; Jensen, E.C.; Lisec, A.D.; Tasto, J.J.; Jahnke, B.; Shoemaker, R.; Dussault, P.; Nickerson, K.W. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 2001, 67, 2982–2992. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, K.W.; Atkin, A.L.; Hornby, J.M. Quorum sensing in dimorphic fungi: Farnesol and beyond. Appl. Environ. Microbiol. 2006, 72, 3805–3813. [Google Scholar] [CrossRef] [PubMed]
- Navarathna, D.H.M.L.P.; Hornby, J.M.; Krishnan, N.; Parkhurst, A.; Duhamel, G.E.; Nickerson, K.W. Effect of farnesol on a mouse model of systemic Candidiasis, determined by use of a DPP3 knockout mutant of Candida albicans. Infect. Immun. 2007, 75, 1609–1618. [Google Scholar] [CrossRef]
- Hargarten, J.C.; Moore, T.C.; Petro, T.M.; Nickerson, K.W.; Atkin, A.L. Candida albicans quorum sensing molecules stimulate mouse macrophage migration. Infect. Immun. 2015, 83, 3857–3864. [Google Scholar] [CrossRef]
- Langford, M.L.; Hasim, S.; Nickerson, K.W.; Atkin, A.L. Activity and toxicity of farnesol towards Candida albicans are dependent on growth conditions. Antimicrob. Agents Chemother. 2010, 54, 940–942. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hornby, J.M.; Nickerson, K.W. Enhanced production of farnesol by Candida albicans treated with four azoles. Antimicrob. Agents Chemother. 2004, 48, 2305–2307. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Henriques, M.; Azeredo, J.; Rocha, S.M.; Coimbra, M.A.; Oliveira, R. Morphogenesis control in Candida albicans and Candida dubliniensis through signaling molecules produced by planktonic and biofilm cells. Eukaryot. Cell 2007, 6, 2429–2436. [Google Scholar] [CrossRef]
- Weber, K.; Sohr, R.; Schulz, B.; Fleischhacker, M.; Ruhnke, M. Secretion of E,E-farnesol and biofilm formation in eight different Candida species. Antimicrob. Agents Chemother. 2008, 52, 1859–1861. [Google Scholar] [CrossRef]
- Machida, K.; Tanaka, T.; Fujita, K.-I.; Taniguchi, M. Farnesol-induced generation of reactive oxygen species via indirect inhibition of the mitochondrial electron transport chain in the yeast Saccharomyces cerevisiae. J. Bacteriol. 1998, 180, 4460–4465. [Google Scholar] [CrossRef]
- Machida, K.; Tanaka, T.; Yano, Y.; Otani, S.; Taniguchi, M. Farnesol-induced growth inhibition in Saccharomyces cerevisiae by a cell cycle mechanism. Microbiology 1999, 145, 293–299. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Semighini, C.P.; Hornby, J.M.; Dumitru, R.; Nickerson, K.W.; Harris, S.D. Farnesol-induced apoptosis in Aspergillus nidulans reveals a possible mechanism for antagonistic interactions between fungi. Mol. Microbiol. 2006, 59, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Semighini, C.P.; Murray, N.; Harris, S.D. Inhibition of Fusarium graminearum growth and development by farnesol. FEMS Microbiol. Lett. 2008, 279, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Abe, S.; Tsunashima, R.; Iijima, R.; Yamada, T.; Maruyama, N.; Hisajima, T.; Abe, Y.; Oshima, H.; Yamazaki, M. Suppression of anti-Candida activity of macrophages by a quorum-sensing molecule, farnesol, through induction of oxidative stress. Microbiol. Immunol. 2009, 53, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Fairn, G.D.; Macdonald, K.; McMaster, C.R. A chemogenomic screen in Saccharomyces cerevisiae uncovers a primary role for the mitochondria in farnesol toxicity and its regulation by the Pkc1 pathway. J. Biol. Chem. 2007, 282, 4868–4874. [Google Scholar] [CrossRef] [PubMed]
- Turrens, J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003, 552, 335–344. [Google Scholar] [CrossRef]
- Wang, K.C.; Ohnuma, S.-I. Isoprenyl diphosphate synthases. BBA Mol. Cell Biol. Lipids 2000, 1529, 33–48. [Google Scholar] [CrossRef]
- Clarke, C.F. New advances in coenzyme Q biosynthesis. Protoplasma 2000, 213, 134–147. [Google Scholar] [CrossRef]
- Ashby, M.N.; Edwards, P. Elucidation of the deficiency in two yeast coenzyme Q mutants. Characterization of the structural gene encoding hexaprenyl pyrophosphate synthetase. J. Biol. Chem. 1990, 265, 13157–13164. [Google Scholar]
- Gin, P.; Clarke, C.F. Genetic evidence for a multi-subunit complex in coenzyme Q biosynthesis in yeast and the role of the Coq1 hexaprenyl diphosphate synthase. J. Biol. Chem. 2005, 280, 2676–2681. [Google Scholar] [CrossRef]
- Ducluzeau, A.-L.; Wamboldt, Y.; Elowsky, C.G.; Mackenzie, S.A.; Schuurink, R.C.; Basset, G.J.C. Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 2012, 69, 366–375. [Google Scholar] [CrossRef]
- Kurtzman, C.; Fell, J. The Yeasts—A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Suzuki, M.; Nakase, T. A phylogenetic study of ubiquinone Q-8 species of the genera Candida, Pichia, and Citeromyces based on 18S ribosomal DNA sequence divergence. J. Gen. Appl. Microbiol. 1999, 45, 239–246. [Google Scholar] [CrossRef][Green Version]
- Suzuki, M.; Nakase, T. A phylogenetic study of ubiquinone-7 species of the genus Candida based on 18S ribosomal DNA sequence divergence. J. Gen. Appl. Microbiol. 2002, 48, 55–65. [Google Scholar] [CrossRef]
- Henriques, M.; Martins, M.; Azeredo, J.; Oliveira, R. Effect of farnesol on Candida dubliniensis morphogenesis. Lett. Appl. Microbiol. 2007, 44, 199–205. [Google Scholar] [CrossRef]
- Jabra-Rizk, M.A.; Shirtliff, M.; James, C.; Meiller, T. Effect of farnesol on Candida dubliniensis biofilm formation and fluconazole resistance. FEMS Yeast Res. 2006, 6, 1063–1073. [Google Scholar] [CrossRef]
- Dumitru, R.; Hornby, J.M.; Nickerson, K.W. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 2004, 48, 2350–2354. [Google Scholar] [CrossRef]
- Sherman, F. Getting started with yeast. Methods Enzymol. 2002, 350, 3–41. [Google Scholar]
- Gillum, A.M.; Tsay, E.Y.; Kirsch, D.R. Isolation of the Candida albicans gene for orotidine-5’-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol. Gen. Genet. MGG 1984, 198, 179–182. [Google Scholar] [CrossRef] [PubMed]
- Slutsky, B.; Staebell, M.; Anderson, J.; Risen, L.; Pfaller, M.; Soll, D.R. “White-opaque transition”: A second high-frequency switching system in Candida albicans. J. Bacteriol. 1987, 169, 189–197. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Junger, W.G. Measurement of oxidative burst in neutrophils. Meth. Mol. Biol. 2012, 844, 115–124. [Google Scholar]
- Okada, K.; Kainou, T.; Matsuda, H.; Kawamukai, M. Biological significance of the side chain length of ubiquinone in Saccharomyces cerevisiae. FEBS Lett. 1998, 431, 241–244. [Google Scholar] [CrossRef]
- Schultz, J.R.; Clarke, C.F. Characterization of Saccharomyces cerevisiae ubiquinone-deficient mutants. Biofactors 1999, 9, 121–129. [Google Scholar] [CrossRef]
- Deveau, A.; Piispanen, A.E.; Jackson, A.A.; Hogan, D.A. Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras-cyclic AMP signaling pathway. Eukaryot. Cell 2010, 9, 569–577. [Google Scholar] [CrossRef]
- Nikolaou, E.; Agrafioti, I.; Stumpf, M.; Quinn, J.; Stansfield, I.; Brown, A.J.P. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol. Biol. 2009, 9, 44. [Google Scholar] [CrossRef]
- Enjalbert, B.; Smith, D.A.; Cornell, M.J.; Alam, I.; Nicholls, S.; Brown, A.J.P.; Quinn, J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Mol. Biol. Cell. 2006, 17, 1018–1032. [Google Scholar] [CrossRef]
- da Silva Dantas, A.; Day, A.; Ikeh, M.; Kos, I.; Achan, B.; Quinn, J. Oxidative stress responses in the human fungal pathogen, Candida albicans. Biomolecules 2015, 5, 142–165. [Google Scholar] [CrossRef] [PubMed]
- Herrero, E.; Ros, J.; Bellí, G.; Cabiscol, E. Redox control and oxidative stress in yeast cells. BBA Gen. Subj. 2008, 1780, 1217–1235. [Google Scholar] [CrossRef]
- Ikner, A.; Shiozaki, K. Yeast signaling pathways in the oxidative stress response. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2005, 569, 13–27. [Google Scholar] [CrossRef]
- Chen, X.J.; Clark-Walker, G.D. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 1999, 194, 197–238. [Google Scholar]
- Søballe, B.; Poole, R.K. Microbial ubiquinones: Multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 1999, 145, 1817–1830. [Google Scholar] [CrossRef]
- Kawamukai, M. Biosynthesis, bioproduction and novel roles of ubiquinone. J. Biosci. Bioeng. 2002, 94, 511–517. [Google Scholar] [CrossRef]
- Nowicka, B.; Kruk, J. Occurrence, biosynthesis and function of isoprenoid quinones. BBA Bioenerg. 2010, 1797, 1587–1605. [Google Scholar] [CrossRef] [PubMed]
- Forsmark-Andrée, P.; Lee, C.-P.; Dallner, G.; Ernster, L. Lipid peroxidation and changes in the ubiquinone content and the respiratory chain enzymes of submitochondrial particles. Free Radic. Biol. Med. 1997, 22, 391–400. [Google Scholar] [CrossRef]
- Katsikas, H.; Quinn, P. The polyisoprenoid chain length influences the interaction of ubiquinones with phospholipid bilayers. BBA Biomembr. 1982, 689, 363–369. [Google Scholar] [CrossRef]
- Sévin, D.C.; Sauer, U. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 2014, 10, 266–272. [Google Scholar] [CrossRef]
- Adamowicz, M.; Conway, T.; Nickerson, K.W. Nutritional complementation of oxidative glucose metabolism in Escherichia coli via pyrroloquinoline quinone-dependent glucose dehydrogenase and the Enter-Doudoroff pathway. Appl. Environ. Microbiol. 1991, 57, 2012–2015. [Google Scholar] [CrossRef]
- Nickerson, K.W.; Atkin, A.L. Deciphering fungal dimorphism: Farnesol’s unanswered questions. Mol. Microbiol. 2017, 103, 567–575. [Google Scholar] [CrossRef]
- Rajagopal, S.; Sudarsan, N.; Nickerson, K.W. Sodium dodecyl sulfate hypersensitivity of clpP and clpB mutants of Escherichia coli. Appl. Environ. Microbiol. 2002, 68, 4117–4121. [Google Scholar] [CrossRef][Green Version]
- Rajagopal, S.; Eis, N.; Bhattacharya, M.; Nickerson, K.W. Membrane derived oligosaccharides (MDOs) are essential for sodium dodecyl sulfate resistance in Escherichia coli. FEMS Microbiol. Lett. 2003, 223, 25–31. [Google Scholar] [CrossRef]







| Yeast Strains | Genotype Description | Source | UQ |
|---|---|---|---|
| C. albicans strains | |||
| SC5314 | Wild type (clinical isolate) | [29] | UQ9 |
| WO-1 | MTLɑ frequent white/opaque switching | [30] | UQ9 |
| S. cerevisiae strains | |||
| BY4741 parent | BY4741 MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 rho+ | [21] | UQ6 |
| ∆coq1::pYES | coq1 knockout transformed with empty vector | [21] | none |
| ∆coq1::pYES + At2g34630 | coq1 knockout transformed with Arabidopsis At2g34630 | [21] | UQ9 |
| ∆coq1::pYES + COQ1 | coq1 knockout transformed native COQ1 gene | [21] | UQ6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pathirana, R.U.; Boone, C.; Nickerson, K.W. Longer Ubiquinone Side Chains Contribute to Enhanced Farnesol Resistance in Yeasts. Microorganisms 2020, 8, 1641. https://doi.org/10.3390/microorganisms8111641
Pathirana RU, Boone C, Nickerson KW. Longer Ubiquinone Side Chains Contribute to Enhanced Farnesol Resistance in Yeasts. Microorganisms. 2020; 8(11):1641. https://doi.org/10.3390/microorganisms8111641
Chicago/Turabian StylePathirana, Ruvini U., Cory Boone, and Kenneth W. Nickerson. 2020. "Longer Ubiquinone Side Chains Contribute to Enhanced Farnesol Resistance in Yeasts" Microorganisms 8, no. 11: 1641. https://doi.org/10.3390/microorganisms8111641
APA StylePathirana, R. U., Boone, C., & Nickerson, K. W. (2020). Longer Ubiquinone Side Chains Contribute to Enhanced Farnesol Resistance in Yeasts. Microorganisms, 8(11), 1641. https://doi.org/10.3390/microorganisms8111641






