Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of DMST-H2
2.2. Acute Oral Toxicity Test
2.3. Genome Sequencing, Assembly, and Bioinformatic Analyses
2.4. Resistance to Simulated Gastrointestinal Conditions
2.5. Preparing for DMST-H2 Materials
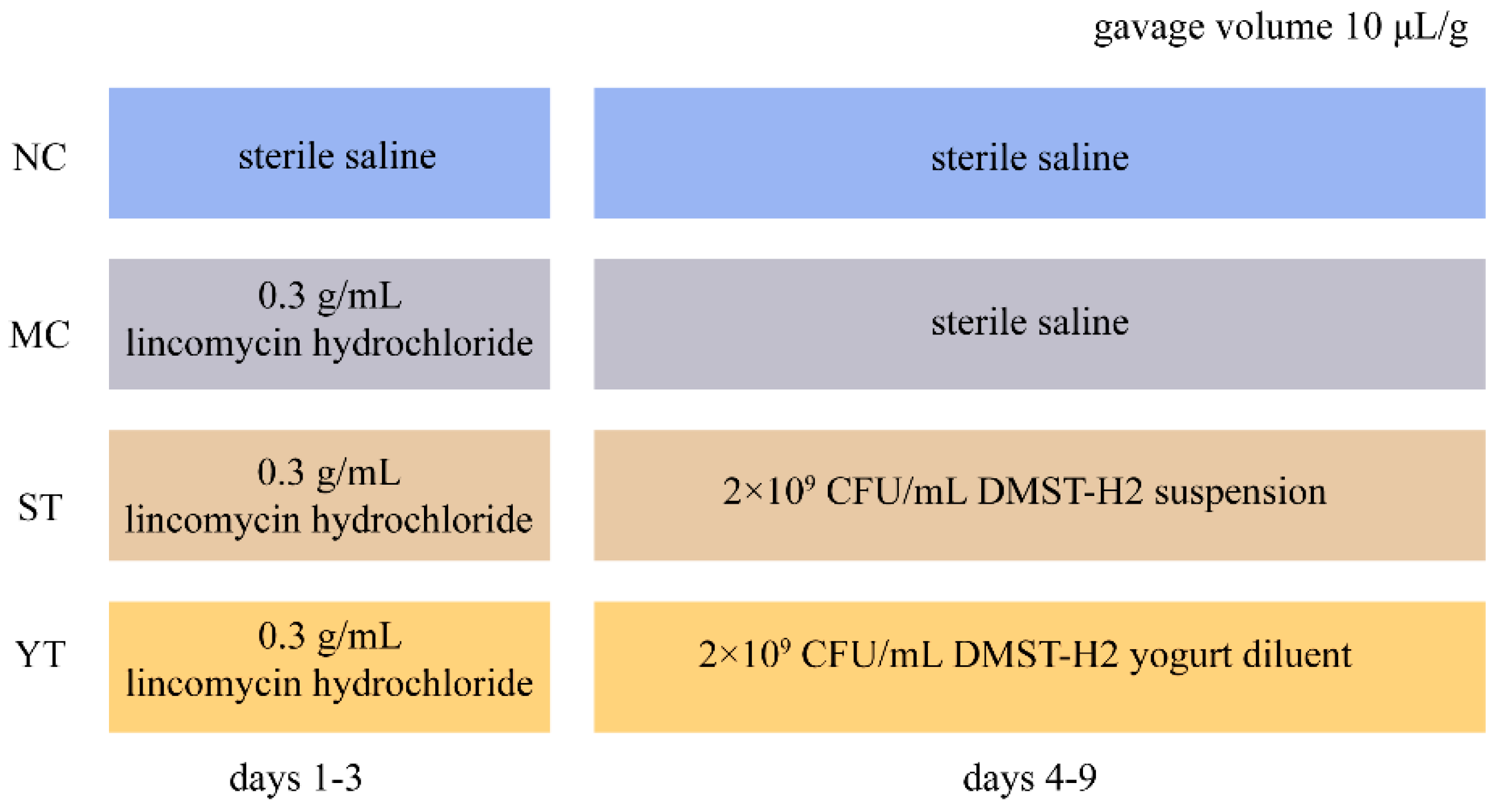
2.6. Animals and the Experiment Design
2.7. Diarrhea Measurement
2.8. Feces Bacterial Culture
2.9. Organ Index and Histological Observation
2.10. Enzyme-Linked Immunosorbent Assays of Serum
2.11. Preparation of Total DNA and High Throughput Sequencing Analysis
2.12. Bioinformatics and Statistical Analysis
3. Results
3.1. Identification and General Genome Features of DMST-H2
3.2. Safety Evaluation of DMST-H2
3.3. Probiotic Potential of DMST-H2
3.4. DMST-H2 Reduces AAD-Related Symptoms
3.5. DMST-H2 Improved AAD-Related Inflammatory Reaction and Tissue Damage
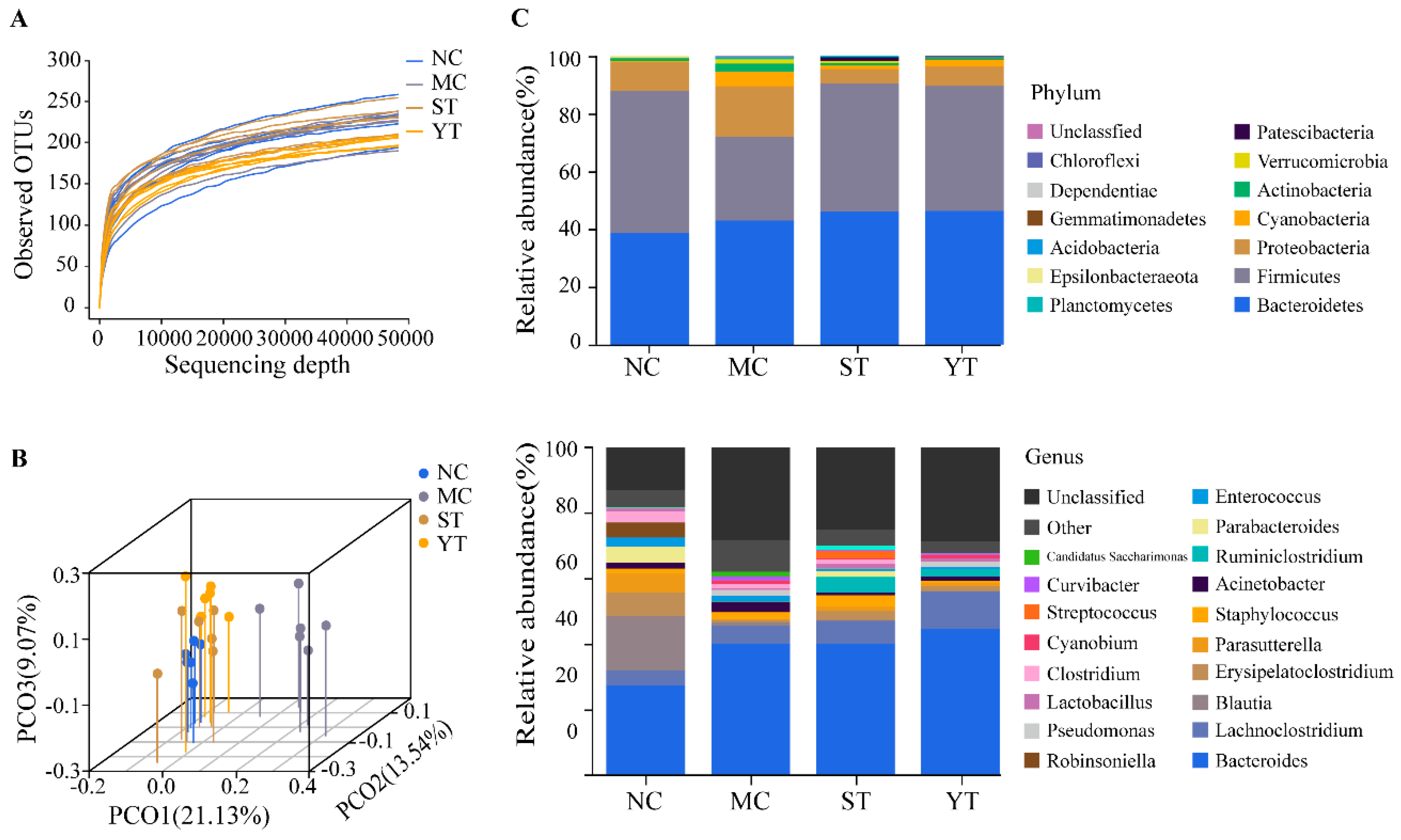
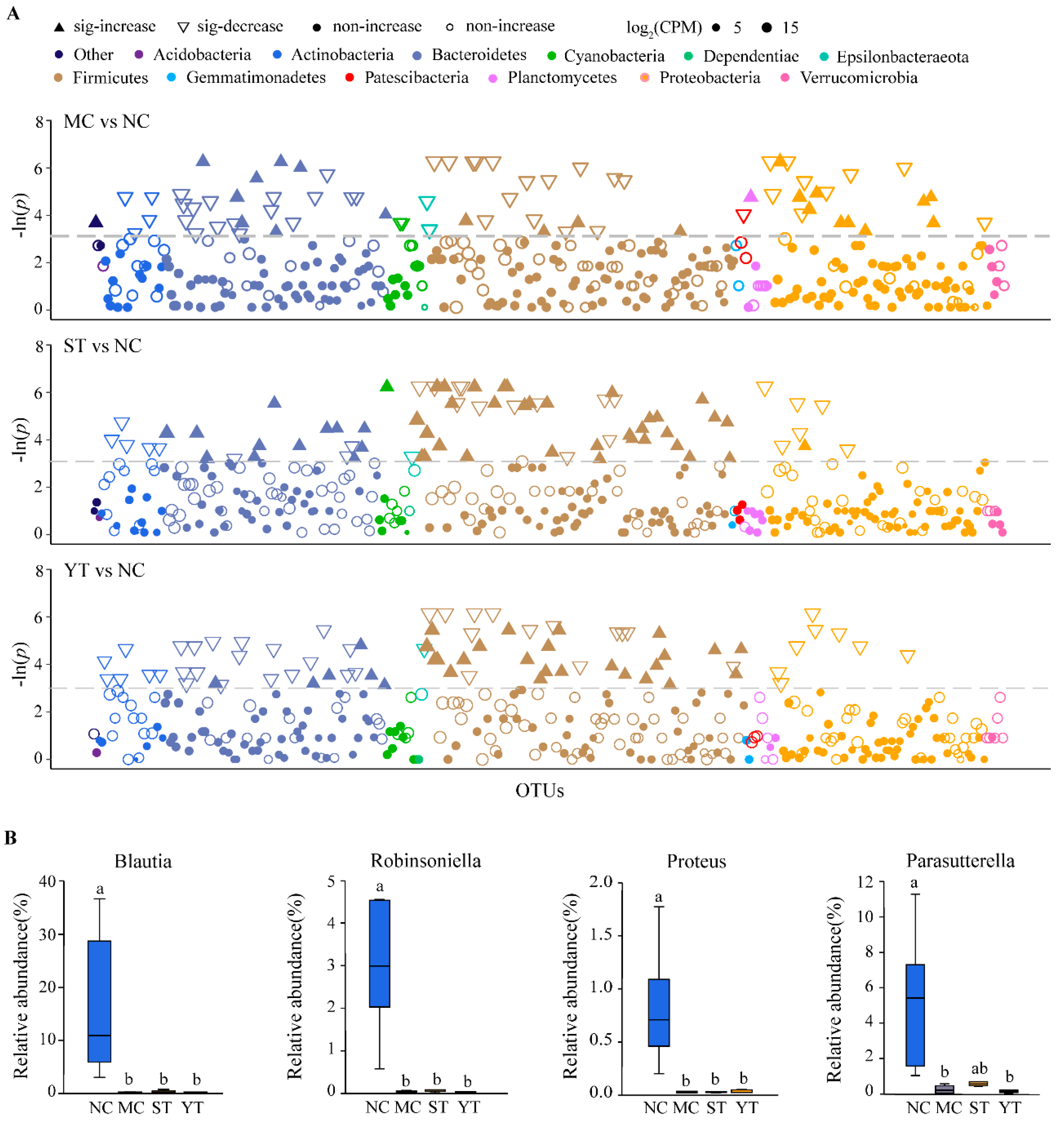
3.6. Composition and Difference Analysis of Gut Microbiota
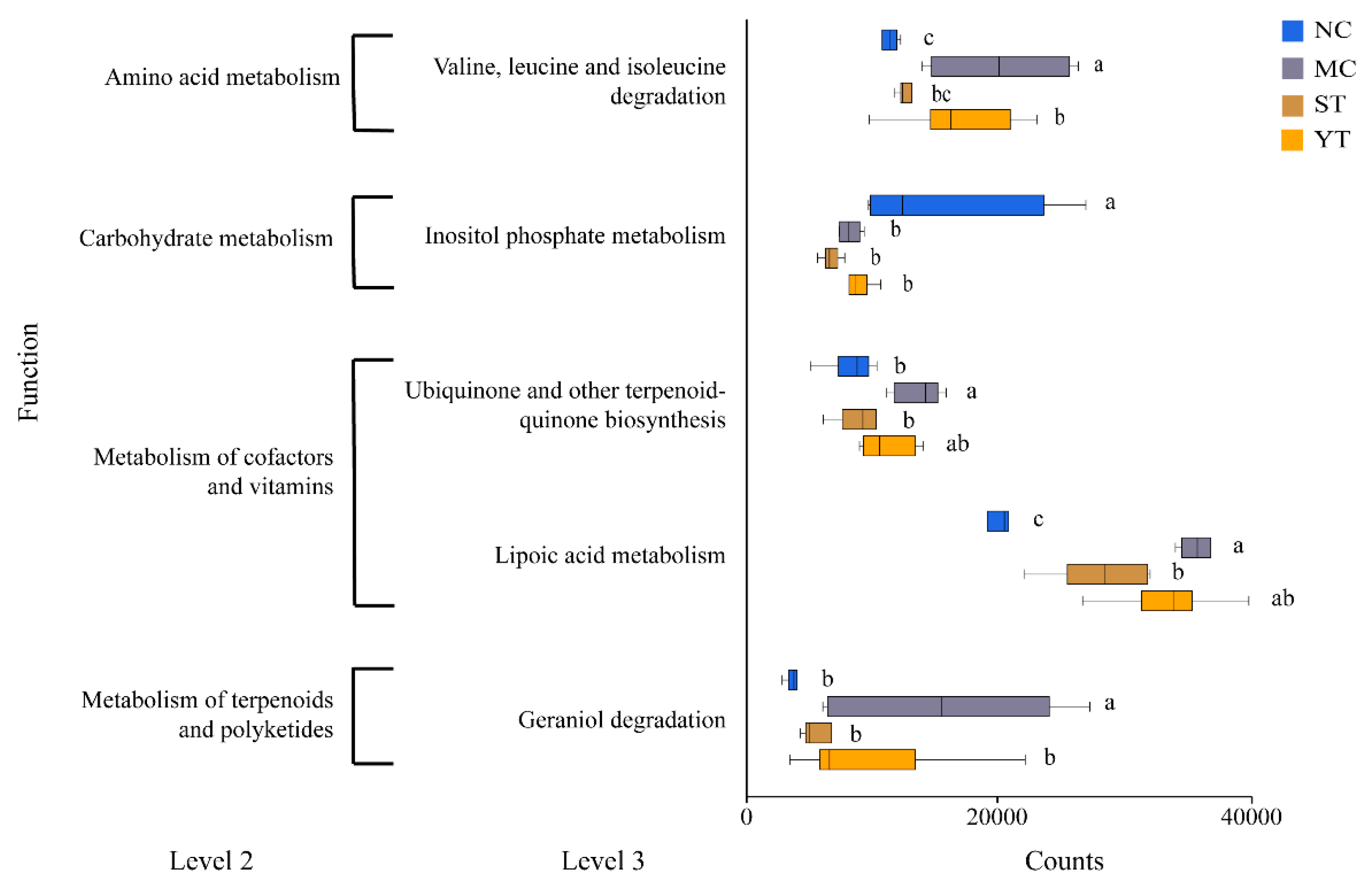
3.7. Functions Predicted
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Agamennone, V.; Krul, C.A.M.; Rijkers, G.; Kort, R. A Practical Guide for Probiotics Applied to the Case of Antibiotic-associated Diarrhea in the Netherlands. BMC Gastroenterol. 2018, 18, 103. [Google Scholar] [CrossRef] [PubMed]
- Mcfarland, L.V. Epidemiology, Risk Factors and Treatments for Antibiotic-Associated Diarrhea. Dig. Dis. 1998, 16, 292–307. [Google Scholar] [CrossRef] [PubMed]
- Blaabjerg, S.; Artai, D.M.; Aabenhus, R. Probiotics for the Prevention of Antibiotic-Associated Diarrhea in Outpatients-A Systematic Review and Meta-Analysis. Antibiotics 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Merenstein, D.; Fraser, C.M.; Marco, M.L. Molecular Mechanisms of Probiotic Prevention of Antibiotic-Associated Diarrhea. Curr. Opin. Biotechnol. 2020, 61, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Willing, B.P.; Russell, S.L.; Finlay, B.B. Shifting the Balance: Antibiotic Effects on Host-Microbiota Mutualism. Nat. Rev. Microbiol. 2011, 9, 233–243. [Google Scholar] [CrossRef]
- Pérez-Cobas, A.E.; Gosalbes, M.J.; Friedrichs, A.; Knecht, H.; Artacho, A.; Eismann, K.; Otto, W.; Rojo, D.; Bargiela, R.; von Bergen, M.; et al. Gut Microbiota Disturbance During Antibiotic Therapy: A Multi-Omic Approach. Gut 2013, 62, 1591–1601. [Google Scholar] [CrossRef]
- Health and Nutritional Properties of Probiotics in Food Including Powder Milk with Live Lactic Acid Bacteria. Available online: Ftp://ftp.fao.org/es/esn/food/wgreport2.pdf (accessed on 30 April 2020).
- Uriot, O.; Denis, S.; Junjua, M.; Roussel, Y.; Dary-Mourot, A.; Blanquet-Diot, S. Streptococcus thermophilus: From Yogurt Starter to A New Promising Probiotic Candidate? J. Funct. Foods 2017, 37, 74–89. [Google Scholar] [CrossRef]
- Jonkers, D.; Stockbrugger, R. Review Article: Probiotics in Gastrointestinal and Liver Diseases. Aliment. Pharmacol. Ther. 2007, 26, 133–148. [Google Scholar] [CrossRef]
- Cai, J.Y.; Zhao, C.Y.; Du, Y.J.; Zhang, Y.Q.; Zhao, M.Y.; Zhao, Q.C. Comparative Efficacy and Tolerability of Probiotics for Antibiotic-Associated Diarrhea: Systematic Review with Network Meta-Analysis. United Eur. Gastroenterol J. 2018, 6, 169–180. [Google Scholar] [CrossRef]
- Corrêa, N.B.O.; Péret, L.A.; Penna, F.J.; Lima, R.L.M.S.; Nicoli, J.R. A Randomized Formula Controlled Trial of Bifidobacterium lactis and Streptococcus thermophilus for Prevention of Antibiotic-Associated Diarrhea in Infants. J. Clin. Gastroenterol. 2005, 39, 385–389. [Google Scholar] [CrossRef]
- Selinger, C.P.; Bell, A.; Cairns, A.; Lockett, M.; Sebastian, S.; Haslam, N. Probiotic VSL#3 Prevents Antibiotic-Associated Diarrhoea in A Double-Blind, Randomized, Placebo-Controlled Clinical Trial. J. Hosp. Infect. 2013, 84, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Khodadad, A.; Farahmand, F.; Najafi, M.; Shoaran, M. Probiotics for the Treatment of Pediatric Helicobacter Pylori Infection: A Randomized Double Blind Clinical Trial. Iran. J. Pediatr. 2013, 23, 79–84. [Google Scholar]
- Kolling, G.L.; Wu, M.; Warren, C.A.; Durmaz, E.; Klaenhammer, T.R.; Timko, M.P.; Guerrant, R.L. Lactic Acid Production by Streptococcus thermophilus Alters Clostridium Difficile Infection and in vitro Toxin A Production. Gut Microbes 2012, 3, 523–529. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.K.; Tan, F.; Liao, W.; Wang, Q.; Mu, J.F.; Zhou, X.R.; Yang, Z.N.; Zhao, X. Isolation and Identification of Lactobacillus plantarum HFY05 from Natural Fermented Yak Yogurt and Its Effect on Alcoholic Liver Injury in Mice. Microorganisms 2019, 7, 530. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis Across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Acute Oral Toxicity Test of National Food Safety Standard. Available online: http://www.cssn.net.cn/cssn/front/listpage.jsp (accessed on 11 May 2020).
- Tarrah, A.; de Castilhos, J.; Rossi, R.C.; Duarte, V.D.; Ziegler, D.R.; Corich, V.; Giacomini, A. In vitro Probiotic Potential and Anti-Cancer Activity of Newly Isolated Folate-Producing Streptococcus thermophilus Strains. Front. Microbiol. 2018, 9, 2214. [Google Scholar] [CrossRef]
- Li, S.S.; Qi, Y.L.; Chen, L.X.; Qu, D.; Li, Z.M.; Gao, K.; Chen, J.B.; Sun, Y.S. Effects of Panax Ginseng Polysaccharides on the Gut Microbiota in Mice with Antibiotic-Associated Diarrhea. Int. J. Biol. Macromol. 2019, 124, 931–937. [Google Scholar] [CrossRef]
- Qi, Y.L.; Chen, L.X.; Gao, K.; Shao, Z.J.; Huo, X.H.; Hua, M.; Liu, S.X.; Sun, Y.S.; Li, S.S. Effects of Schisandra Chinensis Polysaccharides on Rats with Antibiotic-Associated Diarrhea. Int. J. Biol. Macromol. 2019, 124, 627–634. [Google Scholar] [CrossRef]
- Chen, X.H.; Katchar, K.; Goldsmith, J.D.; Nanthakumar, N.; Cheknis, A.; Gerding, D.N.; Kelly, C.P. A Mouse Model of Clostridium difficile-Associated Disease. Gastroenterology 2008, 135, 1984–1992. [Google Scholar] [CrossRef]
- Zhang, W.D.; Zhu, B.; Xu, J.H.; Liu, Y.Y.; Qiu, E.Q.; Li, Z.J.; Li, Z.C.; He, Y.; Zhou, H.W.; Bai, Y. Bacteroides fragilis Protects Against Antibiotic-Associated Diarrhea in Rats By Modulating Intestinal Defenses. Front. Immunol. 2018, 9, 1040. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.L.; Lu, J.M.; Wang, Y.F.; Gu, W.; Yang, X.X.; Yu, J. Intake of Total Saponins and Polysaccharides From Polygonatum kingianum Affects the Gut Microbiota in Diabetic Rats. Phytomedicine 2017, 26, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.J.; Wu, F.H.; Hao, G.G.; Qi, Q.; Li, R.; Li, N.; Wei, L.M.; Chai, T.J. Bacillus subtilis Improves Immunity and Disease Resistance in Rabbits. Front. Immunol. 2017, 8, 354. [Google Scholar] [CrossRef]
- Edgar, R.C. Uparse: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Desantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, A Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. Qiime Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree 2-Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Loaupone, C.; Knight, R. Unifrac: A New Phylogenetic Method for Comparing Microbial Communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community ecology package. R package: 2.5-3. Available online: https://cran.r-project.org (accessed on 5 March 2020).
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Thurber, R.L.V.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S rRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H.; Chang, E.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. R Package Version 3.3.2. Available online: https://cran.r-project.org (accessed on 5 March 2020).
- Wassenaar, T.M.; Zschüttig, A.; Beimfohr, C.; Geske, T.; Auerbach, C.; Cook, H.; Zimmermann, K.; Gunzer, F. Virulence Genes in A Probiotic E. Coli Product with A Recorded Long History of Safe Use. Eur. J. Microbiol. Immunol. 2015, 5, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Salvetti, E.; Orrù, L.; Capozzi, V.; Martina, A.; Lamontanara, A.; Keller, D.; Cash, H.; Felis, G.E.; Cattivelli, L.; Torriani, S.; et al. Integrate Genome-Based Assessment of Safety for Probiotic Strains: Bacillus coagulans GBI-30, 6086 as A Case Study. Appl. Microbiol. Biotechnol. 2016, 100, 4595–4605. [Google Scholar] [CrossRef] [PubMed]
- Li, B.L.; Liu, F.; Tang, Y.R.; Luo, G.W.; Evivie, S.; Zhang, D.Q.; Wang, N.N.; Li, W.; Huo, G.C. Complete Genome Sequence of Lactobacillus Helveticus KLDS1.8701, A Probiotic Strain Producing Bacteriocin. J. Biotechnol. 2015, 212, 90–91. [Google Scholar] [CrossRef] [PubMed]
- Ladero, V.; Linares, D.M.; del Rio, B.; Fernandez, M.; Martin, M.C.; Alvarez, M.A. Draft Genome Sequence Of The Tyramine Producer Enterococcus durans Strain IPLA 655. Genome Announc. 2013, 1, e00265-100213. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Li, B.L.; Du, J.C.; Yu, S.F.; Li, W.; Evivie, S.E.; Guo, L.D.; Ding, X.Y.; Xu, M.; Huo, G.C. Complete Genome Sequence of Enterococcus durans KLDS6.0930, A Strain with Probiotic Properties. J. Biotechnol. 2016, 217, 49–50. [Google Scholar] [CrossRef]
- Hamon, E.; Horvatovich, P.; Izquierdo, E.; Bringel, F.; Marchioni, E.; Aoude-Werner, D.; Ennahar, S. Comparative Proteomic Analysis of Lactobacillus plantarum for the Identification of Key Proteins in Bile Tolerance. BMC Microbiol. 2011, 11, 63. [Google Scholar] [CrossRef]
- Kim, E.; Chang, H.C.; Kim, H.Y. Complete Genome Sequence of Lactobacillus plantarum EM, A Putative Probiotic Strain with the Cholesterol-Lowering Effect and Antimicrobial Activity. Curr. Microbiol. 2020, 77, 1871–1882. [Google Scholar] [CrossRef]
- Senan, S.; Prajapati, J.B.; Joshi, C.G. Whole-Genome Based Validation of the Adaptive Properties of Indian Origin Probiotic Lactobacillus helveticus MTCC 5463. J. Sci. Food Agric. 2015, 95, 321–328. [Google Scholar] [CrossRef]
- Wu, Z.; Wu, J.; Lang, F.X.; Cai, Z.D.; Zeng, X.Q.; Guo, Y.X.; Liu, X.T.; Pan, D.D. Characterization of the Sortase A from Lactobacillus acidophilus ATCC 4356 Involved in Adherence to Intestinal Cells. Future Microbiol. 2020, 15, 485–496. [Google Scholar] [CrossRef]
- Peng, Z.; Vogel, R.F.; Ehrmann, M.A.; Xiong, T. Identification and Characterization of Adhesion Proteins in Lactobacilli Targeting Actin as Receptor. Mol. Cell. Probes 2018, 37, 60–63. [Google Scholar] [CrossRef]
- Singh, M.; Sareen, D. Novel Lant Associated Lantibiotic Clusters Identified by Genome Database Mining. PLoS ONE 2014, 9, e91352. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; van der Donk, W.A. Structural Characterization of Four Prochlorosins: A Novel Class of Lantipeptides Produced by Planktonic Marine Cyanobacteria. Biochemistry 2012, 51, 4271–4279. [Google Scholar] [CrossRef]
- Turroni, F.; Ventura, M.; Buttó, L.F.; Duranti, S.; O’Toole, P.W.; Motherway, M.O.; van Sinderen, D. Molecular Dialogue Between the Human Gut Microbiota and the Host: A Lactobacillus and Bifidobacterium Perspective. Cell. Mol. Life Sci. 2014, 71, 183–203. [Google Scholar] [CrossRef]
- Ling, Z.X.; Liu, X.; Cheng, Y.W.; Luo, Y.Q.; Yuan, L.; Li, L.J.; Xiang, C. Clostridium butyricum Combined with Bifidobacterium infantis Probiotic Mixture Restores Fecal Microbiota and Attenuates Systemic Inflammation in Mice with Antibiotic-Associated Diarrhea. Biomed Res. Int. 2015, 2015, 582048. [Google Scholar] [CrossRef] [PubMed]
- Hendrickx, A.P.A.; Top, J.; Bayjanov, J.R.; Kemperman, H.; Rogers, M.R.C.; Paganelli, F.L.; Bonten, M.J.M.; Willems, R.J.L. Antibiotic-Driven Dysbiosis Mediates Intraluminal Agglutination and Alternative Segregation of Enterococcus faecium From the Intestinal Epithelium. mBio 2015, 6, e01346-15. [Google Scholar] [CrossRef]
- Litvak, Y.; Byndloss, M.X.; Tsolis, R.M.; Bäumler, A.J. Dysbiotic Proteobacteria Expansion: A Microbial Signature of Epithelial Dysfunction. Curr. Opin. Microbiol. 2017, 39, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mehta, H.; Goulet, P.O.; Mashiko, S.; Desjardins, J.; Pérez, G.; Koenig, M.; Senecal, J.L.; Constante, M.; Santos, M.M.; Sarfati, M. Early-Life Antibiotic Exposure Causes Intestinal Dysbiosis and Exacerbates Skin and Lung Pathology in Experimental Systemic Sclerosis. J. Invest. Dermatol. 2017, 137, 2316–2325. [Google Scholar] [CrossRef]
- Glymenaki, M.; Barnes, A.; Hagan, S.O.; Warhurst, G.; McBain, A.J.; Wilson, I.D.; Kell, D.B.; Else, K.J.; Cruickshank, S.M. Stability in Metabolic Phenotypes and Inferred Metagenome Profiles Before the Onset of Colitis-Induced Inflammation. Scientific Rep. 2017, 7, 8836. [Google Scholar] [CrossRef]
- Latvala, S.; Pietilä, T.E.; Veckman, V.; Kekkonen, R.A.; Tynkkynen, S.; Korpela, R.; Julkunen, I. Potentially Probiotic Bacteria Induce Efficient Maturation but Differential Cytokine Production in Human Monocyte-Derived Dendritic Cells. World J. Gastroenterol. 2008, 14, 5570–9327. [Google Scholar] [CrossRef]
- Kekkonen, R.A.; Kajasto, E.; Miettinen, M.; Veckman, V.; Korpela, R.; Julkunen, I. Probiotic Leuconostoc mesenteroides ssp. cremoris and Streptococcus thermophilus Induce Il-12 and IFN-Gamma Production. World J. Gastroenterol. 2008, 14, 1192–1203. [Google Scholar] [CrossRef] [PubMed]
- Latvala, S.; Miettinen, M.; Kekkonen, R.A.; Korpela, R.; Julkunen, I. Lactobacillus rhamnosus GG and Streptococcus thermophilus Induce Suppressor of Cytokine Signalling 3 (SOCS3) Gene Expression Directly and Indirectly via Interleukin-10 in Human Primary Macrophages. Clin. Exp. Immunol. 2011, 165, 94–103. [Google Scholar] [CrossRef]
- Del Carmen, S.; de LeBlanc, A.D.; Martin, R.; Chain, F.; Langella, P.; Bermudez-Humaran, L.G.; LeBlanc, J.G. Genetically Engineered Immunomodulatory Streptococcus thermophilus Strains Producing Antioxidant Enzymes Exhibit Enhanced Antiinflammatory Activities. Appl. Environ. Microbiol. 2014, 80, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Del Piano, M.; Balzarini, M.; Carmagnola, S.; Pagliarulo, M.; Tari, R.; Nicola, S.; Deidda, F.; Pane, M. Assessment of the Capability of A Gelling Complex Made of Tara Gum and the Exopolysaccharides Produced by the Microorganism Streptococcus thermophilus ST10 to Prospectively Restore the Gut Physiological Barrier: A Pilot Study. J. Clin. Gastroenterol. 2014, 48, S56–S61. [Google Scholar] [CrossRef]
- Bailey, J.R.; Probert, C.S.J.; Cogan, T.A. Identification and Characterisation of An Iron-Responsive Candidate Probiotic. PLoS ONE 2011, 6, e26507. [Google Scholar] [CrossRef]
- Bailey, J.R.; Vince, V.; Williams, N.A.; Cogan, T.A. Streptococcus thermophilus NCIMB 41856 ameliorates Signs of Colitis in An Animal Model of Inflammatory Bowel Disease. Benef. Microbes 2017, 8, 605–614. [Google Scholar] [CrossRef]
- Liu, C.; Wu, H.; Liu, S.J.; Chai, S.T.; Meng, Q.X.; Zhou, Z.M. Dynamic Alterations in Yak Rumen Bacteria Community and Metabolome Characteristics in Response to Feed Type. Front. Microbiol. 2019, 10, 1116. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.Z.; Tan, K.; Peng, M.J.; Long, C.X.; Li, D.D.; Tan, Z.J. Bacterial Diversity in Intestinal Mucosa of Antibiotic-Associated Diarrhea Mice. 3 Biotech 2019, 9, 444. [Google Scholar] [CrossRef]
- Grazul, H.; Kanda, L.L.; Gondek, D. Impact of Probiotic Supplements on Microbiome Diversity Following Antibiotic Treatment of Mice. Gut Microbes 2016, 7, 101–114. [Google Scholar] [CrossRef]
- Zhou, D.; Pan, Q.; Xin, F.Z.; Zhang, R.N.; He, C.X.; Chen, G.Y.; Liu, C.; Chen, Y.W.; Fan, J.G. Sodium Butyrate Attenuates High-Fat Diet-Induced Steatohepatitis in Mice by Improving Gut Microbiota and Gastrointestinal Barrier. World J. Gastroenterol. 2017, 23, 60–75. [Google Scholar] [CrossRef]
- Ju, T.T.; Kong, J.Y.; Stothard, P.; Willing, B.P. Defining the Role of Parasutterella, A Previously uncharacterized Member of the Core Gut Microbiota. ISME J. 2019, 13, 1520–1534. [Google Scholar] [CrossRef]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Jarnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A Pyrosequencing Study in Twins Shows That Gastrointestinal Microbial Profiles Vary with Inflammatory Bowel Disease Phenotypes. Gastroenterology 2010, 139, 1844–1854. [Google Scholar] [CrossRef]
- Rastelli, M.; Cani, P.D.; Knauf, C. The Gut Microbiome Influences Host Endocrine Functions. Endocr. Rev. 2019, 40, 1271–1284. [Google Scholar] [CrossRef]





| Sex | Weight (g) | Death Rate (%) | ||
|---|---|---|---|---|
| Day 0 | Day 7 | Day 14 | ||
| Female | 19.08 ± 0.76 | 27.00 ± 1.53 | 32.08 ± 1.54 | 0 |
| Male | 20.22 ± 1.60 | 31.61 ± 2.50 | 38.50 ± 2.80 | 0 |
| Putative Function | Protein Name | Genome Location |
|---|---|---|
| Acid tolerance | F0F1-ATPase | DMST-H2GL000469, DMST-H2GL000470, DMST-H2GL000471, DMST-H2GL000472, DMST-H2GL000473, DMST-H2GL000474, DMST-H2GL000475, DMST-H2GL000476 |
| Tyrosyl-tRNA synthetase | DMST-H2GL001052, DMST-H2GL001867 | |
| Bile salt tolerance | Cyclopropane-fatty-acyl-phospholipid synthase | DMST-H2GL000126 |
| Choloylglycine hydrolase | DMST-H2GL001522 | |
| Oxidative stress | Peptide methionine sulfoxide reductase msrA/msrB | DMST-H2GL001340 |
| Glutathione reductase | DMST-H2GL000398 | |
| Thioredoxin reductase (NADPH) | DMST-H2GL001650 | |
| Superoxide dismutase, Fe-Mn family | DMST-H2GL000768 | |
| Thiol peroxidase, atypical 2-Cys peroxiredoxin | DMST-H2GL001000 | |
| Thioredoxin 1 | DMST-H2GL001797 | |
| Adhesion | Sortase A | DMST-H2GL001272 |
| Elongation factor Tu | DMST-H2GL000478 | |
| Chaperonin GroEL | DMST-H2GL000203 | |
| Competence protein ComGC | DMST-H2GL001860 | |
| Bacteriocin | Lantibiotic biosynthesis protein | DMST-H2GL000092, DMST-H2GL000093, DMST-H2GL000094 |
| Lantibiotic biosynthesis response regulator NisR/SpaR | DMST-H2GL000928 | |
| Lantibiotic biosynthesis sensor histidine kinase NisK/SpaK | DMST-H2GL000929 | |
| Bacteriocin exporter | DMST-H2GL000265, DMST-H2GL000267, DMST-H2GL000268, DMST-H2GL000694, DMST-H2GL000695, DMST-H2GL001682, DMST-H2GL001683 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, J.-S.; Huang, Y.-Y.; Kuang, J.-H.; Yu, J.-J.; Zhou, Q.-Y.; Liu, D.-M. Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea. Microorganisms 2020, 8, 1650. https://doi.org/10.3390/microorganisms8111650
Hu J-S, Huang Y-Y, Kuang J-H, Yu J-J, Zhou Q-Y, Liu D-M. Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea. Microorganisms. 2020; 8(11):1650. https://doi.org/10.3390/microorganisms8111650
Chicago/Turabian StyleHu, Jin-Shuang, Yan-Yan Huang, Jia-Hua Kuang, Jia-Jia Yu, Qin-Yu Zhou, and Dong-Mei Liu. 2020. "Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea" Microorganisms 8, no. 11: 1650. https://doi.org/10.3390/microorganisms8111650
APA StyleHu, J.-S., Huang, Y.-Y., Kuang, J.-H., Yu, J.-J., Zhou, Q.-Y., & Liu, D.-M. (2020). Streptococcus thermophiles DMST-H2 Promotes Recovery in Mice with Antibiotic-Associated Diarrhea. Microorganisms, 8(11), 1650. https://doi.org/10.3390/microorganisms8111650





